Background: O-GlcNAcylation of surface adhesin PsrP catalyzed by GtfA-GtfB complex is involved in the pathogenicity of Streptococcus pneumoniae.
Results: The crystal structure of GtfA reveals a novel O-GlcNAc transferase with a β-meander “add-on” domain.
Conclusion: The add-on domain is crucial for complex formation and acceptor recognition.
Significance: The structure provides insights into the catalytic mechanism of a novel bacterial O-GlcNAc transferase.
Keywords: Crystal Structure, Glycoprotein Biosynthesis, Glycosylation, Glycosyltransferase, Pathogenesis, O-GlcNAc Transferase, Streptococcus pneumoniae, Serine-rich Repeat Glycoprotein
Abstract
Protein glycosylation catalyzed by the O-GlcNAc transferase (OGT) plays a critical role in various biological processes. In Streptococcus pneumoniae, the core enzyme GtfA and co-activator GtfB form an OGT complex to glycosylate the serine-rich repeat (SRR) of adhesin PsrP (pneumococcal serine-rich repeat protein), which is involved in the infection and pathogenesis. Here we report the 2.0 Å crystal structure of GtfA, revealing a β-meander add-on domain beyond the catalytic domain. It represents a novel add-on domain, which is distinct from the all-α-tetratricopeptide repeats in the only two structure-known OGTs. Structural analyses combined with binding assays indicate that this add-on domain contributes to forming an active GtfA-GtfB complex and recognizing the acceptor protein. In addition, the in vitro glycosylation system enables us to map the O-linkages to the serine residues within the first SRR of PsrP. These findings suggest that fusion with an add-on domain might be a universal mechanism for diverse OGTs that recognize varying acceptor proteins/peptides.
Introduction
Protein O-glycosylation is one of the major posttranslational modifications and involves various biological processes (1). Notably, O-GlcNAcylation in vertebrates catalyzed by the O-GlcNAc transferase (OGT)5 plays critical roles in modulating cellular signal transduction, gene transcription, cytoskeletal functions, and cell differentiation (2–4). Aberrant O-GlcNAcylations are correlated with many serious human diseases, including diabetes, cardiovascular disease, neurodegenerative diseases, and cancer (5). To date, the only two structure-known OGTs, human OGT (hOGT) (6, 7) and its close homolog from Xanthomonas campestris (XcOGT) (8, 9), revealed an N-terminal add-on domain of tetratricopeptide repeats (TPR) followed by a conserved catalytic domain of glycosyltransferase type B (GT-B) superfamily (6, 8, 9). The TPR domain is proposed to scaffold interactions with other partner proteins and to determine the substrate specificity (10).
It is currently known that the O-GlcNAcylation is also a common modification in prokaryotes, especially for the pathogenic bacterial strains (8, 9, 11, 12). Most cases of O-linkages identified to date are formed with the Ser/Thr/Tyr residues of bacterial surface-exposed proteins (12). The serine-rich repeat glycoproteins (SRRPs) that exclusively exist in Gram-positive bacteria are a large family of surface adhesins that are important for bacterial adhesion (13), immune evasion, colonization, biofilm formation, and virulence (14–18). The two serine-rich repeat regions (SRR1 and SRR2) of SRRPs are usually subject to heavy glycosylation (18, 19), which is required for the stability of nascent SRRPs in cytoplasm (20) and proper destination in the designated intracellular compartments (19, 21–23). The glycosylation pathway commonly initiates with a step of O-GlcNAcylation catalyzed by an OGT complex of two subunits, which were termed Gtf1 (or GtfA) and Gtf2 (or GtfB) in Streptococcus parasanguinis (or Streptococcus pneumoniae), respectively (24, 25). Inhibition of this initiating OGT complex prevents the glycosylation of SRRPs, which in consequence attenuates the pathogenesis (20, 21). Our previous studies suggested that Gtf1 is the core enzyme, whereas Gtf2 might act as a chaperone (25). Sequence analysis revealed that GtfA consists of an N-terminal domain of unknown function (named DUF1975) and a core catalytic domain of GT-B fold. However, the lack of structural information has been a major impediment to elucidating the molecular mechanism and developing inhibitors against this unique OGT complex.
Here we report the 2.0 Å crystal structure of S. pneumoniae GtfA in complex with GlcNAc and UDP. Structural comparison reveals that GtfA represents a novel OGT with a β-meander add-on domain of DUF1975, which is distinct from the all-α-TPRs of XcOGT. The DUF1975 domain of GtfA involves the recognition of the acceptor PsrP and its co-activator GtfB and thus is critical for the intact OGT activity. Further glycoproteomic analyses provide new insights into the O-GlcNAcylation of serine repeat cluster. Structure-based analysis of this novel OGT may assist in the rational design of novel inhibitors for combating the diseases resulted from the infection of Gram-positive pathogens. Moreover, our findings imply that fusion with an add-on domain might be a universal mechanism adopted by diverse OGTs.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of GtfA, GtfB, and Mutants
The coding regions of GtfA and GtfB were amplified from the genomic DNA of S. pneumoniae TIGR4 and cloned into a pET28a-derived vector with a His6 tag at the N terminus. The Escherichia coli BL21 (DE3) strain was used for the expression of recombinant proteins. The transformed cells were grown at 37 °C in LB culture medium (10 g of NaCl, 10 g of Bacto-Tryptone, and 5 g of yeast extract per liter) containing appropriate antibiotics until the A600 nm reached about 0.6. Protein expression was then induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside by another 16 h at 16 °C. Cells were harvested by centrifugation (6000 × g, 4 °C, 10 min) and resuspended in 40 ml of lysis buffer (50 mm Tris-Cl, pH 8.0, 150 mm NaCl, 5% (v/v) glycerol). After 5 min of sonication and centrifugation at 12,000 × g for 30 min, the supernatant containing the soluble target protein was collected and loaded onto a nickel-nitrilotriacetic acid column (Qiagen, Mississauga ON) equilibrated with the binding buffer (20 mm Tris-Cl, pH 8.0, 150 mm NaCl). The target protein was eluted with 400 mm imidazole and further loaded onto a Superdex 200 column (GE Healthcare) equilibrated with 20 mm Tris-Cl, pH 7.5, 100 mm NaCl. Protein purity was evaluated by electrophoresis, and samples were stored at −80 °C. Before crystallization, the protein sample was concentrated to 6 mg/ml by ultrafiltration (Millipore Amicon).
The selenium-Met (SeMet)-labeled GtfA protein was expressed in E. coli strain B834 (DE3) (Novagen, Madison, WI). Transformed cells were inoculated into LB medium at 37 °C overnight. The cells were harvested and washed twice with the M9 medium. Then the cells were cultured in SeMet medium (M9 medium with 50 mg/liter SeMet and other essential amino acids at 50 mg/liter) to an A600 nm of ∼0.6. Protein expression and purification steps were carried out as described above for the native protein.
Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with the plasmid encoding the wild-type GtfA as a template. The mutant proteins were expressed, purified, and stored in the same manner as the wild-type protein.
Crystallization, Data Collection, and Processing
Crystallization trials of GtfA were done using a Mosquito robot (TTP Labtech) in 96-well plates (Greiner) at 16 °C. The crystals were obtained using the hanging drop vapor-diffusion method, with the initial condition of equilibrating 0.1 μl of 6 mg/ml protein (mixed with UDP and GlcNAc to the final concentration of 10 mm) with and equal volume of the reservoir solution (0.1 m sodium citrate, pH 6.0, 30% PEG 3350). SeMet-substituted protein crystals were obtained at the conditions of 0.1 m Tris-Cl, pH 7.5, 0.2 m Li2SO4, 15% polyethylene glycol 3350. All the crystals were transferred to cryoprotectant (reservoir solution supplemented with 30% glycerol) and flash-cooled with liquid nitrogen. The data were collected at 100 K in a liquid nitrogen stream using beamline 17U with a Q315r CCD (ADSC, MARresearch, Germany) at the Shanghai Synchrotron Radiation Facility (SSRF).
Structure Determination and Refinement
All diffraction data were integrated and scaled with the program HKL2000 (26). The native and SeMet substituted GtfA proteins in the presence of GlcNAc and UDP were crystallized in the space group of P212121. The crystal structure of GtfA was determined using single-wavelength anomalous dispersion phasing (27) method from a single SeMet-substituted protein crystal to a highest resolution of 2.3 Å. The SHELXD program (28) implemented in IPCAS was used to locate the heavy atoms, and the phase was calculated by OASIS (29) and further improved with the programs RESOLVE and Buccaneer (30–32). Electron density maps showed clear features of secondary structural elements. Automatic model building was carried out using Autobuild in PHENIX (33). The initial model was refined using the maximum likelihood method implemented in REFMAC5 (34) as part of CCP4i (35) program suite and rebuilt interactively using the program COOT (36). The structure was refined to an R-factor/R-free of 19.9%/24.8% and was evaluated with the programs MOLPROBITY (37) and PROCHECK (38). Crystallographic parameters are listed in Table 1. All structure figures were prepared with PyMOL (39).
TABLE 1.
Crystal parameters, data collection, and structure refinement
| SeMet-GtfA | GtfA | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Unit cell | 71.88, 125.89, 127.43 | 71.24, 125.63, 127.04 |
| a, b, c (Å) | 90.00 | 90.00 |
| α, β, γ (°) | ||
| Resolution range (Å) | 50.00-2.30 (2.38-2.30)a | 47.41-2.00 (2.11-2.00) |
| Unique reflections | 52,072 (5,144) | 77,663 (11,169) |
| Completeness (%) | 100 (100) | 99.9 (99.8) |
| 〈I/σ(I)〉 | 24.6 (5.9) | 17.0 (4.7) |
| Rmergeb (%) | 12.5 (56.1) | 8.7 (40.3) |
| Average redundancy | 14.7 (14.9) | 8.9 (8.6) |
| Structure refinement | ||
| Resolution range (Å) | 47.41-2.00 | |
| R-factorc/R-freed (%) | 19.9/24.8 | |
| Number of protein atoms | 8,190 | |
| Number of water atoms | 667 | |
| r.m.s.d.e bond lengths (Å) | 0.008 | |
| r.m.s.d. bond angles (°) | 1.237 | |
| Mean B factors (Å2) | 30.9 | |
| Ramachandran plotf (residues, %) | ||
| Most favored (%) | 96.71 | |
| Additional allowed (%) | 3.29 | |
| Outliers (%) | 0 | |
| PDB entry | 4PQG | |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation, and 〈I(hkl)〉 is the mean value for its unique reflection; summations are over all reflections.
c R-factor = Σh‖Fo(h)| − |Fc(h)‖/Σh|Fo(h)|, where |Fo| and |Fc| are the observed and calculated structure-factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e r.m.s.d. from ideal values.
f Categories were defined by Molprobity.
Hydrolytic Activity Assays
The hydrolytic activities of GtfA and/or GtfB were assayed by high performance liquid chromatography (HPLC). All assays were performed at 37 °C in the buffer containing 50 mm Tris, 100 mm NaCl, pH 7.5, 10 mm β-mercaptoethanol with UDP-GlcNAc as the sugar donor and PsrPSRR1 as the acceptor. The donor substrate UDP-GlcNAc (Sigma) was diluted to a series of concentrations from 100 mm stock solution. The reaction was triggered by adding the purified protein solution and terminated by heating at 100 °C for 5 min. All samples were centrifuged at 10,000 × g for 10 min, and the supernatant was subjected to HPLC system (Agilent 1200 Series). The buffer of 100 mm NH4H2PO4, pH 6.2, was used for equilibration of the column (Eclipse XDB-C18 column, 4.6 × 150 mm, Agilent) and separation of the components at a flow rate of 1 ml/min. The product UDP was used as the standard and was quantified by the absorption at 254 nm. The initial velocities and substrate concentrations were used to non-linearly fit the Michaelis-Menten equation plot to calculate the Km and Vmax values. Three independent kinetic determinations were made to calculate the means and standard deviations for the reported Km and kcat values.
In Vitro SRR1 O-GlcNAcylation Assays
The PsrPSRR1 glycosylation assays were performed in a volume of 20 μl with 20 μm recombinant GST-PsrPSRR1 and a series of concentrations of sugar donor UDP-GlcNAc in the buffer containing 50 mm Tris-Cl 100 mm NaCl, pH 7.5, 10 mm β-mercaptoethanol. The glycosylation reaction lasted for 2 h at 37 °C by adding 8 μm GtfA and/or GtfB and was terminated by heating at 100 °C for 5 min with the addition of 5× loading buffer. Then these reaction mixtures were separated in a 12% SDS-PAGE gel and then transferred to polyvinylidene difluoride membranes. GlcNAc-modified GST-PsrPSRR1 proteins were detected on the polyvinylidene difluoride membrane using wheat germ agglutinin-HRP conjugate (WGA-HRP, 20363).
Identification of the O-GlcNAcylation Sites
Completely glycosylated GST-PsrPSRR1 and native GST-PsrPSRR1 were reduced, alkylated, digested with GluC (Sigma), and analyzed by reverse phase liquid chromatography according to a standard quality control method for enzymatic mapping. Using the in-gel digestion protocol (40), the samples were diluted in 50 mm NH4HCO3 and reduced with 10 mm freshly made dithiothreitol at 56 °C for 45 min, carboxyamidomethylated with 55 mm iodoacetamide, and incubated in the dark for 30 min; 200 μl of 30% or 50% ACN (acetonitrile) in 50 mm NH4HCO3 was added to remove the Coomassie stain, and the dried gel was protease-digested overnight at 37 °C. After digestion, the reaction was quenched with 200 μl of 60% ACN, 5% trifluoroacetic acid (TFA). The resulting peptides were suspended in 25 μl of 0.1% formic acid and stored at −20 °C until ready to LTQ Orbitrap Velos (Thermo Fisher) MS analysis.
The data were searched against protein database using the TurboSequest algorithm (Bio-Works, Thermo Fisher). To aid in identification of glycopeptides, we allowed for a mass increase on serine residues looking for the addition of GlcNAc gradually. All remaining spectra were manually evaluated for the presence of glycopeptides and sites of modification and were further validated by TurboSequest searches against the GST-PsrPSRR1 FASTA sequence combined with the TurboSequest common contaminants database.
Construction of a GtfA-GtfB and PsrP Co-expression System and Purification of Recombinant Proteins
An E. coli recombinant system that co-expresses GtfA-GtfB with its acceptor PsrP was used to determine binding of GtfA to PsrP and the function of DUF1975 in the binding. The recombinant E. coli strains were generated as follows. Full-length gtfA-gtfB was amplified from S. pneumoniae TIGR4 genomic DNA using a primer set of gtfA-gtfB-BamHI-1F and gtfA-gtfB-XhoI-1266R. The DNA fragment was digested and cloned into pGEX-6P-1 to construct pGEX-6P-1-gtfA-gtfB in E. coli BL21.1266 bp of 5′ DNA fragment of psrP was amplified and cloned into pET-27b, creating pET-27b-PsrP-His. The resulting plasmid pET-27b-PsrP-His was used as a template to amplify the full expression unit (including the T7 promoter, psrP, a His6 tag, and the transcriptional terminator). The amplified DNA fragment was inserted into the single XhoI site of pGEX-6p-1-GtfA-GtfB, creating the final construct pGEX-6p-1-GtfA-GtfB-PsrP-His in E. coli BL21 (DE3).
Site-directed mutagenesis was used to construct GtfA variants by PCR using a QuikChange mutagenesis kit (Stratagene). In brief, the plasmid pGEX-6p-1-GtfA-GtfB-PsrP-His was used as a template. Mutant alleles were confirmed by sequencing. The resulting plasmids were transformed into E. coli BL21 (DE3) for protein expression.
The interaction between PsrP with GtfA or GtfA variants was determined by co-purification of GST fused GtfA variants and PsrP from recombinant E. coli strains that harbor GST-GtfA variants/GtfB and PsrP. Production of recombinant proteins was carried out as follows. Recombinant E. coli BL21 strains were grown at 37 °C to an optical density of 0.7. Protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.1 mm and incubation overnight at 25 °C before harvesting cell pellets. Cell lysates were prepared from the cell pellets by sonication, and all recombinant proteins were purified by glutathione-Sepharose 4B beads (Amersham Biosciences). Equal amount of purified proteins were subjected to SDS-PAGE analysis followed by Coomassie Brilliant Blue staining and Western blot analysis using anti-GST, anti-His, and anti-PsrP to detect GtfA and PsrP.
RESULTS AND DISCUSSION
Overall Structure of GtfA
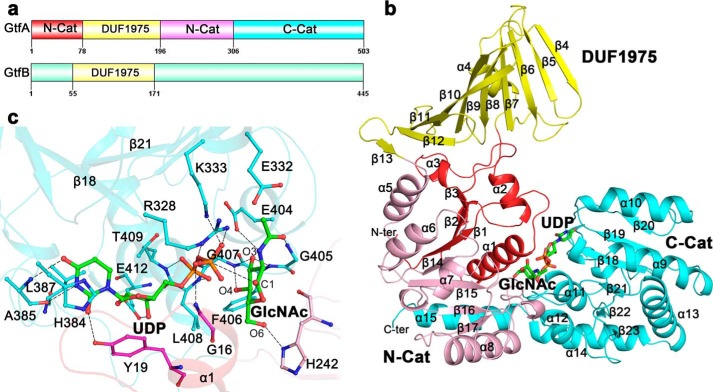
Crystals of the selenium-Met-substituted recombinant GtfA were obtained, and the structure in the presence of UDP and GlcNAc was solved at the resolution of 2.0 Å. Each asymmetric unit of the crystal structure contains two molecules of GtfA with an overall root mean square deviation (r.m.s.d.) of 0.56 Å over 501 Cα atoms. Despite that the crystal packing results in a buried interface of 1600 Å2, both size-exclusion chromatography and ultraperformance liquid chromatography indicated that GtfA exists as a monomer in solution. The molecules of GlcNAc and UDP at the active site could be well defined in the final model. Similar to members in the GT-B superfamily, GtfA comprises a core structure of two Rossmann-like domains: N-Cat and C-Cat (Fig. 1, a and b). The N-Cat is composed of two segments: residues Met-1–Ser-78 and Gly-196–Gly-306 interrupted by DUF1975, whereas the C-Cat (residues Ser-307–Asp-503) has a tail of helix α15 packing on N-Cat (Fig. 1b). Structural homology search using DALI server (41) revealed that the core structure of GtfA resembles GT-B enzymes at a Z-score ≥14.0 despite sharing a sequence identity of 18% (42). The top hits include Bacillus anthracis GT4 glycosyltransferases (PDB code 2JJM, Z score 27.8, r.m.s.d. 3.1 Å over 332 Cα atoms) (43), B. anthracis BaBshA (44), Mycobacteria PimA (45), and Corynebacterium glutamicum MshA (46). Protruding from the N-terminal core structure is an add-on domain of DUF1975 (residues Val-79–Tyr-195), which adopts a β-meander structure of a twisted 10-stranded antiparallel β-sheet (β4-β13). The hydrophobic outer side of DUF1975 is shielded from the solvent by a short helix α4 (Fig. 1b).
FIGURE 1.
Domain organization and overall structure of GtfA. a, three distinct domains of GtfA and two putative domains of GtfB were drawn by Domain Graph, Version 2.0. b, overall structure of GtfA. The four segments are colored red, yellow, pink, and cyan, respectively. The secondary structure elements are numbered sequentially. The UDP and GlcNAc molecules at the active site are shown as sticks. c, the binding sites of UDP and GlcNAc. The binding residues are shown as sticks. The polar interactions are indicated by dashed lines.
The molecules of UDP and GlcNAc are accommodated at the interdomain cleft with most contacts to C-Cat (Fig. 1c). In detail, the uracil base of UDP is stabilized by hydrophobic interactions with His-384 and Leu-387 in addition to hydrogen bonds with Ala-385 and Tyr-19. The ribose ring is stacked by Tyr-19, and its oxygen atoms O2 and O3 are fixed by the carboxylate group of Glu-412 via two conserved hydrogen bonds as found in E. coli NDP-glycosyltransferase MurG (47) and Mycobacterium smegmatis phosphatidylinositol mannosyltransferase PimA (45). The oxygen atoms of the α-phosphate group are stabilized by the backbone amides of Leu-408 and Thr-409, respectively, whereas those of the β-phosphate group interact with basic residues Arg-328 and Lys-333 and with the backbone amide of Gly-16 at the α1-β1 loop (Fig. 1c). This loop at N-Cat usually undergoes significant conformational changes upon sugar-nucleotide binding (47, 48). The GlcNAc molecule is approximately perpendicular to the UDP molecule and adopts a “bent-back” conformation toward the pyrophosphate of UDP (Fig. 1c). This conformation facilitates the anomeric sugar carbon (carbon 1 (C1)) to be exposed for nucleophilic attack (23, 45, 46). GlcNAc is also stabilized by several hydrogen bonds: two of the hydroxyl O3 with the backbone amides of Phe-406 and Gly-407, one of O4 with the amide nitrogen of Gly-407, one of O6 with Nδ1 of His-242, and two of the acetyl group with residues Gly-405 and Glu-404 (Fig. 1c). All these active-site residues, especially the two basic residues Arg-328 and Lys-333, which were proposed to neutralize the negative charge of the phosphate groups of UDP (46), are generally conserved in GT-B enzymes.
Insertion of DUF1975 Makes GtfA a Novel OGT
The N-Cat domain of GtfA is interrupted by DUF1975, a function-unknown domain that is predominantly found in the N-terminal region of various putative bacterial glycosyltransferases. Structural homology search revealed that DUF1975 is similar to a domain or segment of several functionally unrelated proteins, such as Drosophila melanogaster centriolar components dCPAP (PDB code 4BY2, Z score 8.2, r.m.s.d. 3.3 Å over 90 Cα atoms) (49), Borrelia burgdorferi outer membrane protein OspA (PDB code 2FKJ, Z score 7.2, r.m.s.d. 4.4 Å over 96 Cα atoms) (50), and Pseudomonas aeruginosa pyoverdine-iron transporter FpvA (PDB code 2W78, Z score 5.6, r.m.s.d. 6.5 Å over 100 Cα atoms) (51). However, DUF1975 could be only partially superimposed onto these structures, indicating that DUF1975 represents a novel structure.
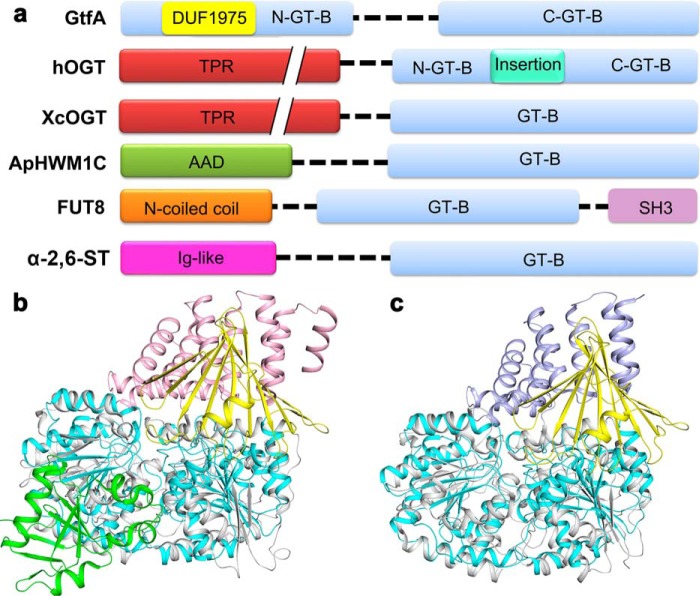
Four different add-on domains have previously revealed in the structures of GT-B glycosyltransferases (Fig. 2a). They are the N-terminal TPR domain of hOGT (PDB code 3PE4) (6) and XcOGT (PDB code 2JLB) (8), the N-terminal all α-domain of N-glucosyltransferase (PDB code 3Q3E) (52), the C-terminal SH3 domain of mammalian α-1,6-fucosyltransferase (PDB code 2DE0) (53), and the N-terminal immunoglobulin like β-sandwich fold domain of α-2,6-sialyltransferase (PDB code 2Z4T) (54). It was assumed that during evolution these add-on domains impart the defined acceptor binding specificity to glycosyltransferases (55). Among these interrupted GT-B glycosyltransferases, GtfA has a molecular function similar to the two OGTs of known structure, hOGT and XcOGT, which have an N-terminal all-α-TPR domain that play an important role in recognizing acceptor and/or partner proteins. Notably, the β-meander DUF1975 domain of GtfA adopts a similar orientation to that of TPRs in hOGT and XcOGT as shown by structural superpositions despite that it possesses a totally different composition of secondary structure elements (Fig. 2, b and c).
FIGURE 2.
Domain organization and structural comparison between GtfA and GT-B enzymes with an add-on domain. a, domain organization of GT-B enzymes with an add-on domain. The proteins are S. pneumoniae GtfA, hOGT, XcOGT, the N-linked glucosyltransferase (ApHWM1C), the mammalian α-1,6-fucosyltransferase (FUT8), and the α-2,6-sialyltransferase (α-2,6-ST). Structural comparisons of GtfA with hOGT (b) and XcOGT (c) are shown. The catalytic and add-on domains of GtfA are colored cyan and yellow, respectively. The catalytic domains of hOGT and XcOGT are colored gray, whereas the add-on domains are colored pink and light blue, respectively. The intervening domain of hOGT is colored green.
DUF1975 Is Necessary for the Formation of an Active GtfA-GtfB Complex
Our previous studies reported that S. parasanguinis Gtf2 functions as a chaperone of Gtf1 for the glycosylation of its acceptor protein Fap1 (24). Accordingly, we hypothesized that S. pneumoniae GtfB may bind to GtfA to form a holoenzyme. Indeed, isothermal titration calorimetry experiments indicated they form a tight complex with a dissociation constant (Kd) of 51.5 nm. Size-exclusion chromatography combined with gel electrophoresis also confirmed that GtfA and GtfB form a stable complex of 1:1 stoichiometry. Moreover, we mapped a region (58LYFNQL63) in the DUF1975 domain of Gtf2 corresponding to the region of 57LYFNQV62 in GtfB, which was critical for the binding to Gtf1 (25). Thus, we determined the Kd value of the individual DUF1975 domain of GtfB (GtfB-DUF1975, residues Met-1–Phe-171) toward GtfA and revealed a Kd of 11.0 μm, suggesting that the DUF1975 domain is necessary but not sufficient for the formation of a stable GtfA-GtfB complex.
In fact, GtfB also functions as a co-activator/chaperone for the full activity of GtfA. Our in vitro hydrolytic assays showed that the GtfA-GtfB complex had an activity (kcat/Km) of 69.1 min−1mm−1 toward UDP-GlcNAc, >100-fold that of the individual GtfA (0.6 min−1mm−1), whereas GtfB did not exhibit any activity (Table 2). In contrast, the complex of GtfA with GtfB-DUF1975 had an activity of 1.1 min−1mm−1, which was at a comparable level with that of individual GtfA (Table 2). Moreover, mutation of either Arg-328 or Lys-333 to alanine completely abolished the activity, suggesting these two UDP binding residues are crucial for the catalysis.
TABLE 2.
Kinetic constants of GtfA towards UDP-GlcNAc
| Enzyme | Ka | Vmax | Kcat | Kcat/Km |
|---|---|---|---|---|
| mm | μm/min | min−1 | mm−1 min−1 | |
| GtfA-GtfB | 1.1 ± 0.2 | 30.6 ± 1.26 | 76.0 ± 3.15 | 69.1 |
| GtfA | 11.8 ± 1.5 | 14.7 ± 0.84 | 7.35 ± 0.42 | 0.63 |
| GtfA-GtfB-DUF1975 | 23.0 ± 1.1 | 13.1 ± 3.03 | 26.2 ± 6.06 | 1.14 |
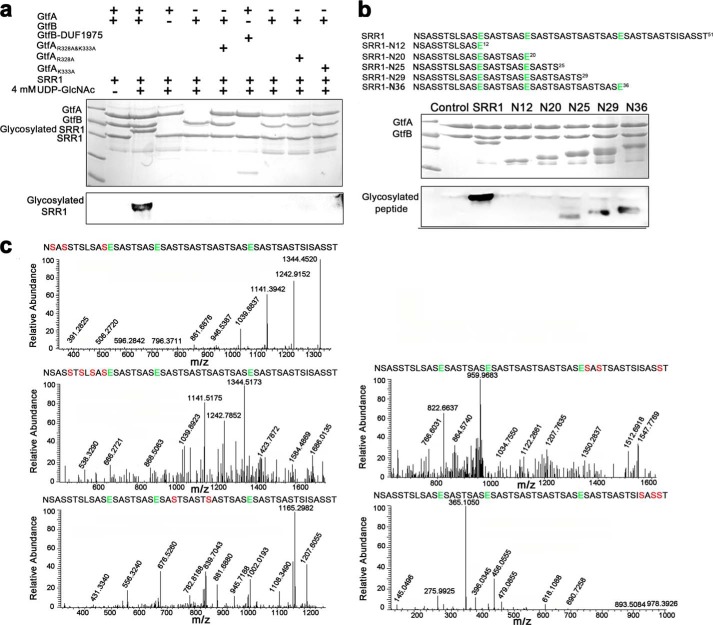
To determine the O-GlcNAcylation activity of GtfA-GtfB toward PsrP, in vitro O-GlcNAcylation assays were performed using UDP-GlcNAc as the sugar donor and SRR1 of PsrP as the peptide acceptor. Neither individual GtfA, GtfB, nor the complex of GtfA with GtfB-DUF1975 could catalyze the glycosylation of SRR1 (Fig. 3a). In contrast, the GtfA-GtfB complex glycosylated SRR1, resulting in a band of O-GlcNAcylated SRR1 of higher molecular weight (Fig. 3a). These results clearly demonstrated that formation of the GtfA-GtfB complex is crucial for the O-GlcNAcylation activity. Notably, no glycosylated SRR1 was detected in GtfA variants of either R328A and/or K333A, in agreement with the results of hydrolytic activity assays.
FIGURE 3.
In vitro O-GlcNAcylation assays. a, in vitro SRR1 O-GlcNAcylation assays. The reaction mixtures were separated by 12% SDS-PAGE (upper columns) and then subjected to Western blot analysis (lower column). The bands corresponding to GtfA, GtfB, GST-SRR1, and glycosylated GST-SRR1 are labeled. b, the O-GlcNAcylation pattern of SRR1. The sequences of the full-length SRR1 and five truncated variants (N12, N20, N25, N29, and N36) are shown. c, LC-MS/MS analyses of the glycosylated SRR1 catalyzed by the GtfA-GtfB complex. The Glu sites subject to GluC digestion are shown in green, and the detected Ser residues to be O-GlcNAcylated are labeled in red.
Despite the growing interests in the study of protein glycosylation of conserved SRRPs, it remains unknown how the SRRs are O-GlcNAcylated. To elucidate the O-GlcNAcylation pattern of PsrP, the full-length SRR1 and five truncated variants (termed SRR1-N12, -N20, -N25, -N29, and -N36, representing the peptides of corresponding N-terminal residues, respectively) were subject to O-GlcNAcylation assays. Only the four peptides of 25 or more residues were O-GlcNAcylated in vitro, whereas no O-GlcNAcylation of either SRR1-N12 or SRR1-N20 was detected (Fig. 3b). These data suggested that a peptide of proper length is necessary for the in vitro O-GlcNAcylation. Furthermore, liquid chromatography mass spectrometry analyses identified that only the Ser, but not Thr residues, were O-GlcNAcylated. In detail, O-GlcNAcylation of serine residues (Ser-2, -4, -5, -7, -9, -11, -23, -29, -37, -39, -47, -49, and -50) within SRR1 were identified by LC-MS (Fig. 3c). Although only half of the Ser residues were detected to be O-GlcNAcylated, it is possible that most or even all Ser residues within SRR1 are modified by O-GlcNAcylation upon the saturation of UDP-GlcNAc, as the molecular mass of the homogenous band of O-GlcNAcylated SRR1 is much greater than the unmodified SRR1 (Fig. 3a).
DUF1975 Is Required for Binding of GtfA to the Acceptor PsrP
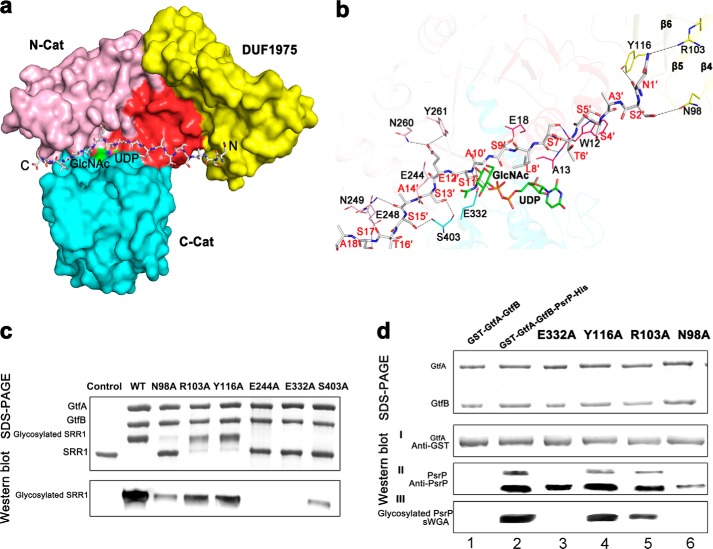
To dissect the acceptor binding site of GtfA, we initially tried co-crystallization of GtfA or GtfA-GtfB complex with various lengths of N-terminal peptide of SRR1 but did not succeed. Alternatively, we docked the 25-residue peptide SRR1-N25 recognized by the complex to our GtfA structure using the HADDOCK program (56). All output models suggested that GtfA only interacts with the N-terminal 18 residues, which was subsequently input to improve the docking model. Among the 12 output clusters, the first cluster of the lowest energy with 47 members satisfies the best interaction restraints and has a largest buried interface area of ∼2160 Å2. The overall backbone r.m.s.d. of 0.8 ± 0.5 Å for the 47 members indicated that the binding model was rather reliable on the theoretical calculation. In the model, the octadecapeptide lies over the UDP and GlcNAc binding sites, well complementary in both shape and charge to the interdomain cleft (Fig. 4a). The peptide chain runs along the cleft with its N terminus proximity to the β4 and β5 of DUF1975 domain. The octadecapeptide is mainly stabilized by residues Trp-12, Ala-13, Glu-18, Glu-244, Glu-248, Asn-249, Asn-260, and Tyr-261 of N-Cat in addition to three residues (Arg-328, Glu-332 and Ser-403) of C-Cat and three residues (Asn-98, Arg-103, and Tyr-116) of the DUF1975 domain (Fig. 4b). Our docking model is also consistent with the previous conclusion that the donor nucleotide-sugar commonly binds to the C-Cat of GT-B enzymes, whereas the acceptor binds to the N-Cat (57). Sequence analysis of these interacting residues suggests that most of them are highly conserved in GtfA homologs from Gram-positive bacteria that encode SRRPs.
FIGURE 4.
A docking model of the N-terminal octadecapeptide of SRR1 binding to GtfA. a, surface presentation of the acceptor peptide-binding site. The four segments of GtfA are colored red, yellow, pink, and cyan, respectively. The octadecapeptide is shown in gray sticks with the N and C termini labeled. The UDP and GlcNAc molecules are colored green. b, a close-up view of the interactions of the octadecapeptide with GtfA. The structure of GtfA was shown as semi-transparent cartoon. The interacting residues and the octadecapeptide are shown as sticks. The polar interactions are indicated as dashed lines. c, in vitro O-GlcNAcylation assays of GtfA and its mutants in the presence of GtfB and GST fused SRR1. The reaction mixtures were separated by electrophoresis and further subjected to Western blot analysis using WGA-HRP conjugate. d, DUF1975 is important for binding of GtfA to the acceptor PsrP. An E. coli recombinant system that co-expresses GtfA or GtfA variants with its acceptor PsrP was used to determine binding of GtfA to PsrP and the function of DUF1975 in the binding. Purified GST fused GtfA and GtfA variants (I),and its interacting partners PsrP (II) were determined by Western blot analysis using anti-GST antibody and anti-PsrP antibody and by Coomassie Brilliant Blue staining. Proteins prepared from GtfA-GtfB alone and co-expression of wild-type GtfA and variants (E322A, Y116A, R106A, and N98A)-GtfB with PsrP were also subjected to wheat germ agglutinin lectin blotting analysis (III).
The docking model represents a pose that Ser-11′ of PsrP is ready for being O-GlcNAcylated. The hydroxyl group of Ser-11′, stabilized by residues Arg-328 and Glu-332, has a distance of 2.9 Å to the β-phosphate oxygen of UDP and 3.1 Å to the anomeric oxygen of GlcNAc (Fig. 4b). The acceptor SRR1 peptide runs over the UDP-GlcNAc binding pocket, similar to the structure of hOGT-UDP-peptide complex (7). This conformation makes UDP-GlcNAc inaccessible to the solvent, consistent with the ordered bi-bi mechanism, in which the sugar donor UDP-GlcNAc binds before the acceptor peptide (58). However, Ser-11′ and UDP are aligned on the same face of GlcNAc plane, indicating GtfA is a retaining OGT, different from the hOGT and XcOGT that both adopt an inverting mechanism. For retaining glycosyltransferases, it has been suggested that the β-phosphate of UDP acts as a general base (59). In the model the hydroxyl group of Ser-11′ of PsrP is 3.5 Å to Glu-332-Oϵ2, and 2.9 Å to the β-phosphate oxygen of UDP, suggesting that Glu-332 of GtfA or UDP may serve as a general base to activate the nucleophilic serine residue. Moreover, Glu-332-Oϵ1 also makes a hydrogen bond with the amide backbone of Ser-11′ of PsrP.
To test the docking model, we first performed the in vitro glycosyltransferase activity assays. Mutation of the highly conserved residue Glu-244 and the putative catalytic residue Glu-332 to alanine completely abolished the O-GlcNAcylation activities (Fig. 4c). Meanwhile, mutation of Ser-403 to alanine significantly diminished the activity (Fig. 4c). Furthermore, we used E. coli in vivo glycosylation system to test the O-GlcNAcylation activity and binding to PsrP. Co-expression of GtfA-GtfB with the recombinant substrate PsrP led to O-GlcNAcylation of PsrP (Fig. 4dIII, second lane), which is also evident by appearance of a upper PsrP band (Fig. 4dII, second lane). PsrP proteins, both glycosylated upper band and non-glycosylated lower band, were readily co-purified with GtfA (Fig. 4dII, second lane), suggesting the in vivo binding between GtfA and PsrP. Mutation of Glu-332 to alanine completely inhibited glycosylation of PsrP but minimally affected the binding (Fig. 4dII, third lane), suggesting the Glu-332 residue is catalytically important as the subtle change has a dramatic impact on the GtfA activity. Importantly site-directed mutagenesis of three residues within DUF1975 reduced binding of GtfA to the acceptor PsrP and concurrently inhibited O-GlcNAcylation of PsrP (Fig. 4d, II and III, fourth-sixth lanes). Notably the binding between GtfA and GtfB were not dramatically changed in all site-directed mutants as indicated by co-purification of GtfA variants and GtfB (Fig. 4dII), demonstrating the unique requirement of DUF1795 for the substrate binding, a new function in addition to its ability to bind to the co-activator GtfB.
Both hOGT (6) and XcOGT (9) contain catalytic domains of GT-B fold after an all-αTPR domain that mediates the recognition to a broad range of proteins (10). Previous studies have proven that TPR, a scaffold for the recognition of partner proteins or acceptor peptides, is crucial for the activity of OGTs (9, 60). Similar to TPR, the DUF1975 domain of GtfA has also been implicated in mediating protein-protein interactions and is essential for the glycosylation of SRRPs (25, 61). In our docking model, we found that the two most N-terminal residues of SRR1, Asn-1′ and Ser-2′, form side-chain hydrogen bonds with residues Asn-98, Arg-103, and Tyr-116 of DUF1975 (Fig. 4b). Mutation of these three residues to alanine reduced the binding of GtfA to the peptide acceptor PsrP and decreased the both in vitro and in vivo glycosyltransferase activity of the GtfA-GtfB complex (Fig. 4, c and d). Thus, despite a totally different overall structure, DUF1975 acts as a domain for recognizing the acceptor, a role similar to TPRs in hOGT and XcOGT.
Taken together, we report here the structure of GtfA, the core subunit of a novel bacterial OGT, which is responsible for the O-GlcNAcylation of alternate serine residues of the serine-rich repeat adhesin PsrP. The β-meander add-on domain DUF1975 of GtfA is critical for the recognition of both co-activator GtfB and acceptor PsrP. The glycoproteomic analysis provides insights into a novel protein O-glycosylation fashion on multiple sites. Structure-based analysis may also assist in the future development of inhibitors against the biogenesis of bacterial SRRPs.
This work was supported, in whole or in part, by National Institutes of Health Grants F33DE022215 and R01DE 017954. This work was also supported by the Ministry of Science and Technology of China Grants 2013CB835300, 2014CB910100, and 2009CB918800 and National Natural Science Foundation of China Grant 31270781.
The atomic coordinates and structure factors (code 4PQG) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- OGT
- O-linked N-acetyl-d-glucosamine (GlcNAc) transferase
- hOGT
- human OGT
- XcOGT
- Xanthomonas campestris OGT
- PsrP
- pneumococcal serine-rich repeat proteins
- UDP-GlcNAc
- uridine diphosphate N-acetylglucosamine
- SeMet
- selenomethionine
- r.m.s.d.
- root mean square deviation
- TPR
- tetratricopeptide repeat
- GT-B
- glycosyltransferase type B
- SRR
- serine-rich repeat
- SRRP
- SRR glycoprotein.
REFERENCES
- 1. Drickamer K., Taylor M. E. (1998) Evolving views of protein glycosylation. Trends Biochem. Sci. 23, 321–324 [DOI] [PubMed] [Google Scholar]
- 2. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaidyanathan K., Durning S., Wells L. (2014) Functional O-GlcNAc modifications: implications in molecular regulation and pathophysiology. Crit Rev. Biochem. Mol. Biol. 49, 140–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torres C. R., Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 5. Yang Y. R., Suh P. G. (2014) O-GlcNAcylation in cellular functions and human diseases. Adv. Biol. Regul. 54, 68–73 [DOI] [PubMed] [Google Scholar]
- 6. Lazarus M. B., Nam Y., Jiang J., Sliz P., Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lazarus M. B., Jiang J., Gloster T. M., Zandberg W. F., Whitworth G. E., Vocadlo D. J., Walker S. (2012) Structural snapshots of the reaction coordinate for O-GlcNAc transferase. Nat. Chem. Biol. 8, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Fleites C., Macauley M. S., He Y., Shen D. L., Vocadlo D. J., Davies G. J. (2008) Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 15, 764–765 [DOI] [PubMed] [Google Scholar]
- 9. Clarke A. J., Hurtado-Guerrero R., Pathak S., Schüttelkopf A. W., Borodkin V., Shepherd S. M., Ibrahim A. F., van Aalten D. M. (2008) Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 27, 2780–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jínek M., Rehwinkel J., Lazarus B. D., Izaurralde E., Hanover J. A., Conti E. (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat. Struct. Mol. Biol. 11, 1001–1007 [DOI] [PubMed] [Google Scholar]
- 11. Shen A., Kamp H. D., Gründling A., Higgins D. E. (2006) A bifunctional O-GlcNAc transferase governs flagellar motility through anti-repression. Genes Dev. 20, 3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nothaft H., Szymanski C. M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 [DOI] [PubMed] [Google Scholar]
- 13. Kukita K., Kawada-Matsuo M., Oho T., Nagatomo M., Oogai Y., Hashimoto M., Suda Y., Tanaka T., Komatsuzawa H. (2013) Staphylococcus aureus SasA is responsible for binding to the salivary agglutinin gp340, derived from human saliva. Infect. Immun. 81, 1870–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheen T. R., Jimenez A., Wang N. Y., Banerjee A., van Sorge N. M., Doran K. S. (2011) Serine-rich repeat proteins and pili promote Streptococcus agalactiae colonization of the vaginal tract. J. Bacteriol. 193, 6834–6842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanchez C. J., Shivshankar P., Stol K., Trakhtenbroit S., Sullam P. M., Sauer K., Hermans P. W., Orihuela C. J. (2010) The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog. 6, e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pyburn T. M., Bensing B. A., Xiong Y. Q., Melancon B. J., Tomasiak T. M., Ward N. J., Yankovskaya V., Oliver K. M., Cecchini G., Sulikowski G. A., Tyska M. J., Sullam P. M., Iverson T. M. (2011) A structural model for binding of the serine-rich repeat adhesin GspB to host carbohydrate receptors. PLoS Pathog 7, e1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King N. P., Beatson S. A., Totsika M., Ulett G. C., Alm R. A., Manning P. A., Schembri M. A. (2011) UafB is a serine-rich repeat adhesin of Staphylococcus saprophyticus that mediates binding to fibronectin, fibrinogen, and human uroepithelial cells. Microbiology 157, 1161–1175 [DOI] [PubMed] [Google Scholar]
- 18. Lizcano A., Sanchez C. J., Orihuela C. J. (2012) A role for glycosylated serine-rich repeat proteins in Gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 27, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou M., Wu H. (2009) Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155, 317–327 [DOI] [PubMed] [Google Scholar]
- 20. Takamatsu D., Bensing B. A., Sullam P. M. (2004) Four proteins encoded in the gspB-secY2A2 operon of Streptococcus gordonii mediate the intracellular glycosylation of the platelet-binding protein GspB. J. Bacteriol. 186, 7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shivshankar P., Sanchez C., Rose L. F., Orihuela C. J. (2009) The Streptococcus pneumoniae adhesin PsrP binds to Keratin 10 on lung cells. Mol. Microbiol. 73, 663–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hegge F. T., Hitchen P. G., Aas F. E., Kristiansen H., Løvold C., Egge-Jacobsen W., Panico M., Leong W. Y., Bull V., Virji M., Morris H. R., Dell A., Koomey M. (2004) Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc. Natl. Acad. Sci. U.S.A. 101, 10798–10803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seepersaud R., Bensing B. A., Yen Y. T., Sullam P. M. (2012) The accessory Sec protein Asp2 modulates GlcNAc deposition onto the serine-rich repeat glycoprotein GspB. J. Bacteriol. 194, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu R., Zhou M., Wu H. (2010) Purification and characterization of an active N-acetylglucosaminyltransferase enzyme complex from Streptococci. Appl. Environ. Microbiol. 76, 7966–7971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu R., Wu H. (2011) A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins. J. Biol. Chem. 286, 34923–34931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromolecular Crystallography 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Brodersen D. E., de La Fortelle E., Vonrhein C., Bricogne G., Nyborg J., Kjeldgaard M. (2000) Applications of single-wavelength anomalous dispersion at high and atomic resolution. Acta Crystallogr. D Biol. Crystallogr. 56, 431–441 [DOI] [PubMed] [Google Scholar]
- 28. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 29. Hao Q., Gu Y. X., Zheng C. D., Fan H. F. (2000) OASIS: a computer program for breaking phase ambiguity in one-wavelength anomalous scattering or single isomorphous substitution (replacement) data. J. Appl. Crystallogr. 33, 980–981 [Google Scholar]
- 30. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laskowski R., Macarthur M., Moss D., Thornton J. (1993) Procheck: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 39. DeLano W. (2002) The PyMOL Molecular Graphics System, De Lano Scientific, San Carlos CA [Google Scholar]
- 40. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 41. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 42. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 43. Ruane K. M., Davies G. J., Martinez-Fleites C. (2008) Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins 73, 784–787 [DOI] [PubMed] [Google Scholar]
- 44. Parsonage D., Newton G. L., Holder R. C., Wallace B. D., Paige C., Hamilton C. J., Dos Santos P. C., Redinbo M. R., Reid S. D., Claiborne A. (2010) Characterization of the N-acetyl-α-d-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis. Biochemistry 49, 8398–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 46. Vetting M. W., Frantom P. A., Blanchard J. S. (2008) Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis. J. Biol. Chem. 283, 15834–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu Y., Chen L., Ha S., Gross B., Falcone B., Walker D., Mokhtarzadeh M., Walker S. (2003) Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc. Natl. Acad. Sci. U.S.A. 100, 845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buschiazzo A., Ugalde J. E., Guerin M. E., Shepard W., Ugalde R. A., Alzari P. M. (2004) Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation. EMBO J. 23, 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cottee M. A., Muschalik N., Wong Y. L., Johnson C. M., Johnson S., Andreeva A., Oegema K., Lea S. M., Raff J. W., van Breugel M. (2013) Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife 2, e01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Makabe K., McElheny D., Tereshko V., Hilyard A., Gawlak G., Yan S., Koide A., Koide S. (2006) Atomic structures of peptide self-assembly mimics. Proc. Natl. Acad. Sci. U.S.A. 103, 17753–17758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Greenwald J., Nader M., Celia H., Gruffaz C., Geoffroy V., Meyer J. M., Schalk I. J., Pattus F. (2009) FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol. Microbiol. 72, 1246–1259 [DOI] [PubMed] [Google Scholar]
- 52. Kawai F., Grass S., Kim Y., Choi K. J., St Geme J. W., 3rd, Yeo H. J. (2011) Structural insights into the glycosyltransferase activity of the Actinobacillus pleuropneumoniae HMW1C-like protein. J. Biol. Chem. 286, 38546–38557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ihara H., Ikeda Y., Toma S., Wang X., Suzuki T., Gu J., Miyoshi E., Tsukihara T., Honke K., Matsumoto A., Nakagawa A., Taniguchi N. (2007) Crystal structure of mammalian α1,6-fucosyltransferase, FUT8. Glycobiology 17, 455–466 [DOI] [PubMed] [Google Scholar]
- 54. Kakuta Y., Okino N., Kajiwara H., Ichikawa M., Takakura Y., Ito M., Yamamoto T. (2008) Crystal structure of Vibrionaceae photobacterium sp. JT-ISH-224 α2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: catalytic mechanism and substrate recognition. Glycobiology 18, 66–73 [DOI] [PubMed] [Google Scholar]
- 55. Qasba P. K., Ramakrishnan B. (2007) Letter to the glyco-forum: catalytic domains of glycosyltransferases with “add-on” domains. Glycobiology 17, 7G–9G [DOI] [PubMed] [Google Scholar]
- 56. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 57. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 58. Gross B. J., Kraybill B. C., Walker S. (2005) Discovery of O-GlcNAc transferase inhibitors. J. Am. Chem. Soc. 127, 14588–14589 [DOI] [PubMed] [Google Scholar]
- 59. Errey J. C., Lee S. S., Gibson R. P., Martinez Fleites C., Barry C. S., Jung P. M., O'Sullivan A. C., Davis B. G., Davies G. J. (2010) Mechanistic insight into enzymatic glycosyl transfer with retention of configuration through analysis of glycomimetic inhibitors. Angew. Chem. Int. Ed. Engl. 49, 1234–1237 [DOI] [PubMed] [Google Scholar]
- 60. Iyer S. P., Hart G. W. (2003) Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 278, 24608–24616 [DOI] [PubMed] [Google Scholar]
- 61. Li Y., Chen Y., Huang X., Zhou M., Wu R., Dong S., Pritchard D. G., Fives-Taylor P., Wu H. (2008) A conserved domain of previously unknown function in Gap1 mediates protein-protein interaction and is required for biogenesis of a serine-rich streptococcal adhesin. Mol. Microbiol. 70, 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]