Background: To understand molecular processes underlying cartilage destruction, we analyzed COMP fragments released into synovial fluid in joint diseases.
Results: Twelve novel COMP neoepitopes have been identified.
Conclusion: The release of COMP neoepitopes provides means for monitoring disease progression.
Significance: Based on the specificity, selectivity, and sensitivity of each neoepitope, a new generation of biomarkers for cartilage destruction can be developed.
Keywords: Arthritis, Biomarker, Cartilage, Mass Spectrometry (MS), Osteoarthritis, COMP, ELISA Development, Affinity Purification, Joint Disease, Neoepitope
Abstract
To identify patients at risk for progressive joint damage, there is a need for early diagnostic tools to detect molecular events leading to cartilage destruction. Isolation and characterization of distinct cartilage oligomeric matrix protein (COMP) fragments derived from cartilage and released into synovial fluid will allow discrimination between different pathological conditions and monitoring of disease progression. Early detection of disease and processes in the tissue as well as an understanding of the pathologic mechanisms will also open the way for novel treatment strategies. Disease-specific COMP fragments were isolated by affinity chromatography of synovial fluids from patients with rheumatoid arthritis, osteoarthritis, or acute trauma. Enriched COMP fragments were separated by SDS-PAGE followed by in-gel digestion and mass spectrometric identification and characterization. Using the enzymes trypsin, chymotrypsin, and Asp-N for the digestions, an extensive analysis of the enriched fragments could be accomplished. Twelve different neoepitopes were identified and characterized within the enriched COMP fragments. For one of the neoepitopes, Ser77, an inhibition ELISA was developed. This ELISA quantifies COMP fragments clearly distinguishable from total COMP. Furthermore, fragments containing the neoepitope Ser77 were released into the culture medium of cytokine (TNF-α and IL-6/soluble IL-6 receptor)-stimulated human cartilage explants. The identified neoepitopes provide a complement to the currently available commercial assays for cartilage markers. Through neoepitope assays, tools to pinpoint disease progression, evaluation methods for therapy, and means to elucidate disease mechanisms will be provided.
Introduction
Destruction of articular cartilage and changes of the underlying bone are key characteristics of joint diseases such as osteoarthritis (OA)2 and rheumatoid arthritis (RA). These pathological conditions resulting in tissue degradation constitute a major medical, social, and economic problem (1). To minimize permanent tissue damage caused by pathological cartilage degeneration, it is important to diagnose such conditions at an early stage (2).
In progressive joint diseases, the degradation of extracellular matrix proteins and proteoglycans leads to irreversible alterations in the properties of the collagen network. In addition, the imbalance in the turnover of matrix proteins often results in increased proteolysis of molecules bound to and exposed at the surface of collagen fibers such as fibromodulin, decorin, and cartilage oligomeric matrix protein (COMP) (3–5). During the last decade, efforts have been made to find suitable biological markers that enable early detection of pathological cartilage degeneration (6, 7).
One such biological marker is COMP. Elevated serum levels of COMP are associated with ongoing joint destruction in rheumatoid arthritis (8–10). COMP is cleaved and released from the cartilage tissue into synovial fluid in OA, RA, and other forms of inflammatory arthritis (6, 10–13), and it is well established that COMP can be used as a marker of cartilage turnover (14).
COMP is primarily found in cartilage (15), but it has also been found in other tissues such as synovium and tendon (16, 17). It is a pentameric protein of 524 kDa (15) in which where the monomers are joined together by a coiled coil domain in the N terminus. Each N-terminal domain is followed by four EGF repeat domains, eight thrombospondin type III domains, and a C-terminal globular domain (18). The C-terminal domain is involved in interactions with other proteins in the extracellular matrix such as collagens I, II, and IX (19–21). Each of the five globular C-terminal domains binds collagen with a KD ≃10−9 and thereby catalyzes collagen fibril assembly (19). Many proteases have been shown to degrade COMP, but the specific cleavage sites within COMP as well as the newly formed N and C termini have so far not been described.
In this work, we identified 12 novel COMP neoepitopes and hereby describe the newly formed N- and C-terminal ends. These neoepitopes were identified through affinity enrichments of knee joint synovial fluids from patients with acute trauma, OA, and RA followed by mass spectrometric identification and characterization of the enriched COMP fragments.
By using an in vitro model of joint disease, we have successfully demonstrated the presence of the COMP neoepitope3 Ser77 as a released fragment from cartilage explants. We have subsequently verified that the same cleavage occurs in vivo by showing the presence of neoepitope Ser77 in the synovial fluid from 16 different patients with acute knee pain. Furthermore, an inhibition ELISA was developed for the neoepitope Ser77 that specifically distinguished and quantified this neoepitope from total COMP.
EXPERIMENTAL PROCEDURES
Materials
Ammonium bicarbonate (NH4HCO3), dithiothreitol (DTT), formic acid, iodoacetamide, N-ethylmaleimide, trifluoroacetic acid (TFA), and Triton X-100 were purchased from Sigma-Aldrich. Anhydrous sodium acetate was purchased from Merck. Sodium chloride (NaCl) was from Scharlab S.L. (Barcelona, Spain). Albumin bovine fraction V, pH 7.0 (BSA) was purchased from SERVA Electrophoresis GmbH (Heidelberg, Germany). The HPLC grade acetonitrile was from Rathburn (Walkerburn, Scotland, UK). Trypsin was purchased from Promega (Madison, WI). Chymotrypsin and Asp-N were from Roche Applied Science. Stage tips were homemade according to Ref. 22 from 47-mm Empore C18 extraction discs (3M, Minneapolis, MN).
Human Clinical Samples
Synovial fluid was aspirated from knees of patients with acute knee pain with or without acute trauma (seeking care at the emergency room within the 1st week), established OA, and established RA (Table 1). The use of patient samples was approved by The Ethical Committee in Lund, Sweden (411/2005). The samples were stored at −80 °C prior to analysis.
TABLE 1.
Cohort demographics
| AT | Knee pain | OA | RA | |
|---|---|---|---|---|
| n | 19 | 16 | 20 | 20 |
| Mean age (years) | 31.2 | 43.9 | 67.7 | 64.3 |
| Age range (years) | 13–65 | 16–65 | 55–85 | 41–83 |
Human donor knee cartilage from the tibial plateau (19-year-old male, modified Collins scale (23) grade 0 (normal)) was obtained from the Gift of Hope Organ and Tissue Donor Network (Elmhurst, IL). The procedure was approved by the Office of Research Affairs at Rush-Presbyterian-St. Luke's Medical Center and the Committee on Use of Humans as Experimental Subjects at the Massachusetts Institute of Technology.
Antibodies
The mouse monoclonal antibody (2D3) toward the N-terminal domain of COMP was described previously (24, 25). The mouse monoclonal antibody toward the thrombospondin type III domain (TSP-III) was developed in house (26). The C-terminal antibody used in Western blots was a peptide antibody toward the last 15 amino acids in human COMP raised in goat (27).
Neoepitope antibodies were raised using immunogenic peptides containing the 5-mer neoepitope cleavage site followed by three glycine residues and one cysteine coupled to keyhole limpet hemocyanin (GenScript, Piscataway, NJ). For antibody affinity purification, the peptide CDACGMQQS77 (internal cysteine was alkylated) was immobilized to SulfoLink coupling resin according to the manufacturer's protocol (Thermo Scientific, Rockford, IL). The column was equilibrated with HBS (10 mm HEPES, 150 mm NaCl, pH 7.4) prior to incubation for 1 h at room temperature with crude neoepitope sera. The column was washed with 15 ml of HBS and 2 ml of HBS with 1 m NaCl before elution of antibodies using 3 m potassium thiocyanate. The eluate was immediately desalted on a PD10 column (GE Healthcare) equilibrated with HBS.
Affinity Enrichment of COMP from Synovial Fluid
For affinity enrichments, mouse monoclonal antibodies toward the N-terminal domain (2D3) or toward the TSP-III domain were coupled to MiniLeak gel according to the manufacturer's protocol (KemEnTec, Denmark). Synovial fluid from a patient with acute trauma was diluted with 2 volumes of PBS I (20 mm phosphate, 150 mm NaCl, pH 7.4) containing 0.8% (w/v) SDS and incubated for 2 h at room temperature. Excess SDS was neutralized by addition of 1 volume of 4% Triton X-100 in PBS I and incubated overnight at room temperature. In small scale enrichment of synovial fluid without SDS, the same fragments were enriched (data not shown), and thus SDS was omitted. Synovial fluids from patients with OA and RA were incubated with 5 mm N-ethylmaleimide prior to centrifugation for 20 min at 1000 × g at room temperature. To diminish unspecific binding to the MiniLeak gel, the synovial fluid samples were first passed through a column containing MiniLeak gel without any bound antibody. The flow-through was then applied to the affinity column with the N-terminal antibody, and subsequently the flow-through from the N-terminal affinity column was applied to the affinity column with the TSP-III domain antibody. The columns were washed with HBS, HBS with 0.5 m NaCl, and finally HBS. Bound proteins were eluted using 0.1 m citrate, pH 3 and immediately neutralized with 1.5 m Tris, pH 8.8. Eluted fractions were precipitated with ethanol overnight at 4 °C and collected by centrifugation (13,200 × g, 4 °C, 30 min). The pellets were reprecipitated in ethanol with 50 mm sodium acetate for 4 h at −20 °C and collected by centrifugation (13,200 × g, 4 °C, 30 min) prior to SDS-PAGE.
SDS-PAGE and Western Blot
The precipitate pellets were dissolved in SDS-PAGE sample buffer (28) without reducing agent and separated on 4–16% gradient SDS-polyacrylamide gels. Triplicate bands from each eluate were digested with trypsin, chymotrypsin, and Asp-N as described below.
In screening synovial fluids, samples were diluted 10× with non-reducing sample buffer and separated by 4–16% SDS-PAGE. Following electrophoresis, proteins were transferred to PVDF membranes (Thermo Scientific) at 100 V for 1 h in 25 mm Tris, 192 mm glycine, 10% methanol. Membranes were blocked for 1 h at room temperature in 3% (w/v) BSA in T-TBS (10 mm Tris, 150 mm NaCl, 0.05% Tween 20, pH 7.4). Primary antibodies were diluted in T-TBS with 3% (w/v) BSA (monoclonal antibodies, 10,000× dilution; polyclonal antibodies, 1000× dilution) and incubated either for 1 h at room temperature or overnight at 4 °C. As secondary antibody either goat anti-mouse or donkey anti-rabbit was used (Jackson ImmunoResearch Laboratories, West Grove, PA); both were diluted 10,000× in T-TBS with 3% BSA. Immunoblots were visualized using SuperSignal® WestDura Extended Duration Substrate (Thermo Scientific) on a luminescence image analyzer (Fuji Film LAS-1000).
Mass Spectrometry Sample Preparation
After staining overnight with blue silver stain (29), bands of interest were excised and reduced with 10 mm DTT at 56 °C for 30 min and alkylated with 50 mm iodoacetamide for 30 min at room temperature (dark). Bands were digested overnight at 37 °C with either 20 ng/μl trypsin in 25 mm NH4HCO3, 50 ng/μl chymotrypsin in 25 mm NH4HCO3, or 40 ng/μl Asp-N in 50 mm phosphate buffer, pH 8. Peptides were extracted consecutively with 1% TFA, twice with 50% acetonitrile in 0.1% TFA, and 100% acetonitrile. After drying, the extracted peptides were dissolved in 10 μl of 0.1% TFA and purified on C18 stage tips (22).
Mass Spectrometry
Purified peptide samples were redissolved in 0.2% formic acid and analyzed on an Esquire HCT IonTrap (Bruker Daltonik, Bremen, Germany) as described (30). Database searches were performed using Mascot (version 2.1) MS/MS Ions Search in the UniProtKB (2010_09) database. Mascot search parameters included carbamidomethylation of cysteine as a fixed modification and deamidation (Asn and Gln) and oxidation (Met) as variable modifications. Other Mascot search parameters were monoisotopic masses, ±0.4-Da peptide mass tolerance, ±0.4-Da fragment mass tolerance, maximum of three missed cleavages, minimum ion score of 20, and taxonomy of Homo sapiens. Database searches using semi-style cleavages were used to identify peptides containing a neoepitope end. The peptide ends not representing a tryptic, chymotryptic, or Asp-N cleavage are referred to as the neoepitopes. To remove false positive neoepitopes caused by unspecificity of the proteases used, recombinant COMP was digested with trypsin, chymotrypsin, and Asp-N. Only for chymotrypsin were three neoepitopes found in the control samples (data not shown), and these were removed from the resulting summary of neoepitopes.
Neoepitope Ser77 Inhibition ELISA and Total COMP ELISA
An inhibition ELISA was developed for the neoepitope Ser77 and was used to quantify synovial fluid samples from patients with RA, OA, and acute trauma. The peptide CDACGMQQS77 (internal cysteine was alkylated) was cross-linked to BSA using m-maleimidobenzoyl-N-hydroxysuccinimide ester (Thermo Scientific) according to the manufacturer's protocol. All incubations were performed at room temperature. Coating concentrations and buffers were optimized (data not shown), and binding plates were prepared by coating 96-well microtiter plates (Nunc-Immuno plates, Maxisorp, Nunc Intermed Ltd., Copenhagen, Denmark) with 50 μl of BSA-cross-linked peptide (12.5 ng/ml in PBS II (150 mm NaCl, 10 mm potassium phosphate, pH 7.4) overnight.
Coated wells were washed with 0.15 m sodium chloride, 0.05% (w/v) Tween 20 and blocked with 120 μl of 2 mg/ml BSA in PBS II for 1 h. A protein fragment containing the N-terminal part of COMP (amino acids 20–77) and ending at neoepitope Ser77 was a generous gift provided by Dr. Tobias Gustavsson (The Novo Nordisk Foundation, Center for Protein Research, University of Copenhagen, Copenhagen, Denmark). The protein fragment, labeled NT-Ser77, was reduced and alkylated as described above prior to use. Standard NT-Ser77 (2 μg/ml to 15.6 ng/ml) and synovial fluids (diluted 40×) were diluted in a solution of 0.5% BSA, 0.8% (w/v) SDS, 10 mm EDTA in incubation buffer (0.14 m NaCl, 8 mm sodium phosphate, 1.5 mm potassium phosphate, 2.7 mm KCl, pH 7.4) and incubated overnight in a Sterilin plate (Bibby Sterilin Ltd., UK). One volume of affinity-purified neoepitope Ser77 antibody (0.1 ng/ml) in 4% Triton X-100 in 10 mm sodium phosphate, pH 7.4 was added to the Sterilin plates. After 1-h preincubation, 50 μl of the mixture was added to the coated wells of the Nunc plate. The plate was incubated for 1 h prior to washing, and bound antibodies were detected by the addition of 50 μl of swine anti-rabbit IgG-alkaline phosphatase (Dako A/S, Denmark) diluted 1:1000 in 2 mg/ml BSA, incubation buffer with 0.05% Tween 20. After 1-h incubation, the plates were washed, and 100 μl of substrate was added (1 mg/ml p-nitrophenyl phosphate in 1 m diethanolamine, pH 9.8 containing 0.5 m MgCl2). The absorbance of each sample and standard was measured at 405 nm in duplicate by a microplate reader (Expert96, AsysHitech, Austria). The Mikrowin 200 software program (AsysHitech) was used to plot the calibration curve and to calculate the amount of COMP neoepitope in the samples. Total COMP was measured in the same synovial fluid samples (diluted 40× or 80×) according to the manufacturer's protocol (AnaMar AB, Lund, Sweden).
Cartilage Explants
Full thickness human knee cartilage explants (3-mm diameter, ∼1.5 mm thick) were obtained using a dermal punch. Disks were maintained in high glucose DMEM supplemented with 10 mm HEPES buffer, 1% insulin-transferrin-selenium (10 μg/ml, 5.5 μg/ml, and 5 ng/ml, respectively), 0.1 mm nonessential amino acids, 0.4 mm proline, 20 μg/ml ascorbic acid, 100 units/ml penicillin G, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B in a 37 °C, 5% CO2 incubator prior to treatment. After 2 days of equilibration, groups of cartilage explants were cultured with an added cytokine mixture consisting of 100 ng/ml TNF-α, 50 ng/ml IL-6, and 250 ng/ml sIL-6R as described previously (31). Control explants were maintained in culture medium as above with no cytokines added. Medium changes were carried out every 3 days, and used medium was collected for analysis at each time point between day 3 and day 21 during the 21-day culture.
Statistical Methods
Data from neoepitope Ser77 ELISA did not show a Gaussian distribution so all statistical tests used were non-parametric. Data are presented as median values with interquartile range. A p value <0.05 was considered to be statistically significant. Analyses were performed using unpaired Mann-Whitney U test in GraphPad Prism Version 6 (GraphPad Software Inc., La Jolla, CA).
RESULTS
Identification and Characterization of COMP Neoepitopes in Synovial Fluids
Using mouse monoclonal antibodies toward the N-terminal coiled coil domain and the thrombospondin type III domain, affinity enrichments of synovial fluids from patients with joint disease were performed. Mass spectrometric characterization of the enriched COMP fragments resulted in the identification of 12 novel neoepitopes (Table 2). Peptides ending with an amino acid other than those formed by the proteases used (trypsin, chymotrypsin, and Asp-N) are referred to as neoepitopes. The ending amino acids of the neoepitopes are superscripted.
TABLE 2.
COMP neoepitopes identified in synovial fluid from patients with AT, OA, and RA
The end of the identified peptide that corresponds to a neoepitope is superscripted. The highest MS/MS ion score is presented, and the number of bands in which the neoepitope was identified is shown in parentheses. CT, chymotrypsin.
| Peptide identified by MS | Synovial fluid | Digesting enzyme | Neoepitope amino acid | Ion score (n) |
|---|---|---|---|---|
| NTVMECDACGMQQS77↓VR | AT | Trypsin | Ser77 | 64 (1) |
| LH↓91CAPGFCFPGVACIQTESGAR | OA | Trypsin | 91Cys | 33 (1) |
| IQ↓105TESGARCGPCPAGF | RA | CT | 105Thr | 41 (2) |
| PN↓195SVCINTRGSF | RA | CT | 195Ser | 32 (4) |
| QVCTDINECETGQHNCVPNSVCINTRG202↓SF | OA | Trypsin | Gly202 | 22 (1) |
| QCGPCQPGFVGDQASGCQRRAQ226↓RF | RA | CT | Gln226 | 48 (3) |
| PE↓523NAEVTLTDFR | OA | Trypsin | 523Asn | 57 (1) |
| IDVCPENAEVTLTDF531↓RA | AT | Trypsin | Phe531 | 52 (1) |
| VL↓555NQGREIVQTMNS | RA, OA | Asp-N | 555Asn | 43 (3) |
| EIVQTMNSDPGLAVGY574↓TA | OA | Trypsin | Tyr574 | 38 (2) |
| EIVQTMNSDPGLAVGYTAF577↓NG | OA | Trypsin | Phe577 | 38 (2) |
| HT↓658GDTESQVRLLWK | RA | Asp-N | 658Gly | 26 (1) |
In summary, two neoepitopes were identified in synovial fluid from a patient with acute trauma (Ser77 and Phe531), six were identified in synovial fluid from a patient with OA (91Cys, G202, 523Asn, 555Asn, Tyr574, and Phe577), and five were identified in synovial fluid from a patient with RA (Thr105, 195Ser, Gln226, 555Asn, and 658Gly). Six of the neoepitopes contain a new C-terminal end (Ser77, Gly202, Gln226, Phe531, Tyr574, and Phe577), and six contain a new N-terminal end (91Cys, 105Thr, 195Ser, 523Asn, 555Asn, and 658Gly). Furthermore, six of the neoepitopes (Ser77, 91Cys, 105Thr, 195Ser, Gly202, and Gln226) are located between the N-terminal and the thrombospondin type III domains, whereas the other six neoepitopes (523Asn, Phe531, 555Asn, Tyr574, Phe577, and 658Gly) are located within or in close proximity to the C-terminal globular domain.
A representative SDS-polyacrylamide gel and Western blot of enriched COMP fragments from OA synovial fluid with the excised bands used for identification of the neoepitopes are shown in Fig. 1. COMP was detected with high sequence coverage (18–54%) in all bands except the very weak band G, which only contained keratin. In the 300-kDa band (labeled C), the neoepitopes Gly202, Tyr574, and Phe577 were identified. The 70-kDa band (labeled L) resulted in identification of neoepitopes 91Cys, 523Asn, Tyr574, and Phe577. As can be seen, some of the neoepitopes (Tyr574 and Phe577) were identified in both bands C and L. In band L, it was clearly shown that this fragment does not contain the N-terminal domain as seen by the enrichment of this fragment on the TSP-III column but not the N-terminal domain column and by the Western blot of the eluted fractions (Fig. 1). The most N-terminally detected peptide in this band contained the neoepitope 91Cys, indicating that the N terminus has been cleaved off at or before this site.
FIGURE 1.
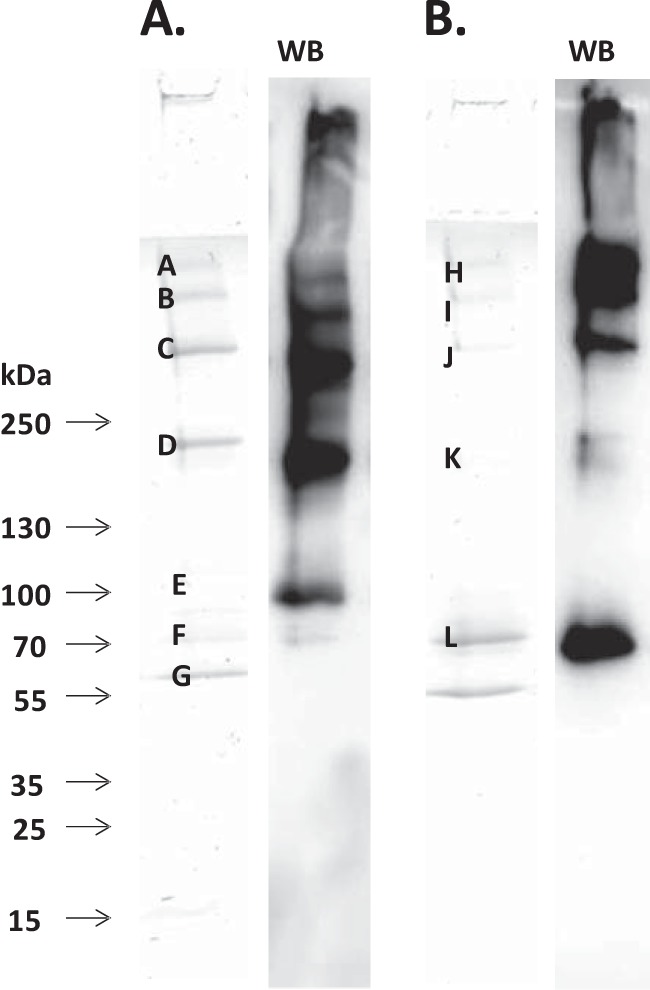
Representative image of COMP fragments in OA synovial fluid. COMP fragments from OA synovial fluid were affinity-enriched using mouse monoclonal antibodies toward the N-terminal domain (A) and toward the TSP-III of COMP (B). Eluates were separated by non-reducing 4–16% SDS-PAGE and either stained with blue silver (29) or blotted to PVDF membranes (marked WB) followed by visualization using the N-terminal antibody for the N terminus-enriched eluate or using the TSP-III antibody for the TSP-III-enriched eluate. In-gel digests of the enriched fragments were analyzed with mass spectrometry as described under “Experimental Procedures.” Bands are labeled A–L, and neoepitopes were identified in bands labeled C and L.
Only one neoepitope, 555Asn, was identified in both OA and RA. However, that all neoepitopes could occur in all diseases but at different levels depending on the disease state cannot be ruled out.
Synovial Fluids from Patients with Acute Knee Pain Contain the Neoepitope Ser77
Synovial fluids from 16 patients were analyzed by Western blots using neoepitope antibody Ser77. No difference was seen in terms of the volume of aspirated synovial fluid (ranging from 20 to 135 ml). A band of ∼40 kDa is detectable in all samples (Fig. 2). This band corresponds to cleavage at Ser77 within all five monomers of the COMP pentamer. Because the gels and blots were analyzed under non-reducing conditions, the N-terminal coiled coil domain remains intact and migrates as a cleaved pentamer of 40 kDa. For several of the samples, a band at 100 kDa, corresponding to fragmentations at Ser77 at four of five monomers within the COMP pentamer, is also visible. A band at 15 kDa is also detected for some of the samples; these bands probably reflect further fragmentations within the coiled coil domain resulting in reduced molecular weight of the migrating molecule.
FIGURE 2.
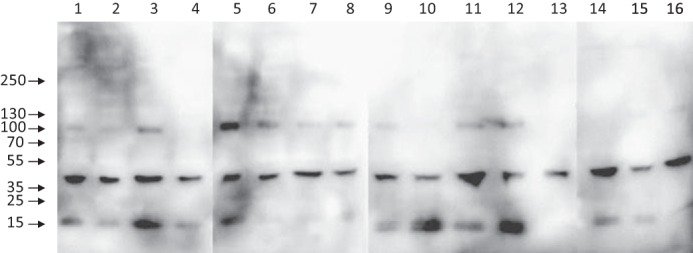
The neoepitope Ser77 is present in synovial fluid from patients with acute knee pain. Western blots of synovial fluid from knee pain patients using COMP neoepitope antibody Ser77 are shown. The presence of neoepitope Ser77 is clearly visible in all analyzed samples (1–16) as a band migrating at 40 kDa. For several of the samples, bands of 100 and 15 kDa are also visible. Samples are distributed based on aspirated synovial fluid volume ranging from 20 (sample 1) to 135 ml (sample 16). Samples 3, 7, and 11 had an acute trauma.
Quantification of Neoepitope Ser77 and Total COMP in Synovial Fluid from Patients with Different Joint Diseases
An inhibition ELISA was developed to measure the amounts of neoepitope Ser77 in synovial fluid of patients with acute trauma (n = 19), OA (n = 20), and RA (n = 20) (Fig. 3A). The median values of neoepitope Ser77 for the patient groups were 8.41 μg/ml (interquartile range, 5.76–18.12) for acute trauma, 4.52 μg/ml (interquartile range, 3.42–9.92) for OA, and 1.23 μg/ml (interquartile range, 0.70–1.62) for RA.
FIGURE 3.
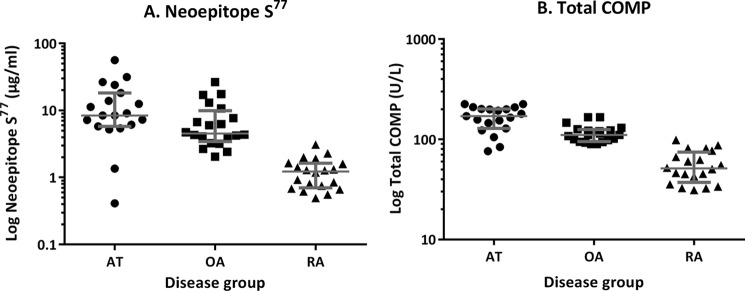
ELISA quantification of neoepitope Ser77 and total COMP in synovial fluid from patients with joint disease. A, an inhibition ELISA developed for neoepitope Ser77 was used to quantify the amounts of neoepitope present in synovial fluids from knee patients with AT, OA, and RA. Logarithmic median values with whiskers at the interquartile range are presented. Mann-Whitney p values between the groups were: AT versus OA, p = 0.0377; OA versus RA, p = < 0.0001; AT versus RA, p = < 0.0001. B, total amount of COMP was measured using the AnaMar COMP assay. Mann-Whitney p values between the groups were: AT versus OA, p = 0.0005; OA versus RA, p = < 0.0001; AT versus RA, p = 0.0001.
Total COMP as measured using the AnaMar assay also was determined (Fig. 3B). The median values of total COMP for the patient groups were 170.5 units/liter (interquartile range, 128.0–200.7) in acute trauma, 110.3 units/liter (interquartile range, 95.12–125.4) for OA, and 51.15 units/liter (interquartile range, 37.23–74.16) for RA.
By comparing the median values of neoepitope versus total COMP among the three patients groups, we could see that the neoepitope assay was distinguishable from the total COMP assay. Using the total COMP assay, the highest levels were present in the acute trauma (AT) group (∼3.3- and 1.5-fold higher than the RA and OA groups, respectively). There were also 2.2-fold higher levels in the OA versus the RA group.
With the neoepitope Ser77 assay, the AT group also had the highest levels (6.9- and 1.9-fold higher than the RA and OA groups respectively). There were also 4.5-fold higher levels in the OA versus the RA group. The coefficients of variation for the total COMP levels were 28 (AT), 20 (OA), and 36% (RA), and for the neoepitope assay, they were 98 (AT), 85 (OA), and 53% (RA).
COMP Neoepitope Ser77 Fragments Are Released from Cytokine-stimulated Human Cartilage Explants
Human cartilage explant plugs were incubated with cytokines (TNF-α and IL-6/sIL-6R), and the COMP fragments released into the incubation media were analyzed by Western blots using antibodies toward the N-terminal domain, the C-terminal domain, or the neoepitope Ser77 (Fig. 4). The use of antibodies toward both the N-terminal and the C-terminal domains shows how intact the released molecules are, whereas the neoepitope Ser77 antibody shows the presence of this fragment. This experiment was repeated on three additional plugs taken from different locations within the same normal tibial plateau, and all showed similar results.
FIGURE 4.
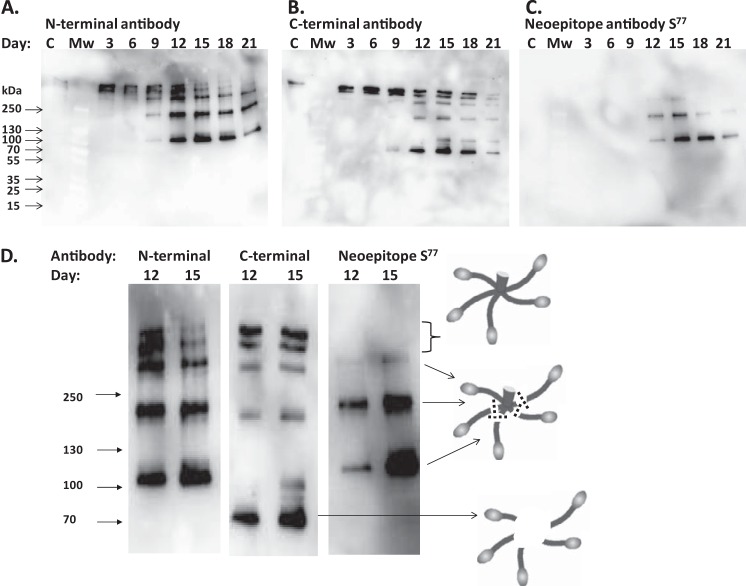
Identification of COMP fragmentations in cytokine-stimulated human cartilage explants. Explants were incubated with cytokines (TNF-α and IL-6/sIL-6R), and the COMP fragments released to the incubation media were analyzed with Western blots using antibodies toward the N-terminal domain (A), the C-terminal domain (B), and the neoepitope Ser77 (C). Samples labeled C represent pooled control media collected at all time points. Molecular mass markers (kDa) are indicated. D, enlargement of the results from days 12 and 15 with a schematic image representing the cleavages within the COMP pentamer (variable numbers of cleavages represent the different sizes).
The control sample contains a pool of media without added cytokines collected from all time points during the 21-day culture (Fig. 4, labeled C). The release of COMP from cartilage explants into the surrounding medium is very limited during normal conditions. However, two weak high molecular mass bands above 250 kDa are detected with both the N-terminal and the C-terminal antibodies.
For cytokine-stimulated cartilage, the release of intact COMP is detected at days 3 and 6 with both N- and C-terminal antibodies as double bands above 250 kDa (Fig. 4, A and B). At day 9, N-terminal fragments are seen at ∼300, 200, and 100 kDa. These fragments start to appear at day 9 and persist until day 21 but show strongest staining, indicating highest amount released, at day 12. It is also clearly visible that the intact high molecular weight COMP double bands show the strongest staining at day 12 and then decrease. This is also seen with the C-terminal antibody where intact double high molecular weight bands are seen at days 3–9 with highest abundance at day 9 and then decrease. At day 9, a C terminus-containing fragment is detected at 70 kDa that increases until day 15 and then starts to decrease. At day 12, two C terminus-containing bands appear at 300 and 200 kDa that remain until day 21.
Using the neoepitope antibody Ser77, two fragments at 200 and 100 kDa are clearly visible from day 12 to day 21 (Fig. 4C). The size of the fragments indicates that the COMP pentamer has been cleaved at position Ser77 in three of five monomers for the 200-kDa band and in four of five monomers for the 100-kDa band. A weak band at 300 kDa that could represent cleavage in two of the five monomers is indicated at day 15. The results from days 12 and 15 are depicted in Fig. 4D with a schematic image representing the cleavages within the COMP pentamer. The glycosaminoglycan release was also measured using the dimethyl methylene blue assay, which showed that ∼30–50% was released before the demonstrated release of the Ser77 epitope.
Structural Effects on the COMP Pentamer Caused by Cleavage at the Neoepitope Sites
Six of the identified neoepitopes (Ser77, 91Cys, 105Thr, 195Ser, Gly202, and Gln226) are located between the N-terminal and the thrombospondin type III domains. These neoepitopes could result in fragmentations of COMP that disrupt its pentamer organization. The other six identified neoepitopes (523Asn, Phe531, 555Asn, Tyr574, Phe577, and 658Gly) are located within or in close proximity to the C-terminal globular domain of COMP. Cleavages at the neoepitope sites could interfere with several of the interactions between COMP and other proteins because these interactions mainly occur with the C-terminal globular domain (19) (Fig. 5). The disulfide bridge between Cys520 and Cys741 in the C-terminal domain of COMP complicates the involvement of cleavages within the C-terminal domain because cleavages can occur between these two amino acids without affecting the migration properties of the COMP molecule as analyzed by non-reducing SDS-PAGE. However, the advantage of analyzing released COMP fragments under non-reducing conditions is that it gives an indication of how intact the released COMP molecules are.
FIGURE 5.
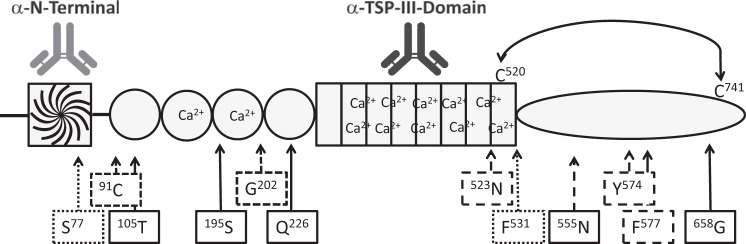
Schematic presentation of the identified neoepitopes in COMP. Shown is a monomeric view of the domains in COMP: the N-terminal coiled coil domain, the four EGF domains, the eight thrombospondin type III domains, and the C-terminal globular domain. Epitopes for the antibodies used for affinity enrichments (α-N-terminal and α-TSP-III domain) are illustrated on top of the monomer. The cysteine bridge between Cys520 and Cys740 within the C-terminal domain is marked. Boxes showing the neoepitope amino acid and arrows indicating locations are marked. Neoepitopes identified in AT are boxed and marked with dotted lines, those in OA are marked with broken lines, those in RA are marked with lines, and that identified in both OA and RA is marked with a thick box. The ending amino acids of the neoepitopes are superscripted; Ser77 represents the newly formed C terminus, whereas 91Cys represents a newly formed N terminus.
DISCUSSION
Using affinity enrichments, we have identified and characterized COMP fragments present in synovial fluids of patients with joint disease. We describe the presence of 12 novel neoepitopes within these COMP fragments.
It is a well established fact that COMP fragments are present in synovial fluids in a variety of joint disease conditions (6, 10, 12). Characteristics of these COMP fragments have previously been described using mouse monoclonal epitope mapping (32, 33). However, the present report for the first time defines ending amino acids, neoepitopes, within these COMP fragments.
By identifying the COMP neoepitopes, we could raise peptide antibodies specific for these neoepitopes. This gives us the unique possibility to quantify these neoepitopes in synovial fluids using inhibition ELISA. By removing the requirement of a capture antibody, novel fragmentations present in synovial fluids can be determined. Furthermore, the characterization of these neoepitopes provides important information regarding the specificity of the degrading enzymes causing these cleavages.
We have developed an inhibition ELISA toward one of these neoepitopes, Ser77. This neoepitope is unique as compared with both other human thrombospondins and COMP from different species, making it suitable as a biomarker specific to human cartilage degradation.
The highest amounts of both total COMP and neoepitope Ser77 were found in acute trauma synovial fluids followed by OA and finally RA. The increased variability (coefficient of variation) found with the neoepitope assay could reflect that this assay is more sensitive for various stages of disease, making it a strong candidate for further biomarker evaluation.
The release of COMP fragments containing neoepitope Ser77 was furthermore determined in human knee cartilage explants. Within this experiment, this neoepitope is clearly prominent in the cytokine-stimulated medium. This was also verified through an extensive mass spectrometric analysis of the cartilage explant medium comparing untreated controls with trauma-stimulated cartilage explants.4 These results indicate that the protease causing the neoepitope Ser77 cleavage is activated or up-regulated by the presence of the cytokines TNF-α and IL-6/sIL-6R. This suggests the possibility of selective inhibition trials to prevent the cleavage at this neoepitope site.
From the present data on synovial fluids, we cannot yet confirm whether the proteolytic processing of COMP occurred within the cartilage or after release of total COMP into the synovial fluid where accumulated proteases could then act. However, based on the kinetics of the release of neoepitope Ser77 demonstrated in the cartilage explant studies (Fig. 4), it is most likely that degradation of COMP into fragments is initiated within the cartilage tissue, and then release into the medium occurs.
Our data clearly show that COMP is released both as intact pentamer and fragments from cytokine-stimulated human cartilage explants. In the search for biomarkers of joint disease involving the detection of released fragments from cartilage, the presence of neoepitope Ser77 represents a strong candidate because it clearly is distinguishable from intact COMP.
A eukaryotic expression system used for purification of pentameric recombinant COMP has previously shown the presence of a 100-kDa fragment starting at amino acid 78Val. These authors suggest that a protease present either within the cells or in the culture medium is able to cleave COMP at this amino acid (21). This is consistent with the cleavage at amino acid Ser77 observed in the present study.
Purified COMP has been shown to be a substrate for several MMPs such as MMP-1 (interstitial collagenase), MMP-3 (stromelysin 1), MMP-9 (gelatinase B), and MMP-13 (collagenase 3) (34). COMP fragments containing the N-terminal domain or the EGF domain have previously been shown in synovial fluids from patients with OA, RA, and anterior cruciate ligament injury as bands of 80 and 100 kDa on Western blots (35). Also ADAMTS-7 (36) and ADAMTS-12 (37) have been shown to cleave COMP by binding to the EGF domains, forming fragments of 100 kDa. Furthermore, IL-1α stimulation of bovine nasal cartilage shows release of COMP fragments of 110 kDa (38). Although many proteases have been shown to degrade COMP, the specific cleavage sites within COMP have not been identified. Our identified COMP neoepitopes can be of use for determination of the proteases involved in cartilage degradation and their involvement in the disease progression. The involvement of the released COMP neoepitopes and their role in disease propagation are also of great importance (1).
The main focus of analyzing COMP fragments in joint disease was to identify and characterize protein neoepitopes created by the pathological process that can be separated from normal tissue turnover. Assays for quantifying these neoepitopes can be used in diagnostic approaches, disease monitoring, and evaluation of treatment and therapy.
Acknowledgments
We thank Dr. Tobias Gustavsson (The Novo Nordisk Foundation, Center for Protein Research, University of Copenhagen, Copenhagen, Denmark) for the generous gift of the protein used in the inhibition ELISA. Yang Li and Yang Wang (Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, MA) harvested, prepared, and cultured the human cartilage explant samples. We thank Dr. Susan Chubinskaya of Rush University Medical Center for providing human tissue, Gift of Hope and the donor's family for human cartilage, and Dr. Arkady Margulis for tissue procurement. We thank the Inga-Britt and Arne Lundberg Foundation as well as the Crafoord foundation for funding the mass spectrometers used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grant AR045779. This work was also supported by the European Commission Seventh Framework Program for Research, NanoDiaRA.
For neoepitopes, the superscript number before the residue indicates a new N-terminal end, and the superscript number following the residue indicates a new C-terminal end.
P. Önnerfjord and A. Grodzinsky, unpublished data.
- OA
- osteoarthritis
- AT
- acute trauma
- RA
- rheumatoid arthritis
- COMP
- cartilage oligomeric matrix protein
- sIL-6R
- soluble IL-6 receptor
- TSP-III
- thrombospondin type III domain
- HBS
- HEPES-buffered saline
- MMP
- matrix metalloproteinase.
REFERENCES
- 1. Heinegård D., Saxne T. (2011) The role of the cartilage matrix in osteoarthritis. Nat. Rev. Rheumatol. 7, 50–56 [DOI] [PubMed] [Google Scholar]
- 2. Kraus V. B., Burnett B., Coindreau J., Cottrell S., Eyre D., Gendreau M., Gardiner J., Garnero P., Hardin J., Henrotin Y., Heinegård D., Ko A., Lohmander L. S., Matthews G., Menetski J., Moskowitz R., Persiani S., Poole A. R., Rousseau J. C., Todman M. (2011) Application of biomarkers in the development of drugs intended for the treatment of osteoarthritis. Osteoarthritis Cartilage 19, 515–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saxne T., Månsson B., Heinegård D. (2006) in Rheumatoid Arthritis: New Frontiers in Pathogenesis and Treatment (Firestein G., Panayi G., Wollheim F., eds) pp. 301–313, Oxford University Press, Oxford [Google Scholar]
- 4. Heinegård D. (2009) Proteoglycans and more—from molecules to biology. Int. J. Exp. Pathol. 90, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heathfield T. F., Onnerfjord P., Dahlberg L., Heinegård D. (2004) Cleavage of fibromodulin in cartilage explants involves removal of the N-terminal tyrosine sulfate-rich region by proteolysis at a site that is sensitive to matrix metalloproteinase-13. J. Biol. Chem. 279, 6286–6295 [DOI] [PubMed] [Google Scholar]
- 6. Tseng S., Reddi A. H., Di Cesare P. E. (2009) Cartilage oligomeric matrix protein (COMP): a biomarker of arthritis. Biomark. Insights 4, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lotz M., Martel-Pelletier J., Christiansen C., Brandi M. L., Bruyère O., Chapurlat R., Collette J., Cooper C., Giacovelli G., Kanis J. A., Karsdal M. A., Kraus V., Lems W. F., Meulenbelt I., Pelletier J. P., Raynauld J. P., Reiter-Niesert S., Rizzoli R., Sandell L. J., Van Spil W. E., Reginster J. Y. (2013) Value of biomarkers in osteoarthritis: current status and perspectives. Ann. Rheum. Dis. 72, 1756–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andersson M. L., Svensson B., Petersson I. F., Hafström I., Albertsson K., Forslind K., Heinegård D., Saxne T. (2013) Early increase in serum-COMP is associated with joint damage progression over the first five years in patients with rheumatoid arthritis. BMC Musculoskelet. Disord. 14, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindqvist E., Eberhardt K., Bendtzen K., Heinegård D., Saxne T. (2005) Prognostic laboratory markers of joint damage in rheumatoid arthritis. Ann. Rheum. Dis. 64, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Månsson B., Carey D., Alini M., Ionescu M., Rosenberg L. C., Poole A. R., Heinegård D., Saxne T. (1995) Cartilage and bone metabolism in rheumatoid arthritis. Differences between rapid and slow progression of disease identified by serum markers of cartilage metabolism. J. Clin. Investig. 95, 1071–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neidhart M., Hauser N., Paulsson M., DiCesare P. E., Michel B. A., Häuselmann H. J. (1997) Small fragments of cartilage oligomeric matrix protein in synovial fluid and serum as markers for cartilage degradation. Br. J. Rheumatol. 36, 1151–1160 [DOI] [PubMed] [Google Scholar]
- 12. Saxne T., Heinegård D. (1992) Cartilage oligomeric matrix protein: a novel marker of cartilage turnover detectable in synovial fluid and blood. Br. J. Rheumatol. 31, 583–591 [DOI] [PubMed] [Google Scholar]
- 13. Sharif M., Saxne T., Shepstone L., Kirwan J. R., Elson C. J., Heinegård D., Dieppe P. A. (1995) Relationship between serum cartilage oligomeric matrix protein levels and disease progression in osteoarthritis of the knee joint. Br. J. Rheumatol. 34, 306–310 [DOI] [PubMed] [Google Scholar]
- 14. Williams F. M., Spector T. D. (2008) Biomarkers in osteoarthritis. Arthritis Res. Ther. 10, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hedbom E., Antonsson P., Hjerpe A., Aeschlimann D., Paulsson M., Rosa-Pimentel E., Sommarin Y., Wendel M., Oldberg A., Heinegård D. (1992) Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J. Biol. Chem. 267, 6132–6136 [PubMed] [Google Scholar]
- 16. Di Cesare P. E., Carlson C. S., Stollerman E. S., Chen F. S., Leslie M., Perris R. (1997) Expression of cartilage oligomeric matrix protein by human synovium. FEBS Lett. 412, 249–252 [DOI] [PubMed] [Google Scholar]
- 17. DiCesare P., Hauser N., Lehman D., Pasumarti S., Paulsson M. (1994) Cartilage oligomeric matrix protein (COMP) is an abundant component of tendon. FEBS Lett. 354, 237–240 [DOI] [PubMed] [Google Scholar]
- 18. Oldberg A., Antonsson P., Lindblom K., Heinegård D. (1992) COMP (cartilage oligomeric matrix protein) is structurally related to the thrombospondins. J. Biol. Chem. 267, 22346–22350 [PubMed] [Google Scholar]
- 19. Halász K., Kassner A., Mörgelin M., Heinegård D. (2007) COMP acts as a catalyst in collagen fibrillogenesis. J. Biol. Chem. 282, 31166–31173 [DOI] [PubMed] [Google Scholar]
- 20. Rosenberg K., Olsson H., Mörgelin M., Heinegård D. (1998) Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 273, 20397–20403 [DOI] [PubMed] [Google Scholar]
- 21. Thur J., Rosenberg K., Nitsche D. P., Pihlajamaa T., Ala-Kokko L., Heinegård D., Paulsson M., Maurer P. (2001) Mutations in cartilage oligomeric matrix protein causing pseudoachondroplasia and multiple epiphyseal dysplasia affect binding of calcium and collagen I, II, and IX. J. Biol. Chem. 276, 6083–6092 [DOI] [PubMed] [Google Scholar]
- 22. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 23. Muehleman C., Bareither D., Huch K., Cole A. A., Kuettner K. E. (1997) Prevalence of degenerative morphological changes in the joints of the lower extremity. Osteoarthritis Cartilage 5, 23–37 [DOI] [PubMed] [Google Scholar]
- 24. Geng H., Nandakumar K. S., Pramhed A., Aspberg A., Mattsson R., Holmdahl R. (2012) Cartilage oligomeric matrix protein specific antibodies are pathogenic. Arthritis Res. Ther. 14, R191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geng H., Nandakumar K. S., Xiong L., Jie R., Dong J., Holmdahl R. (2013) Incomplete B cell tolerance to cartilage oligomeric matrix protein in mice. Arthritis Rheum. 65, 2301–2309 [DOI] [PubMed] [Google Scholar]
- 26. Heinegård D. (February 7, 2012) U. S. Patent 8,110,663
- 27. Hesselstrand R., Kassner A., Heinegård D., Saxne T. (2008) COMP: a candidate molecule in the pathogenesis of systemic sclerosis with a potential as a disease marker. Ann. Rheum. Dis. 67, 1242–1248 [DOI] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. Candiano G., Bruschi M., Musante L., Santucci L., Ghiggeri G. M., Carnemolla B., Orecchia P., Zardi L., Righetti P. G. (2004) Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 25, 1327–1333 [DOI] [PubMed] [Google Scholar]
- 30. Önnerfjord P., Khabut A., Reinholt F. P., Svensson O., Heinegård D. (2012) Quantitative proteomic analysis of eight cartilaginous tissues reveals characteristic differences as well as similarities between subgroups. J. Biol. Chem. 287, 18913–18924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sui Y., Lee J. H., DiMicco M. A., Vanderploeg E. J., Blake S. M., Hung H. H., Plaas A. H., James I. E., Song X. Y., Lark M. W., Grodzinsky A. J. (2009) Mechanical injury potentiates proteoglycan catabolism induced by interleukin-6 with soluble interleukin-6 receptor and tumor necrosis factor alpha in immature bovine and adult human articular cartilage. Arthritis Rheum. 60, 2985–2996 [DOI] [PubMed] [Google Scholar]
- 32. Lai Y., Yu X. P., Zhang Y., Tian Q., Song H., Mucignat M. T., Perris R., Samuels J., Krasnokutsky S., Attur M., Greenberg J. D., Abramson S. B., Di Cesare P. E., Liu C. J. (2012) Enhanced COMP catabolism detected in serum of patients with arthritis and animal disease models through a novel capture ELISA. Osteoarthritis Cartilage 20, 854–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vilím V., Vobůrka Z., Vytásek R., Senolt L., Tchetverikov I., Kraus V. B., Pavelka K. (2003) Monoclonal antibodies to human cartilage oligomeric matrix protein: epitope mapping and characterization of sandwich ELISA. Clin. Chim. Acta 328, 59–69 [DOI] [PubMed] [Google Scholar]
- 34. Ganu V., Goldberg R., Peppard J., Rediske J., Melton R., Hu S. I., Wang W., Duvander C., Heinegård D. (1998) Inhibition of interleukin-1α-induced cartilage oligomeric matrix protein degradation in bovine articular cartilage by matrix metalloproteinase inhibitors: potential role for matrix metalloproteinases in the generation of cartilage oligomeric matrix protein fragments in arthritic synovial fluid. Arthritis Rheum. 41, 2143–2151 [DOI] [PubMed] [Google Scholar]
- 35. Vilim V., Lenz M. E., Vytasek R., Masuda K., Pavelka K., Kuettner K. E., Thonar E. J. (1997) Characterization of monoclonal antibodies recognizing different fragments of cartilage oligomeric matrix protein in human body fluids. Arch. Biochem. Biophys. 341, 8–16 [DOI] [PubMed] [Google Scholar]
- 36. Liu C. J., Kong W., Ilalov K., Yu S., Xu K., Prazak L., Fajardo M., Sehgal B., Di Cesare P. E. (2006) ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 20, 988–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu C. J., Kong W., Xu K., Luan Y., Ilalov K., Sehgal B., Yu S., Howell R. D., Di Cesare P. E. (2006) ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J. Biol. Chem. 281, 15800–15808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dickinson S. C., Vankemmelbeke M. N., Buttle D. J., Rosenberg K., Heinegård D., Hollander A. P. (2003) Cleavage of cartilage oligomeric matrix protein (thrombospondin-5) by matrix metalloproteinases and a disintegrin and metalloproteinase with thrombospondin motifs. Matrix Biol. 22, 267–278 [DOI] [PubMed] [Google Scholar]