Background: RNA granules are associated with translational control and neurodegenerative disease.
Results: Nuclear factor associated with dsRNA 2 (NFAR2) enhances RNA granule assembly through two domains, one of which is antagonized by nuclear factor 45 (NF45).
Conclusion: NFAR2 and NF45 play novel regulatory roles in RNA granule assembly and disassembly.
Significance: Mechanisms of RNA granule assembly and disassembly are the basis for understanding translational control and neurodegenerative disease.
Keywords: Protein Assembly, Protein Domain, Protein Kinase RNA-activated (PKR), Proteomics, Ribonuclear Protein (RNP), Low Complexity Sequence Domain, NF45, NFAR2/NF110, RNA Granule, RNG105/caprin1
Abstract
RNA granules are large messenger ribonucleoprotein complexes that regulate translation and mRNA translocation to control the timing and location of protein synthesis. The regulation of RNA granule assembly and disassembly is a structural basis of translational control, and its disorder is implicated in degenerative disease. Here, we used proteomic analysis to identify proteins associated with RNA granule protein 105 (RNG105)/caprin1, an RNA-binding protein in RNA granules. Among the identified proteins, we focused on nuclear factor (NF) 45 and its binding partner, nuclear factor associated with dsRNA 2 (NFAR2), and we demonstrated that NF45 promotes disassembly of RNA granules, whereas NFAR2 enhances the assembly of RNA granules in cultured cells. The GQSY domain of NFAR2 was required to associate with messenger ribonucleoprotein complexes containing RNG105/caprin1, and it was structurally and functionally related to the low complexity sequence domain of the fused in sarcoma protein, which drives the assembly of RNA granules. Another domain of NFAR2, the DZF domain, was dispensable for association with the RNG105 complex, but it was involved in positive and negative regulation of RNA granule assembly by being phosphorylated at double-stranded RNA-activated kinase sites and by association with NF45, respectively. These results suggest a novel molecular mechanism for the modulation of RNA granule assembly and disassembly by NFAR2, NF45, and phosphorylation at double-stranded RNA-activated kinase PKR sites.
Introduction
RNA granules are large macromolecular complexes composed of mRNA, ribosomes, translation factors, and RNA-binding proteins regulating gene expression by stalling translation (1). RNA granules are also involved in mRNA transport. Translation is repressed during transport, but it is de-repressed by releasing the mRNA and other components at their destination (2–5). Thus, RNA granules are in equilibrium between assembly and disassembly to regulate translation, but in neurodegenerative disease such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration, they are prone to conversion into irreversible pathological aggregates (6, 7). Recent studies proposed that degenerative disease-associated mutations in RNA granule components, e.g. TAR DNA-binding protein 43 (TDP-43), fused in sarcoma/translocated in sarcoma (FUS2/TLS), heterogeneous nuclear ribonucleoprotein (hnRNP) A2B1 and hnRNPA1 enhance their incorporation into RNA granules and promote RNA granule aggregation (6–8). These proteins contain prion-like low complexity (LC) sequence domains, which are responsible for RNA granule assembly under normal conditions and the formation of pathological aggregates in their mutant forms (6–9).
Different types of RNA granules have been described, including stress granules (SGs), germ granules, and neuronal RNA granules. SGs are induced by several kinds of stress, such as oxidative stress and virus infections that induce eIF2α phosphorylation by heme-regulated eIF2α kinase and double-stranded RNA (dsRNA)-activated kinase (PKR), and are implicated in cellular defense against stress (10, 11). Neuronal RNA granules are another type of RNA granule that plays central roles in mRNA transport and local translation in dendrites, and they are responsible for synapse formation, plasticity, and long term memory (12–14). Several RNA-binding proteins are shared between SGs and neuronal RNA granules, e.g. fragile X mental retardation protein, staufen, RasGAP SH3 domain-binding protein (G3BP), and RNA granule protein 105 (RNG105)/caprin1 (1, 15–18). Expression of RNG105/caprin1 or G3BP that interacts with RNG105/caprin1 (18, 19), in cultured A6, 293T, Cos, and HeLa cells, induces the formation of TIA-1-containing SG-like RNA granules in the absence of stressors (18, 20–22). In neurons, RNG105/caprin1 plays a role in the transport of specific mRNAs into dendrites, and the loss of RNG105/caprin1 results in the degeneration of dendrites and neuronal networks (23). Mice with gene knockouts of RNG105/caprin1 and G3BP exhibit similar phenotypes in terms of fetal growth retardation, cell death in the brain, and neonatal lethality with respiratory failure (23, 24).
Nuclear factor associated with dsRNA 1 (NFAR1)/nuclear factor (NF) 90 and NFAR2/NF110 are splice variants transcribed from a single interleukin enhancer binding factor 3 (ILF3) gene (25, 26). NFAR1 and NFAR2 were initially identified in a complex with NF45/ILF2 as a transcription activation factor (27, 28). Besides transcription regulation, NFAR1 and NFAR2 bind to AU-rich elements in the 3′-untranslated region of mRNAs and regulate their stabilization, localization, and translation. For example, NFAR1/2 bind to and stabilize interleukin-2 and mitogen-activated protein kinase phosphatase-1 mRNAs (29, 30) and bind to the 3′-untranslated region of Tau mRNA, which is known as an element required for its transport to the axon in neurons (31). NFAR1/2 also inhibit global translation as well as bind to viral RNA and prevent its translation (32, 33). For the prevention of viral RNA translation, NFAR1/2 are phosphorylated on Thr-188 and Thr-315 by PKR and exported from the nucleus to the cytoplasm to be retained on polysomes (33).
NFAR1 and NFAR2 have dsRNA-binding motifs and an Arg-Gly-Gly motif (RGG box) known as an RNA-binding motif. Their zinc finger nucleic acid binding domains (DZF domains) include the PKR phosphorylation sites and form heterodimers with the DZF domain of NF45 (33, 34). NFAR2, but not NFAR1, has an additional C-terminal domain, which is enriched with Gly, Gln, Ser, and Tyr residues (GQSY domain), whose function is not well understood except that it can enhance gene expression from specific promoters (35).
In this study, using proteomic analysis, NF45 was identified as a protein associated with RNG105- and G3BP-containing messenger ribonucleoprotein (mRNP) complexes in mitosis when RNA granules disappeared. Expression of NF45 in cultured cells promoted disassembly of RNA granules, whereas its binding partner NFAR2 enhanced RNA granule assembly. Two domains of NFAR2, the GQSY domain similar to the LC sequence domain of FUS and the DZF domain containing PKR phosphorylation sites and that binds to NF45, were involved in the regulation of RNA granule assembly. These results suggest a novel mechanism of RNA granule assembly and disassembly regulated by NFAR2 phosphorylation by PKR and NF45.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
A6 cells were cultured in 10% fetal bovine serum (FBS) and 50% Leivobitz's L-15 medium (Invitrogen) at 23 °C without a CO2 atmosphere. HeLa cells were cultured in 10% FBS and DMEM (Sigma) at 37 °C with CO2. Mitotic A6 cells were collected selectively by pipetting with a 5-ml plastic pipette after culturing in medium containing 1 μg/ml nocodazole for 2 h. Cells remaining on the culture plates after pipetting were used as interphase cells (36). For microscopy experiments, cells were grown on glass coverslips or glass-bottomed dishes. RNG105-GFP and RNG105-monomeric red fluorescent protein 1 (mRFP1) transfectants were produced previously (17, 20). Transfections were performed using Lipofectin (Invitrogen) for A6 cells and Lipofectamine 2000 (Invitrogen) for HeLa cells in accordance with the manufacturer's protocols. Transfection of polyinosinic-polycytidylic acid (poly(I-C)) (Sigma) into HeLa cells was performed at a concentration of 2 μg/ml. After transfection, cells were cultured for 4 h, then fixed, and immunostained. Stable transfectants were selected in the presence of 0.7 mg/ml geneticin (Invitrogen) followed by picking up fluorescent colonies using a CKX41 microscope (Olympus, Tokyo, Japan) equipped with an epifluorescence module. Sodium arsenite (Wako Pure Chemical, Osaka, Japan) was added to culture medium at concentrations indicated in the figure legends. Imidazolo-oxindole PKR inhibitor C16 (Sigma) was added at 1 μm.
Plasmid Construction
pEGFP-N1 vector (Clontech) was used to produce GFP-tagged proteins. To produce mRFP1-tagged proteins, the GFP coding sequence in the pEGFP-N1 vector was replaced with the mRFP1 coding sequence as described previously (17). cDNAs for NFAR1, NFAR2, NF45, and FUS were obtained by reverse transcription-PCR from mouse brain RNA using primers 5′-gtcgacatgcgtcccatgagaattttt-3′ (NFARs-F) and 5′-ggatcccctccgccgcccccggaagccccaaaatcatgat-3′ for NFAR1, NFARs-F and 5′-ggatcccccccgccccctcctctgtactggtagttcatgctgtg-3′ (NFAR2-R) for NFAR2, 5′-aagcttatgaggggtgacagaggac-3′ and 5′-gtcgacccaccgccacctccctcctgagtctccatgctttct-3′ for NF45, and 5′-gcgtcgacatggcttcaaacgactataccc-3′ and 5′-cgggatccccgccgccacccccatatggcctctccctgcg-3′ (FUS-R) for FUS. The cDNAs were cloned into the SalI/BamHI sites or the HindIII/SalI sites of the vectors.
FUSΔLC was amplified by PCR using primers 5′-gcgtcgacatggcttcaaactatggccaagatcagtcc-3′ and FUS-R and cloned into the SalI/BamHI sites of the pEGFP-N1 vector. To produce GQSY-FUSΔLC, GQSY and FUSΔLC fragments were amplified by PCR using primers 5′-gcgtcgacatggcttcatccagctacagctcctactaccaa-3′ (GQSY-F) and 5′-ctccactgctacttctgtactggtagttcatgctgtg-3′, and 5′-taccagtacagaagtagcagtggaggtggtgga-3′ and FUS-R, respectively. The fragments were ligated by PCR using primers GQSY-F and FUS-R, and then cloned into the SalI/BamHI sites of the pEGFP-N1 vector. To produce NFAR2ΔDZF, N- and C-terminal fragments of NFAR2 were amplified by PCR using primers NFARs-F and 5′-ttctttggtttcttgggcatcctagtcatgtgttccgcctt-3′, and 5′-aaggcggaacacatgactaggatgcccaagaaaccaaagaa-3′ and NFAR2-R, respectively. The fragments were ligated by PCR using primers NFARs-F and NFAR2-R, and cloned into the SalI/BamHI sites of the pEGFP-N1 vector.
Thr-188 (ACG) and Thr-315 (ACA) of NFAR2 were changed to Ala (TA mutant) or Asp (TD mutant) by mutating (ACG, ACA) to (GCG, GCA) or (GAT, GAT), respectively, using QuikChange Lightning multisite-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA). Plasmids for RNG105-GFP (17), RNG105-mRFP1 (20), and G3BP-GFP (23) were produced previously.
Immunoprecipitation (IP)
IP was performed using agarose-conjugated anti-GFP (Medical and Biological Laboratories, Nagoya, Japan) as described previously (17). Briefly, cells were homogenized in 0.25 m sucrose, 0.7% Triton X-100, 0.1 mm DTT, protease inhibitors (10 μg/ml leupeptin, pepstatin, aprotinin, and 1 mm PMSF), and 1,000 units/ml RNase inhibitor (Takara Bio, Shiga, Japan), and then centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant was added to 1:10 volume of 10× PBS followed by 1:20 volume of anti-GFP-agarose beads. After rocking for 2 h at 4 °C, the beads were washed three times in PBS containing 0.1 mm DTT, protease inhibitors, and 100 units/ml RNase inhibitor. IP in the presence of RNase was performed in the continuous presence of 0.2 mg/ml RNase A (Wako Pure Chemical) without RNase inhibitor in the cell extracts and the wash buffer. Immunoprecipitates were analyzed by SDS-PAGE using a two-dimensional Silver Stain II kit (Cosmo Bio, Tokyo, Japan), Western blotting, or mass spectrometry.
Mass Spectrometry
Immunoprecipitates with the anti-GFP antibody were separated by SDS-PAGE and stained using the Silver Stain MS kit (Wako Pure Chemical). After bands were cut out from the gel, they were destained with 15 mm K3(Fe(CN)6) and 50 mm Na2S2O3 for 10 min, washed with H2O, dehydrated with 50% acetonitrile in 25 mm NH4HCO3 for 5 min, and dried in a vacuum desiccator. The gel slices were deoxidized in 10 mm DTT in 25 mm NH4HCO3 at 56 °C for 1 h, washed with 25 mm NH4HCO3, alkylated with 55 mm iodoacetamide in 25 mm NH4HCO3 at room temperature for 45 min, dehydrated, and dried again. After the gel slices were rehydrated with 10 μg/ml trypsin in 50 mm NH4HCO3 on ice for 30 min, excess solution was removed, and the gel slices were incubated at 37 °C for 12 h for in-gel digestion. Digested peptides were extracted with 50% acetonitrile and 5% CF3COOH at room temperature for 1 h. After freeze-drying, the peptides were dissolved in 30% acetonitrile and 0.1% formic acid and then analyzed with Q-TOF Premier (Waters, Milford, MA).
Western Blotting
Western blotting was performed on polyvinylidene difluoride membranes with primary antibodies, anti-NF45/ILF2 (LS-B3952, Lifespan BioSciences, Seattle, WA), anti-GFP (GF200, Nacalai Tesque, Kyoto, Japan), and anti-RFP antibodies (PM005, Medical and Biological Laboratories). Biotinylated secondary antibodies (GE Healthcare) and alkaline phosphatase-conjugated streptavidin (GE Healthcare) were used for detection with bromochloroindolyl phosphate/nitro blue tetrazolium solution.
Immunofluorescence Staining
Cultured cells were fixed with 3.7% formaldehyde in PBS for 10 min. After washing with PBS, the cells were treated with 0.5% Triton X-100 in PBS and then washed with PBS. Specimens were blocked with 10% fetal bovine serum and then incubated with the following primary antibodies: anti-RNG105 (17); anti-G3BP (23); anti-ILF3 (NFAR1/2) (H3609-B01P, Abnova, Taipei City, Taiwan); anti-phospho-eIF2α (Ser-51) (119A11, Cell Signaling Technology, Danvers, MA), and anti-puromycin (3RH11, KeraFAST Inc., Boston). After washing with PBS, the reacted proteins were labeled with cyanine 3- or 5-conjugated anti-rabbit IgG, DyLight 649-conjugated anti-goat IgG (Jackson ImmunoResearch, West Grove, PA), and Alexa 488-conjugated anti-mouse IgG (Invitrogen).
Fluorescence Microscopy and Image Analysis
Fluorescence images were acquired using a DeltaVision optical sectioning microscope (Applied Precision, Issaquah, WA) equipped with an IX70 microscope (Olympus) with a PlanApo 60× or a UApo 40× oil objective lens or using an A1 confocal microscope equipped with a Ti-E inverted microscope (Nikon Instruments, Melville, NY) with a PlanApo VC60× oil objective lens. Time-lapse imaging of A6 cells was performed using the optical sectioning microscope with the 40× oil objective lens at room temperature.
The images were converted into binary images and quantified for the area of cells, nuclei, and RNA granules using Adobe Photoshop software. To analyze the fluorescence intensity of RNA granules and nuclei, original images were multiplied by the binary images and then the mean pixel intensity in each area was measured using Photoshop software. To analyze the fluorescence intensity of the cytoplasm, original images were multiplied by inverted binary images to mask nuclei and RNA granules, and then the mean pixel intensity was measured. Mean background fluorescence was subtracted from the mean intensity of the target areas.
Sucrose Density Gradient Centrifugation
Cultured cells were incubated with 100 μg/ml cycloheximide in culture medium for 15 min, washed with ice-cold PBS, and then lysed with cell lysis buffer (20 mm Hepes-KOH, pH 7.4, 15 mm MgCl2, 200 mm KCl, 1% Triton X-100, 100 μg/ml cycloheximide, 2 mm DTT, 1 mg/ml heparin, and protease inhibitors). After centrifugation at 14,000 × g for 5 min at 4 °C, the supernatant was overlaid onto 7–47% w/w linear sucrose gradients in the cell lysis buffer lacking Triton X-100 and heparin, which were prepared using a gradient gel-making device (ATTO, Tokyo, Japan) and then centrifuged at 100,000 × g for 4 h at 4 °C in an SW41Ti swing rotor (Beckman Coulter, Brea, CA). The sample was fractionated in 20 fractions, and their absorbance at 254 nm was measured using NanoDrop (Thermo Fisher Scientific, Waltham, MA).
Cell Survival Analysis
Cultured cells were treated with several concentrations of arsenite for 3 h. After washing twice with the culture medium, the cells were cultured for an additional 21 h in the absence of arsenite. Phase contrast images of the cells were acquired using the CKX41 microscope with an LCAch N 20× objective lens and an E-620 digital single lens reflex camera (Olympus) to count the number of cells attached to and detached from the culture plates. Detached cells were collected by pipetting and treated with 0.2% trypan blue in PBS for 15 min to stain nonviable cells. Viable and nonviable cells were counted using the CKX41 microscope.
Ribopuromycilation Assay
Ribopuromycilation assay was conducted as described previously with a modification (22). Briefly, cells were pulse-labeled with 50 μg/ml puromycin and 100 μg/ml cycloheximide for 10 min at 23 °C and washed with 100 μg/ml cycloheximide in PBS for 3 min on ice. After permeabilization with 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 25 mm KCl, 100 μg/ml cycloheximide, 10 units/ml RNase inhibitor, protease inhibitors, and 0.015% digitonin for 1 min on ice, the cells were fixed by adding 1:10 volume of ice-cold 37% formaldehyde for 5 min on ice. After postfixation with 3.7% formaldehyde in PBS for 10 min at room temperature, the cells were subjected to immunofluorescence staining with the anti-puromycin antibody.
Statistical Analysis and Informatics
Data are expressed as mean ± S.E. Two samples were compared using an unpaired t test. More than two samples were compared using one-way analysis of variance followed by Tukey-Kramer test. Pearson's product-moment correlation coefficients were calculated to measure the relationship between two variables. χ2 test was used to test the difference in the proportion of living and dead cells between two samples.
Prediction of protein structural disorder was performed using the DISOPRED2 predictor (37) and the PONDR-FIT and VSL2B predictors (38, 39). Disorder-based potential binding sites were predicted using the ANCHOR and the MoRFpred predictors (40, 41).
RESULTS
Identification of NF45 in mRNP Complexes Containing RNG105 and G3BP
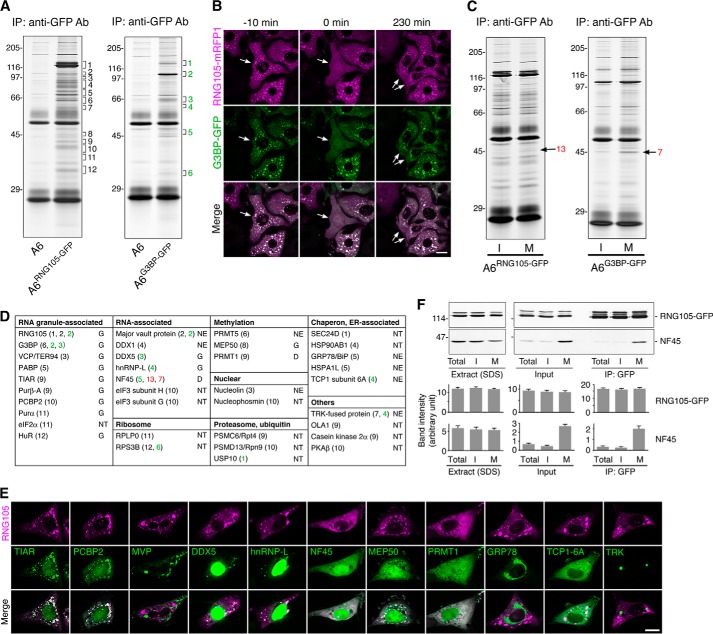
Using anti-GFP antibody-conjugated beads, we performed IP from A6 cells stably expressing RNG105-GFP or G3BP-GFP. SDS-PAGE of the immunoprecipitates showed that the GFP fusion proteins and their associated proteins were immunoprecipitated from the transfectants (Fig. 1A). Analysis of these proteins by mass spectrometry identified several proteins already known to be localized to RNA granules and other proteins, including RNA-binding proteins, proteasome-associated proteins, chaperon proteins, and proteins involved in methylation (Fig. 1D). The identified proteins were fused to GFP and expressed in A6 cells with RNG105-mRFP1, which found proteins that co-localized with RNA granules and protein N-methyltransferase 1 that induced the disassembly of RNG105-localized RNA granules (Fig. 1, D and E).
FIGURE 1.
Identification of RNG105- and G3BP-associated proteins. A, immunoprecipitates with an anti-GFP antibody (Ab) from control A6 cells and RNG105-GFP-expressing A6 cells (left panel) or G3BP-GFP-expressing A6 cells (right panel) were silver-stained after SDS-PAGE. Numbers on the left indicate molecular mass (kDa), and numbers on the right indicate gel regions cut and analyzed by mass spectrometry. B, fluorescence images of A6 cells co-expressing RNG105-mRFP1 and G3BP-GFP. Arrows indicate a cell undergoing mitosis at 0 min and then dividing into two daughter cells. Note that the cells were cultured in the absence of stressors. Scale bar, 10 μm. C, immunoprecipitates with the anti-GFP antibody from interphase (I) and mitotic (M) cells were silver-stained after SDS-PAGE. Left panel, RNG105-GFP-expressing A6 cells; right panel, G3BP-GFP-expressing A6 cells. Arrows indicate proteins whose amounts were higher in mitotic co-immunoprecipitates than in interphase. D, gel regions indicated in A and C were cut and analyzed by mass spectrometry. Identified proteins were classified according to their functions. Numbers correspond to the region numbers in A and C. The proteins were fused to GFP and expressed in A6 cells with RNG105-mRFP1 to test their co-localization with RNA granules and effects on RNA granules. The results are indicated on the right side. G, co-localized with RNA granules; D, promotes disassembly of RNA granules; NE, no effect on RNA granules; NT, not tested. E, representative images of A6 cells co-expressing the identified proteins fused to GFP and RNG105-mRFP1. Scale bar, 10 μm. F, input extracts, and the immunoprecipitates with the anti-GFP antibody from total, interphase (I), or mitotic (M) A6 cells expressing RNG105-GFP were immunoblotted with anti-GFP and anti-NF45 antibodies. Extracts with SDS-PAGE sample buffer were also blotted. Bottom panels show quantification of the band intensity of immunoblotting (n = 3).
During the observation of A6 cells expressing RNG105-GFP or G3BP-GFP by fluorescence microscopy, we noticed that RNA granules disappeared in mitotic cells. Time-lapse observation of A6 cells co-expressing RNG105-mRFP1 and G3BP-GFP revealed that RNG105 and G3BP were co-localized in RNA granules in interphase cells, but these RNA granules disappeared within a few minutes after nuclear envelope breakdown and reappeared after nuclear formation following cell division (Fig. 1B). This led to the idea that the protein composition of mRNP complexes containing RNG105 and G3BP may change between interphase and mitotic cells. IP of RNG105-GFP or G3BP-GFP from RNG105-GFP- or G3BP-GFP-expressing interphase and mitotic cells showed that the SDS-PAGE pattern of the immunoprecipitates was largely unchanged, but a few proteins changed in abundance between interphase and mitotic cells (Fig. 1C). A major change was an increase in the amount of ∼45-kDa proteins in the mitotic immunoprecipitates both from RNG105-GFP- and G3BP-GFP-expressing cells (Fig. 1C). Mass spectrometric analysis of these proteins identified them as NF45 (Fig. 1D). Western blotting of the immunoprecipitates confirmed that the association of NF45 with the RNG105-GFP mRNP complex was increased in mitotic cells, which was because of the increase of NF45 in input extracts (Fig. 1F). Because the total amount of NF45 was not increased in mitosis, the increase may be caused by release of NF45 from the nucleus after nuclear envelope breakdown in mitosis.
NF45 Disassembles RNA Granules and Renders Cells Susceptible to Stress
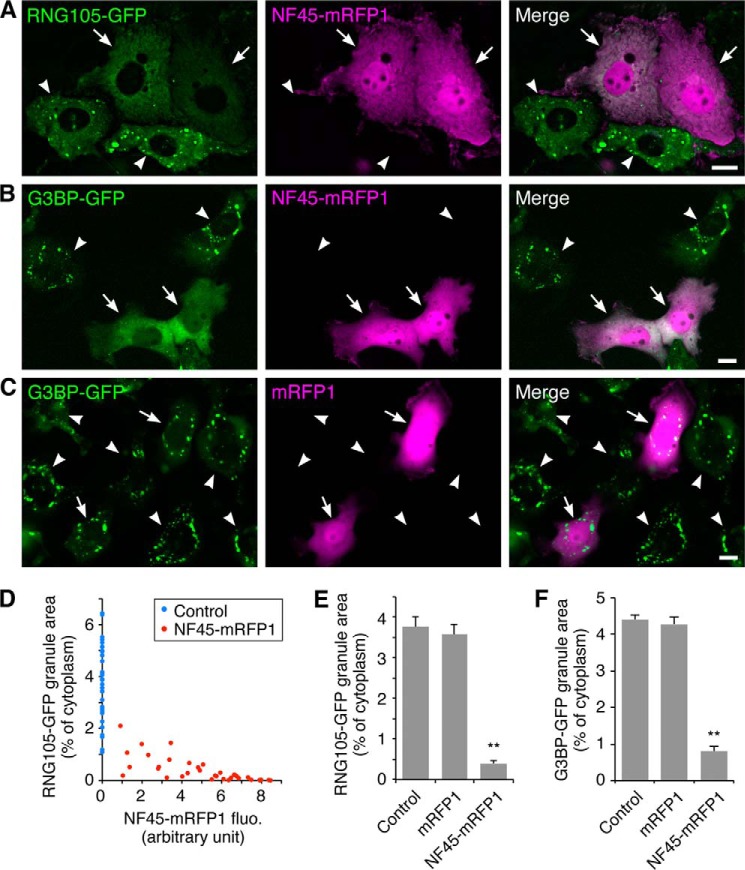
Expression of NF45 as a GFP fusion protein in A6 cells with RNG105-mRFP1 showed that NF45 induced the disassembly of RNG105-localizing RNA granules (Fig. 1, D and E). Similarly, expression of NF45-mRFP1 in A6 cells stably expressing RNG105-GFP resulted in the disassembly of RNG105-localizing RNA granules (Fig. 2, A and E). The extent of disassembly was dependent on the expression level of NF45 (Fig. 2D). Expression of NF45-mRFP1 also induced the disassembly of G3BP-GFP RNA granules, but expression of control mRFP1 did not (Fig. 2, B, C, and F). These results indicated that high level expression of NF45 disassembled RNA granules containing RNG105 and G3BP.
FIGURE 2.
NF45 promotes RNA granule disassembly. A–C, A6 cells stably expressing RNG105-GFP (A) or G3BP-GFP (B and C) were transfected with NF45-mRFP1 (A and B) or mRFP1 (C). Arrows and arrowheads denote NF45-mRFP1- or mRFP1-transfected and untransfected cells, respectively. The cells were cultured in the absence of stressors. Scale bars, 10 μm. D, dose-dependent effect of NF45-mRFP1 expression on RNG105-GFP granule disassembly. Control, untransfected neighboring cells. E and F, quantification of RNG105-GFP or G3BP-GFP granule area. n = 35; **p < 0.01, Tukey-Kramer test.
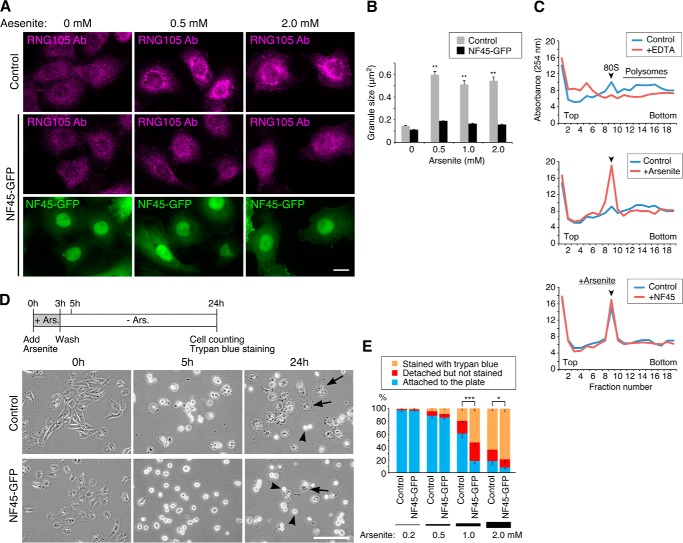
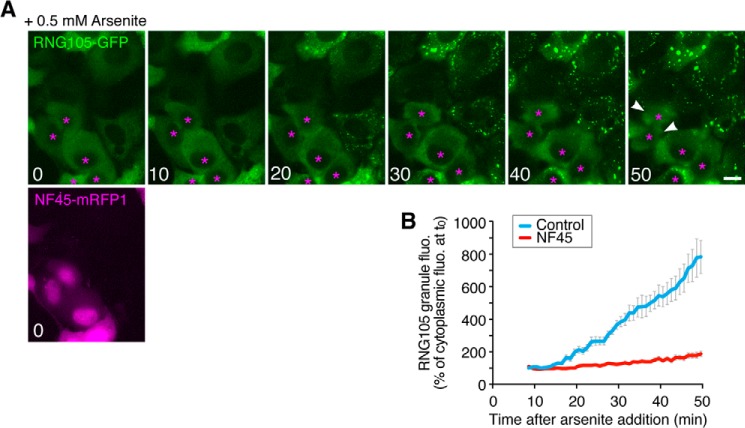
Because RNA granules (SGs) are implicated in cellular defense against stress, we examined whether expression of NF45 affected cell resistance to stress. First, A6 cells stably expressing NF45-GFP were generated, and the effect of NF45 expression on stress-induced SG formation was analyzed. In control A6 cells, SGs were induced by arsenite treatment, as judged by immunostaining with an anti-RNG105 antibody. In contrast, SG formation was inhibited in NF45-GFP-expressing cells (Fig. 3, A and B). Inhibition of SG formation by transient NF45 expression was also observed in time-lapse imaging of RNG105-GFP-expressing cells after arsenite treatment (Fig. 4). The inhibition of SG formation by NF45 was not mediated by restoring arsenite-induced translation inhibition because polysome profiles after arsenite treatment were indistinguishable between control and NF45-expressing cells (Fig. 3C). Next, NF45-GFP stable transfectants and control cells were cultured in the presence of arsenite for 3 h and then further cultured in the absence of arsenite (Fig. 3D). Most of the cells were rounded up after treatment with arsenite at concentrations higher than 1 mm, but some cells survived and reattached to the culture plate after culturing without arsenite. Expression of NF45 reduced the number of cells that reattached to the culture plate (Fig. 3, D and E). Furthermore, the number of dead cells was increased by NF45 expression (Fig. 3E). These results indicated that the expression of NF45 rendered cells susceptible to arsenite stress and suggested the link between SG assembly and cellular defense against stress.
FIGURE 3.
NF45 expression renders cells susceptible to stress. A, control A6 cells and stable transfectants with NF45-GFP were cultured with arsenite at the indicated concentrations for 1 h. The cells were immunostained with an anti-RNG105 antibody. Scale bar, 10 μm. B, mean size of RNG105 granules in A; n = 30; ** indicates p < 0.01, Tukey-Kramer test between 0 mm arsenite-treated control cells and indicated cells. C, polysome profiles analyzed by sucrose density gradient centrifugation. Shown are the absorbance profiles of fractions at 254 nm. Top panels, A6 cell extracts were treated without or with EDTA, which dissociated 80 S ribosomes into subunits and disassembled polysomes. Middle panels, A6 cells were cultured in the absence or presence of 0.5 mm arsenite for 1 h, which repressed translation and disassembled polysomes. Ribosomes dissociated from mRNA accumulated in the 80 S fractions. Bottom panels, control cells and NF45-GFP stable transfectants were cultured in the presence of 0.5 mm arsenite for 1 h. Polysome disassembly was not affected by expression of NF45-GFP. Arrowheads denote the peak of 80 S ribosomes. D, control A6 cells and NF45-GFP stable transfectants were cultured with 1.0 mm arsenite for 3 h. After removing arsenite, the cells were cultured for an additional 21 h. Phase contrast images were taken at 0, 5, and 24 h after arsenite addition. Arrows and arrowheads indicate cells attached to and detached from the culture plate, respectively. Scale bar, 100 μm. E, quantification of cell survival at 24 h after arsenite addition in D. Dead cells detached from the culture plates were detected by staining with trypan blue. n = 5 experiments; *, p < 0.05; ***, p < 0.005; χ2 test was performed on total cell numbers from the five experiments (>500 cells each).
FIGURE 4.
NF45 reduces the assembly of arsenite-induced RNG105-localizing SGs. A, A6 cells stably expressing a low level of RNG105-GFP, which did not form RNA granules in basal condition, were transfected with NF45-mRFP1. The cells were stressed with 0.5 mm arsenite and viewed using a time-lapse fluorescence microscope. Numbers indicate time (minutes) after arsenite addition. Asterisks indicate cells transfected with NF45-mRFP1. Other cells are untransfected cells. Arrowheads denote cells detached from the coverslip. Scale bar, 10 μm. B, quantification of arsenite-induced assembly of RNG105-localizing SGs in A. Control, untransfected cells; NF45, NF45-mRFP1-transfected cells. n = 15.
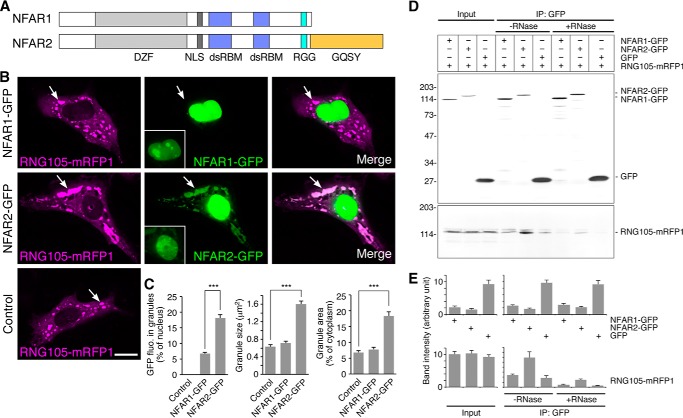
NFAR2, but Not NFAR1, Localizes with and Enlarges RNA Granules
Because NF45 forms heterodimers with NFAR1 and NFAR2 (27, 42), we investigated whether NFAR1 and NFAR2 also have effects on RNA granule assembly. NFAR1 and NFAR2 were fused to GFP and expressed with RNG105-mRFP1 in A6 cells. NFAR1 was located in the nucleus and was not concentrated in the RNG105-localizing RNA granule area (Fig. 5, B and C). In contrast, NFAR2 was located predominantly in the nucleus but also co-localized with RNG105 RNA granules (Fig. 5, B and C). These results indicated that the C-terminal GQSY domain of NFAR2 was required for its localization with RNA granules because only NFAR2, but not NFAR1, has the GQSY domain (Fig. 5A). In addition to co-localization with RNA granules, NFAR2 increased the size of RNG105 RNA granules (Fig. 5, B and C). IP analysis of transfectants using an anti-GFP antibody showed that RNG105-mRFP1 was co-immunoprecipitated with NFAR2-GFP but not with NFAR1-GFP (Fig. 5, D and E). RNase treatment reduced the amount of RNG105-mRFP1 co-precipitates but still left small interaction with NFAR2-GFP (Fig. 5, D and E), suggesting that NFAR2 can associate with RNG105-containing mRNP complexes through protein-protein interaction, and the mRNP complexes may be disassembled by RNase treatment. Taken together, these results suggested that NFAR2 is associated with RNG105-localizing RNA granules through the GQSY domain and enhances the assembly of RNA granules.
FIGURE 5.
NFAR2, but not NFAR1, co-localizes with and enlarges RNA granules. A, schematic diagram of the domain structure of NFAR1 and NFAR2. DZF, zinc finger nucleic acid binding domain; NLS, nuclear localization sequence; dsRBM, dsRNA-binding motif; RGG, Arg-Gly-Gly motif responsible for RNA binding; GQSY, Gly-, Gln-, Ser-, Tyr-rich domain. B, A6 cells were co-transfected with RNG105-mRFP1 and NFAR1-GFP (top panels), RNG105-mRFP1 and NFAR2-GFP (middle panels), or RNG105-mRFP1 alone (bottom panels). Arrows indicate RNG105-mRFP1 granules. The brightness of pictures was adjusted to show RNA granules distinctly, and the original images around nuclei used to quantify fluorescence intensity in C are shown in the insets. Scale bar, 10 μm. C, quantification of RNA granules in B. Fluorescence intensity of NFAR1-GFP and NFAR2-GFP in granules was normalized to that in the nucleus of the same cell. NFAR2-GFP was co-localized with granules and increased the mean size and the total area of granules in the cytoplasm. n = 30 cells; ***, p < 0.001, t test. D, IP from A6 cells co-expressing RNG105-mRFP1 and NFAR1-GFP, NFAR2-GFP, or GFP with the anti-GFP antibody in the presence or absence of RNase. Immunoprecipitates were immunoblotted with anti-GFP and anti-RFP antibodies. E, quantification of the band intensity in D. n = 3.
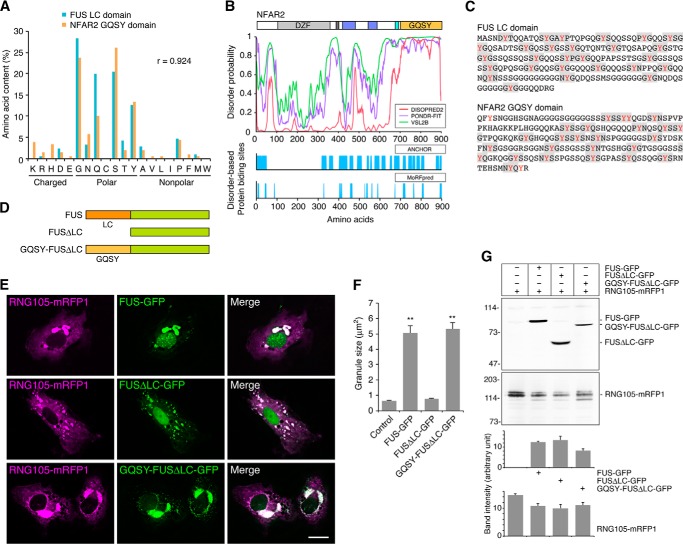
GQSY Domain of NFAR2 Is Similar to the LC Sequence Domain of FUS
We then analyzed the structural and functional features of the GQSY domain. The amino acid composition of the GQSY domain has little diversity, i.e. the domain is highly enriched with Gly, Gln, Ser, and Tyr residues, but other charged or nonpolar amino acids are infrequent (Fig. 6A). Because such LC sequences are suggested to characterize natively disordered polypeptides (43), the NFAR2 sequence was subjected to predictive analysis of native disorder. The disorder probability of the GQSY domain was high (Fig. 6B), supporting the notion that the GQSY domain includes an LC sequence. In addition, the GQSY domain was predicted to contain multiple regions that bind to protein partners through disorder-based binding (Fig. 6B). It has been reported that the LC sequence domain of FUS can undergo phase transition from a soluble phase to a hydrogel-like state in vitro, and the domain is required for localization to SGs in cells (9, 44). The FUS LC sequence domain contains 27 variants of tripeptide sequence (G/S)Y(G/S) repeats, which are required for hydrogel formation and localization to SGs (9, 44). Comparison of the amino acid compositions of the NFAR2 GQSY domain and the FUS LC sequence domain revealed that the two domains share a high level of similarity (Fig. 6A). Furthermore, the NFAR2 GQSY domain contains 24 variants of (G/S)Y(G/S) repeats (Fig. 6C). Thus, the GQSY domain of NFAR2 is structurally similar to the LC sequence domain of FUS.
FIGURE 6.
Structural and functional relation between the GQSY domain of NFAR2 and the LC sequence domain of FUS. A, high correlation of the amino acid composition between the mouse FUS LC sequence domain and the NFAR2 GQSY domain. Both domains are enriched with amino acids with polar side chains, especially Gly, Gln, Ser, and Tyr residues. B, top panel, structural disorder of the GQSY domain predicted by DISOPRED2, PONDR-FIT, and VSL2B predictors. Bottom panel, disorder-based potential binding sites predicted by ANCHOR and MoRFpred predictors. C, amino acid sequences of the FUS LC sequence domain and the NFAR2 GQSY domain. (G/S)Y(G/S) and similar motifs are highlighted. D, schematic diagram of the domain structure of FUS, FUSΔLC, and FUSΔLC fused to NFAR2 GQSY. E, A6 cells were co-transfected with RNG105-mRFP1 and FUS-GFP, FUSΔLC-GFP, or GQSY-FUSΔLC-GFP. Scale bar, 10 μm. F, quantification of RNG105-mRFP1 granule size in E. n = 30 cells; **p < 0.01; Tukey-Kramer test between control and indicated cells. G, immunoblotting of FUS-GFP proteins and RNG105-mRFP1 in the transfectants in E. Transfection efficiency was not significantly different among the cells as follows: 16.6 ± 1.3, 15.4 ± 1.2, 16.4 ± 2.4, and 17.6 ± 1.0% for control, FUS-GFP, FUSΔLC-GFP, and GQSY-FUSΔLC-GFP transfectants, respectively. Bottom panels show quantification of the band intensity of immunoblotting (n = 3).
Next, we examined whether the GQSY domain is functionally similar to the LC sequence domain of FUS. Full-length FUS, LC sequence domain deletion mutant (FUSΔLC), and a chimeric FUS with its LC sequence domain replaced by the NFAR2 GQSY domain (GQSY-FUSΔLC) were constructed (Fig. 6D). These constructs were fused to GFP and expressed in A6 cells with RNG105-mRFP1 (Fig. 6E). FUS co-localized with RNG105 and increased the size of RNG105-localizing RNA granules (Fig. 6, E and F). In contrast, although FUSΔLC co-localized with RNG105, it lost the ability to increase the size of RNA granules, indicating that the LC sequence domain was required for the assembly of large RNA granules (Fig. 6, E and F). Expression of GQSY-FUSΔLC resulted in large RNG105 RNA granules, which indicated that the GQSY domain restored the ability to assemble large RNA granules (Fig. 6, E and F). The restoration was not because of higher expression of GQSY-FUSΔLC than FUSΔLC or increased expression of RNG105 in the GQSY-FUSΔLC transfectants (Fig. 6G). These results indicated that the GQSY domain of NFAR2 was functionally exchangeable with the LC sequence domain of FUS in terms of the ability to assemble large RNA granules.
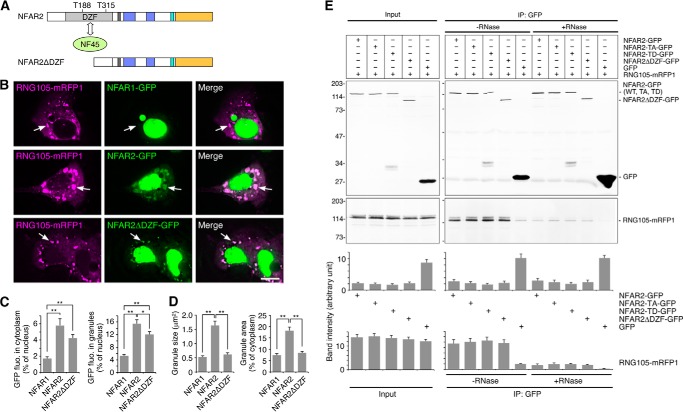
DZF Domain of NFAR2 Is Required to Enhance RNA Granule Assembly
The DZF domains of NFAR1 and NFAR2 are phosphorylated at Thr-188 and Thr-315 by PKR, which causes their retention on ribosomes and translational inhibition (Fig. 7A) (33). In addition to being phosphorylated by PKR, the DZF domain is bound by NF45 to form a heterodimer (Fig. 7A) (27, 42). We then examined the effect of the DZF domain of NFAR2 on RNA granule assembly. We constructed NFAR2 lacking the DZF domain (NFAR2ΔDZF), fused it to GFP, and introduced it to A6 cells with RNG105-mRFP1 (Fig. 7, A and B). The distribution pattern of NFAR2ΔDZF was similar to that of wild-type NFAR2, and it co-localized with RNG105 RNA granules (Fig. 7, B and C). IP from the transfectants also indicated that NFAR2ΔDZF-GFP associated with the RNG105-mRFP1 complex at the same level as wild-type NFAR2-GFP (Fig. 7E). However, the expression of NFAR2ΔDZF did not induce large RNG105-containing RNA granules, as compared with wild-type NFAR2, which suggested that the DZF domain was required to enhance the assembly of RNA granules (Fig. 7, B and D). Together, these results suggested that the DZF domain is involved in the assembly of RNA granules without affecting the association between the GQSY domain and RNG105 mRNP complexes.
FIGURE 7.
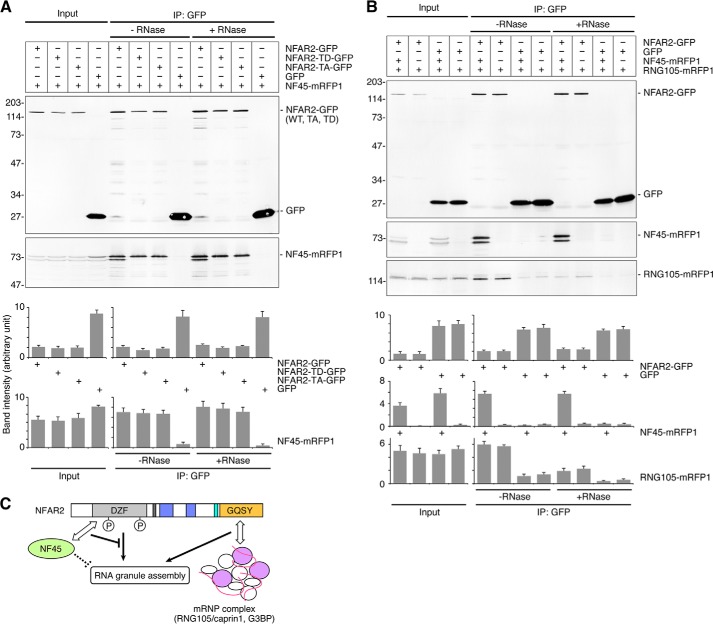
NFAR2ΔDZF co-localizes with RNG105 RNA granules but loses the ability to enlarge RNA granules. A, schematic diagram of NFAR2ΔDZF. The DZF domain includes PKR phosphorylation sites (Thr-188 and Thr-315) and binds to NF45. B, A6 cells were co-transfected with RNG105-mRFP1 and NFAR1-GFP, NFAR2-GFP, or NFAR2ΔDZF-GFP. Arrows indicate RNG105-mRFP1 granules. Scale bar, 10 μm. C, quantification of cytoplasmic and granule localization of NFAR1-GFP, NFAR2-GFP, and NFAR2ΔDZF-GFP in B. D, quantification of the mean size and total area of RNG105-mRFP1 granules in B. n = 25; *, p < 0.05; **, p < 0.01, Tukey-Kramer test. E, IP from A6 cells co-expressing RNG105-mRFP1 and NFAR2-GFP, NFAR2-TA-GFP, NFAR2-TD-GFP, NFAR2ΔDZF-GFP, or GFP using the anti-GFP antibody in the presence or absence of RNase. Immunoprecipitates were immunoblotted with anti-GFP and anti-RFP antibodies. Bottom panels show quantification of the band intensity (n = 3).
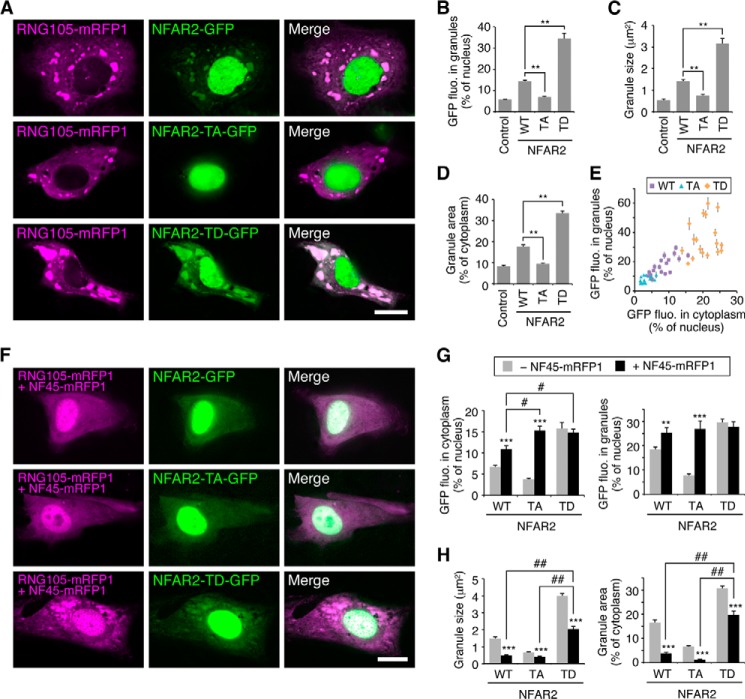
Phosphomimetic Mutations of NFAR2 at PKR Sites in the DZF Domain Enhances Cytoplasmic Retention, RNA Granule Localization, and Assembly
To examine the effect of NFAR2 phosphorylation by PKR on its localization to, and the assembly of, RNA granules, phosphodeficient (TA) and phosphomimetic (TD) mutants of NFAR2 were constructed and expressed as GFP fusion proteins in A6 cells (Figs. 7E, 8, and 9A). When the NFAR2-TA mutant was expressed with RNG105-mRFP1, NFAR2-TA reduced its co-localization with RNG105 and also reduced the size of RNG105 RNA granules compared with wild-type NFAR2 (Fig. 8, A–D). In contrast, the NFAR2-TD mutant increased its co-localization with RNG105 and induced larger RNA granules than wild-type NFAR2 (Fig. 8, A–D). Because the number of RNG105-localizing RNA granules was not increased by expression of wild-type NFAR2 or NFAR2-TD, the increase in the total area of RNA granules was attributed to the increase in the size of RNA granules (Fig. 8, C and D). Quantitative analysis showed that increased localization of NFAR2-TD to RNA granules was correlated with the increase in its cytoplasmic localization, and reduced localization of NFAR2-TA to RNA granules was correlated with its reduced localization in the cytoplasm (Fig. 8E). These results suggested that phosphorylation of the DZF domain of NFAR2 by PKR increases its cytoplasmic retention, which increases its association with RNA granules to enhance the assembly of RNA granules.
FIGURE 8.
Regulation of RNA granule assembly and disassembly through the DZF domain of NFAR2, phosphorylation mutants and NF45 association. A, both Thr-188 and Thr-315 in the DZF domain of NFAR2 were mutated to Ala or Asp in phosphodeficient (TA) or phosphomimetic (TD) mutants, respectively. A6 cells were co-transfected with RNG105-mRFP1 and NFAR2-GFP, NFAR2-TA-GFP, or NFAR2-TD-GFP. Scale bar, 10 μm. B–D, quantification of RNG105-mRFP1 granules in A. The TA mutant reduced its ability to localize to and enhance the assembly of RNG105 granules, whereas the TD mutant increased the ability compared with wild-type (WT) NFAR2. n = 25; **, p < 0.01, Tukey-Kramer test. E, correlation between cytoplasmic localization and granule localization of NFAR2-GFP and its mutants. r = 0.87, p = 2.68 × 10−20. F, same experiments as A were performed except that the cells were additionally co-transfected with NF45-mRFP1. Although RNG105 and NF45 were visualized in the same color, RNG105 granules were distinguishable because of their bright signals. Essentially the same results were obtained by using NF45 without fluorescent protein tags. Scale bar, 10 μm. G, quantification of cytoplasmic and granule localization of NFAR2-GFP and its mutants in A and F. H, quantification of the mean size and total area of RNG105-mRFP1 granules in A and F. n = 25; **, p < 0.01; ***, p < 0.001, t test between NF45-mRFP1-transfected and -untransfected cells. #, p < 0.05; ##, p < 0.01, Tukey-Kramer test among NF45-mRFP1-transfected cells.
FIGURE 9.
Binding of NF45 to NFAR2 does not affect the association between NFAR2 and RNG105 mRNP complexes. A, IP from A6 cells co-expressing NF45-mRFP1 and NFAR2-GFP, NFAR2-TD-GFP, NFAR2-TA-GFP, or GFP using the anti-GFP antibody in the presence or absence of RNase. Immunoprecipitates were immunoblotted with anti-GFP and anti-RFP antibodies. Lower bands of NF45-mRFP1 were detected in cell lysates (input) and the NFAR2-GFP immunoprecipitates, although the reasons are not known. B, IP with the anti-GFP antibody from A6 cells co-expressing RNG105-mRFP1 and NFAR2-GFP or GFP, which were additionally transfected with or without NF45-mRFP1. C, a model for the role of the GQSY and DZF domains of NFAR2 and NF45 in RNA granule assembly.
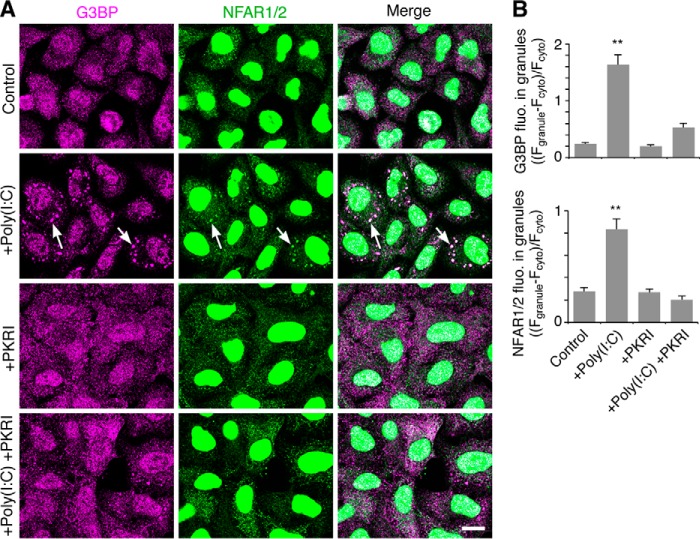
Next, we tested whether the increase in the cytoplasmic and RNA granule localization of NFAR2 was dependent on PKR activity. It is reported that PKR is activated by diverse stress signals, including arsenite through PKR activating protein (PACT) (45, 46), although heme-regulated eIF2α kinase is a more favorable target of arsenite (47). PKR is more strongly activated by dsRNA (poly(I-C)), which is demonstrated to phosphorylate NFAR1 and NFAR2 (33). In addition, PKR is activated by G3BP-induced RNA granule formation (22). First, we examined the effect of poly(I-C)-induced PKR on NFAR2 localization to RNA granules (SGs). In this analysis, HeLa cells were used because poly(I-C) transfectants were not distinguished from nontransfectants, and transfection efficiency of HeLa cells was much higher (>90%) than A6 cells (<20%). Without poly(I-C), SGs were not apparently formed in the cytoplasm as detected by immunostaining for G3BP (Fig. 10A). When poly(I-C) was introduced in cells, it induced the formation of G3BP-containing SGs, which were immunostained with an anti-NFAR1/2 antibody. SG formation by poly(I-C) was reduced by PKR inhibition, and concomitantly, SG staining with the NFAR1/2 antibody was reduced (Fig. 10, A and B). Thus, NFAR1/2 was localized to PKR-induced SGs.
FIGURE 10.
PKR-dependent RNA granule (SG) localization of NFAR1/2. A, HeLa cells were transfected with poly(I-C) to activate PKR and then cultured for 4 h in the absence or presence of PKR inhibitor (PKRI). The cells were co-immunostained for G3BP and NFAR1/2. Arrows denote SGs. Scale bar, 10 μm. B, quantification of the accumulation of G3BP and NFAR1/2 in SGs. Fluorescence intensity of G3BP and NFAR1/2 in SGs is normalized to that in the cytoplasm. n = 50; **, p < 0.01, Tukey-Kramer test between control and indicated cells.
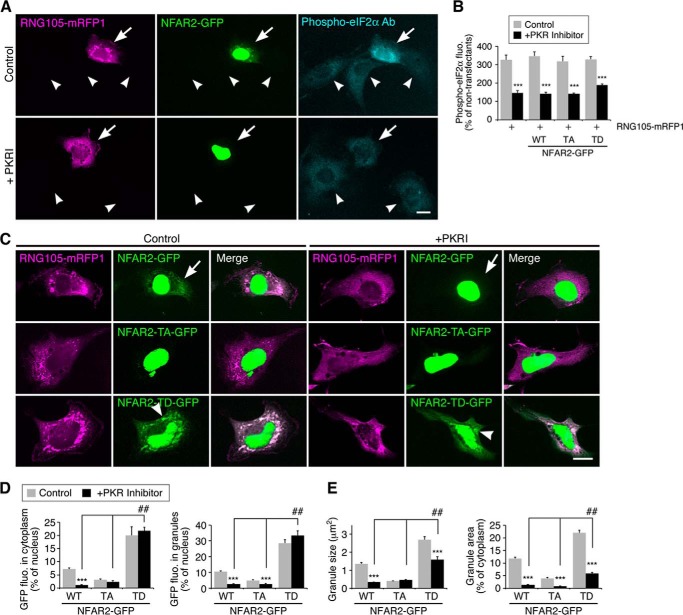
Next, we reverted to using the A6 transfectants and examined the effect of PKR inhibition on NFAR2 localization to the cytoplasm and RNA granules. As reported previously (18), expression of RNG105 increased the phosphorylation level of eIF2α (Fig. 11, A and B). This phosphorylation was reduced by PKR inhibition, indicating that PKR was activated by RNG105 expression (Fig. 11, A and B), which was similar to G3BP expression (22). NFAR2 co-expressed with RNG105 was localized mainly in the nucleus and also in the cytoplasm and RNA granules (Fig. 11C). However, when PKR was inhibited, NFAR2 localization to the cytoplasm and RNA granules was markedly reduced, and it was restricted to the nucleus (Fig. 11, C and D). In contrast, localization of NFAR2-TD in the cytoplasm and RNA granules was not changed even when PKR was inhibited (Fig. 11, C and D). These results indicated that the increase of NFAR2 in the cytoplasm and RNA granules was PKR-dependent and that Thr-188 and Thr-325 were the target sites of PKR. Although the localization of NFAR2-TD was not affected by PKR inhibition, RNA granule formation was impaired by PKR inhibition (Fig. 11, C and E). This may be because other endogenous pathways necessary for the formation RNA granules, e.g. eIF2α phosphorylation, were inhibited. However, NFAR2-TD still induced larger RNA granules than wild-type NFAR2 or NFAR2-TA even in the presence of PKR inhibitor, suggesting that phosphomimetic mutations overcame the effect of PKR inhibition (Fig. 11, C and E). Taken together, these results supported the notion that NFAR2 phosphorylation by PKR enhances its cytoplasmic retention, localization to RNA granules, and the assembly of RNA granules.
FIGURE 11.
PKR inhibition reduces the cytoplasmic and RNA granule localization of NFAR2, but not of NFAR2-TD. A, A6 cells co-transfected with RNG105-mRFP1 and NFAR2-GFP were cultured in the presence or absence of PKR inhibitor and stained with an anti-phospho-eIF2α antibody (Ab). Arrows and arrowheads indicate transfected and untransfected cells, respectively. Scale bar, 10 μm. B, quantification of phospho-eIF2α fluorescence intensity in transfectants expressing RNG105-mRFP1 with or without NFAR2-GFP or its mutants. Fluorescence intensity was normalized to that in untransfected neighboring cells. C, A6 cells co-expressing RNG105-mRFP1 and NFAR2-GFP or its mutants were cultured without (left panels) or with (right panels) PKR inhibitor. Arrows indicate cytoplasmic and granule localization of NFAR2-GFP, which was reduced by PKR inhibition. Arrowheads indicate cytoplasmic and granule localization of NFAR2-TD-GFP, which was not significantly affected by PKR inhibition. Scale bar, 10 μm. D, quantification of cytoplasmic and granule localization of NFAR2-GFP and its mutants in C. E, quantification of granules in the cells in C. n = 30; ***, p < 0.001, t test between control and PKR inhibitor-treated cells. ##, p < 0.01, Tukey-Kramer test among PKR inhibitor-treated cells.
NF45 Antagonizes RNA Granule Assembly by NFAR2
It is known that NF45 binds directly to NFAR1/2 (27, 42). Co-expression of NFAR2-GFP and NF45-mRFP1 in A6 cells and subsequent IP with an anti-GFP antibody confirmed that NFAR2 and NF45 formed a complex independently on RNA (Fig. 9A). The amount of NF45 bound to NFAR2 was not significantly changed by mutations in the PKR phosphorylation sites (Fig. 9A). Next, we examined the effect of NF45 binding to NFAR2 on RNA granule assembly. A6 cells were co-transfected with RNG105-mRFP1 and NFAR2-GFP or its phosphorylation site mutants and additionally with or without NF45-mRFP1 (Fig. 8, A and F). Expression of NF45 changed the distribution pattern of wild-type NFAR2 and NFAR2-TA, i.e. their cytoplasmic localization was increased by NF45 expression (Fig. 8, A, F, and G). This may be because cytoplasmic localization of NF45-mRFP1 was higher than that of NFAR2-GFP or NFAR2-TA-GFP (cytoplasmic NF45-mRFP1 was 47.7 ± 3.6% of nuclear NF45-mRFP1; cf. Fig. 8G) and was accompanied by NFAR2-GFP or NFAR2-TA-GFP that bound to NF45-mRFP1. Concomitantly, their localization to RNG105 RNA granules was also increased in NF45-expressing cells (Fig. 8G). In contrast, the distribution pattern of NFAR2-TD was not affected by NF45 expression (Fig. 8, A, F, and G). This may be because cytoplasmic localization of NFAR2-TD was relatively high even without NF45 expression, or NF45 may have little effect on the localization of phosphorylated NFAR2.
Although cytoplasmic and RNA granule localization of wild-type NFAR2 and NFAR2-TA was increased by NF45 co-expression, the assembly of RNG105-containing RNA granules was reduced by NF45 co-expression (Fig. 8, A, F, and H). NF45 expression also reduced the enhancement of RNA granule assembly by NFAR2-TD (Fig. 8, A, F, and H). These results suggested that NF45 antagonized the ability of NFAR2 to enhance RNA granule assembly in the cytoplasm. Nevertheless, RNA granule assembly was enhanced more by NFAR2-TD than by wild-type NFAR2 or NFAR2-TA, even when NF45 was co-expressed (Fig. 8, F and H). Thus, RNA granule assembly was balanced by negative regulation by NF45 binding to NFAR2 and positive regulation by phosphorylation at PKR sites on NFAR2. Phosphorylation of NFAR2 contributes not only to its cytoplasmic retention but also to increasing its activity to assemble large RNA granules by overcoming NF45 in the cytoplasm.
Although NF45 binding to NFAR2 reduced the assembly of RNG105-containing RNA granules, IP analysis indicated that NF45 binding to NFAR2 did not change the association between NFAR2 and the RNG105 mRNP complex (Fig. 9B). Similarly, although phosphorylation site mutations in NFAR2 influenced the assembly of RNG105-containing RNA granules, these mutations did not change the association between NFAR2 and the RNG105 mRNP complex (Fig. 7E). These results were consistent with the results that the deletion of the DZF domain did not change the association between NFAR2 and the RNG105 mRNP complex (Fig. 7E). Taken together, these results suggested that the DZF domain regulates RNA granule assembly positively and negatively by being phosphorylated at PKR sites and by association with NF45, respectively, not through regulating the interaction between the GQSY domain and the RNG105 mRNP complex.
Translational Repression by NFAR1/2 and De-repression by NF45
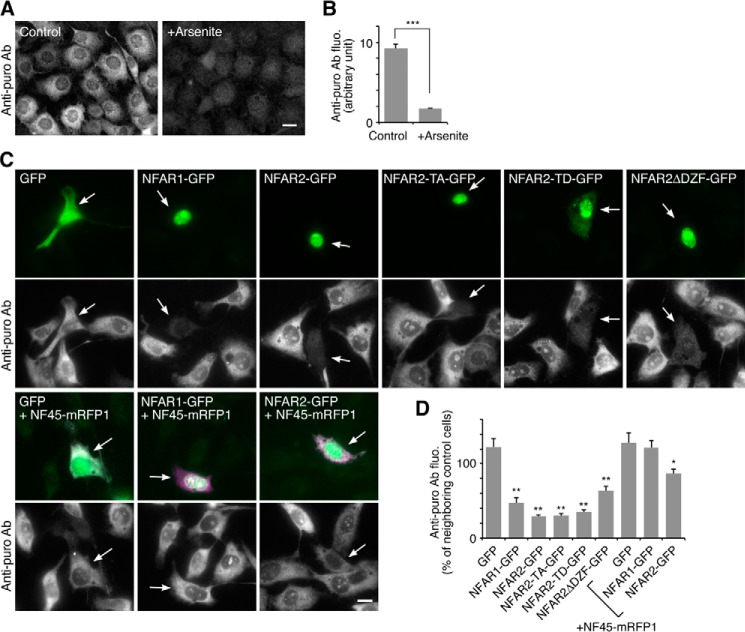
RNA granule assembly and disassembly are often linked to translational repression and activation. Then we examined the effect of the expression of NFAR1, NFAR2, and NF45 on translation in cells. Because stable transfectants of NFAR1 or NFAR2 were not obtained, we conducted ribopuromycilation assay (22) instead of sucrose density gradient centrifugation. Treatment of cells with arsenite reduced anti-puromycin antibody staining (Fig. 12, A and B), which confirmed the reproducibility of this method. Expression of NFAR1, NFAR2, or its mutants significantly reduced the staining with the antibody, suggesting that NFAR2 inhibited translation independently on its ability to localize to RNA granules (Fig. 12, C and D). Co-expression of NF45 antagonized the translational repression by NFAR1 and NFAR2 (Fig. 12, C and D), suggesting linkage between the roles of NF45 in RNA granule disassembly and translational de-repression. But the translational de-repression by NF45 may be rather specific for NFAR1 and NFAR2 because polysome dissociation induced by arsenite, which is mediated by eIF2α phosphorylation, was not affected by NF45 (Fig. 3C). Thus, RNA granule assembly and disassembly modulated by NFAR2 and NF45 were not likely to be strictly linked to translation regulation, providing a novel regulatory mechanism for RNA granule assembly and disassembly. Nevertheless, as NFAR2 and eIF2α are both the targets of PKR, RNA granule assembly by NFAR2/NF45 and translation initiation are simultaneously regulated under the control of PKR.
FIGURE 12.
Translational repression by NFAR1/2 and de-repression by NF45. A, A6 cells were analyzed by ribopuromycilation assay to measure translation in cells. Staining with an anti-puromycin antibody was reduced by treatment with 0.5 mm arsenite for 30 min. Scale bar, 10 μm. B, quantification of the cells in A. n = 32; ***, p < 0.001, t test. C, A6 cells expressing indicated proteins were analyzed by ribopuromycilation assay. Arrows denote transfected cells. Scale bar, 10 μm. D, quantification of anti-puromycin antibody staining of the transfectants in C. n = 30; *, p < 0.05; **, p < 0.01, Tukey-Kramer test between GFP-expressing cells (leftmost bar) and indicated cells.
DISCUSSION
This study identified NFAR2 and NF45 as positive and negative regulatory proteins, respectively, of RNA granule assembly. Although it has been unclear whether the two NFAR splice variants, NFAR1 and NFAR2, have different properties from one another, we demonstrated that NFAR2, but not NFAR1, has the ability to localize to and enhance the assembly of RNA granules. This ability is attributed to the C-terminal GQSY domain, which interacts with mRNP complexes containing RNG105 and has similar structural and functional features to the FUS LC sequence domain. Another domain of NFAR2 responsible for the assembly of RNA granules is the DZF domain, whose ability to enhance RNA granule assembly is balanced by positive regulation by phosphorylation at PKR sites and negative regulation by binding with NF45. The results provide new insights into the molecular mechanism by which NFAR2, its phosphorylation by PKR, and NF45 regulate RNA granule assembly.
Previous proteomic analyses suggested that NFAR1 and/or NFAR2 were contained in mRNP complexes. IP of RNA granule components such as Argonaute proteins or TDP-43 identified NFAR1/2 together with other RNA granule components such as fragile X mental retardation protein, HuR, poly-A-binding protein, staufen, FUS, G3BP, and RNG105/caprin1 (48, 49). However, it remained unclear whether NFAR proteins are in fact components of RNA granules. We demonstrated that NFAR2, but not NFAR1, was a component of RNA granules.
NFAR2, but not NFAR1, has the GQSY domain that is similar to the FUS LC sequence domain, a subtype of intrinsically disordered regions. Intrinsically disordered proteins often bind to their specific partners via disorder-to-order transition, playing roles in regulating the function of their binding partners and in promoting the assembly of macromolecular complexes (50). Intrinsically disordered regions are commonly found in proteins that nucleate SGs and other SG-associated proteins involved in cellular signaling (51). Intrinsically disordered proteins are also associated with numerous human diseases, including neurodegeneration and cancer (50).
The phase transition of FUS LC sequence domains from a soluble phase to a hydrogel-like state is proposed to be the molecular principle driving RNA granule assembly (9, 44), which is consistent with our results that FUS and GQSY-FUSΔLC, but not FUSΔLC, increased the size of RNA granules (Fig. 6). Serine residues in the (G/S)Y(G/S) motif of FUS LC sequence domain are reportedly phosphorylated by DNA-dependent protein kinase (DNA-PK), which reduces the binding of FUS to the hydrogel, offering a means of dynamic control of RNA granule assembly (44). NFAR1/2 are also reported to form a complex with and are phosphorylated by DNA-PK in the nucleus (52). Because RNA granules are formed not in the nucleus but in the cytoplasm, it will be of interest to investigate in the future whether the phosphorylation of NFAR2 by DNA-PK controls RNA granule assembly and disassembly in the cytoplasm and nucleus.
The GQSY domain was required for the association with RNG105 mRNP complexes, and the binding of RNG105 to NFAR2 included both RNase-resistant and sensitive fractions. This suggests that a fraction of RNG105 was associated with the GQSY domain of NFAR2 through protein-protein interactions, and other fractions were associated in an RNA-dependent manner. RNG105 mRNP complexes may contain multiple RNG105 molecules and other proteins, which are bridged by mRNAs, and associate with the GQSY domain of NFAR2 (Fig. 9C).
Immunoprecipitates with RNG105 from mitotic cells included multiple types of proteins whose composition was largely unchanged from interphase cells, although RNA granules disappeared in mitotic cells (Fig. 1, B and C). These results suggest that small RNG105 mRNP complexes are maintained even when large RNA granules are disassembled in mitosis. We hypothesize that RNG105 mRNP complexes may exist as small units, and the units are connected to each other when RNA granules are formed. The connection may be released by proteins like NF45 because the association of NF45 with RNG105 mRNP complexes was increased in mitosis, and expression of NF45 in cells led to disassembly of RNA granules. Because the increased association of NF45 with RNG105 mRNP complexes was likely because of the release of NF45 from the nucleus in mitosis, its binding partner NFAR2 may also be released from the nucleus and increase the association with RNG105 mRNP complexes in mitosis. The NF45-NFAR2 dimer could compete with other factors for the binding to RNG105 mRNP complexes and induce RNA granule disassembly, which may be supported by the results that co-expression of NF45 and NFAR2 led to disassembly of RNG105-induced RNA granules (Fig. 8F, cf. 5B, control).
The DZF domain of NFAR2 was dispensable for the association with RNG105 mRNP complexes but responsible for RNA granule assembly. This suggests that the DZF domain forms complexes independently on the RNG105 mRNP complex. Formation of the DZF domain-including complex may be enhanced by phosphorylation with PKR and inhibited by binding with NF45, neither of which change the association between NFAR2 and RNG105 (Figs. 7E, and 9, B and C). NFAR2 may function as a connector of RNG105 mRNP complexes through its multivalent domains as follows: the GQSY domain that associates with RNG105 mRNP complexes and the DZF domain. When the DZF domain is bound by NF45, the connector function could be reduced, which may explain the mechanism of how NF45 disassembles RNA granules. This model does not exclude another possibility that proteins such as FUS also function as connector proteins, proteins such as protein N-methyltransferase 1 disassemble RNA granules in a post-translational modification-dependent manner, and NF45 targets other proteins in addition to NFAR2 (Fig. 9C).
Phosphorylation of NFAR2 at PKR sites induced a shift in its localization from the nucleus to the cytoplasm, which led to the enhancement of RNA granule assembly (Fig. 8, A–E). Although there are opposing results regarding the binding between phosphorylated NFAR2 and NF45, i.e. the current results showed that phosphorylated NFAR2 kept the binding to NF45 but a previous report showed that it did not (33), the net result was the same; phosphorylated NFAR2 increased its retention in the cytoplasm. Cytoplasmic retention of NFAR2 and NFAR2-TA was also increased by the expression of NF45 to almost the same level as that of NFAR2-TD, but they enhanced the assembly of RNA granules far less than NFAR2-TD (Fig. 8, F–H). These results suggested that phosphorylation of NFAR2 contributes not only to its cytoplasmic retention but also to overcoming RNA granule disassembly by NF45. Thus, when PKR is inactive, NFAR2 is localized in the nucleus and suppressed by NF45 even if a fraction of NFAR2 is exported to the cytoplasm. However, when PKR is activated, NFAR2 is phosphorylated, retained in the cytoplasm, and overcomes NF45 to enhance the assembly of RNA granules. The interplay between the positive and negative regulation of NFAR2 may function in the abrupt switching between the appearance and disappearance of RNA granules.
RNA granules (SGs) are induced by stress-responsive kinases, including PKR, which is activated by several kinds of stress such as viral dsRNA infection and peroxide or arsenite treatment (33, 45, 46). RNA granules contribute to translational arrest and cellular defense against stress. In this study, we showed that expression of NF45 disassembled RNA granules and rendered cells susceptible to arsenite stress, but arsenite-induced translational arrest was not affected (Fig. 3). This sustained translational arrest may be because NF45 could not affect the translational initiation pathway, including eIF2α, although NF45 antagonizes translational repression by NFAR1/2 (Fig. 12). These results suggest that translational arrest is not sufficient for the protection of cells from stress and that RNA granule assembly has additional functions in cell survival, e.g. sequestration and nullification of stress-responsive apoptosis-promoting signaling molecules (53, 54, 10, 11).
It was reported that knockdown of NF45 increases multinucleated cells, suggesting that NF45 is required for the proper progression of mitosis (42). Our present results that the association of NF45 with RNG105 mRNP complexes was increased in mitosis suggest a possible involvement of the regulation of RNA granules by NF45 in mitotic processes. Recently, RNA granule disassembly during meiosis was reported (55). RNA granules storing dormant cyclin B1 mRNA are formed in immature oocytes, but they are disassembled concomitantly with germinal vesicle (nuclear envelope) breakdown, which activates cyclin B1 mRNA translation. This RNA granule disassembly is reminiscent of our present result that RNG105-localizing RNA granules disappeared in mitosis. Whether NF45 is involved in the disassembly of RNA granules storing cyclin B1 mRNA and the progression of cyclin B1-dependent oocyte maturation and mitotic events are interesting issues to be solved in the future.
NFAR1 is ubiquitously expressed in several organs, but expression of NFAR2 is organ-specific (56). In particular, brain, testis, and skeletal muscle show high expression levels of NFAR2, which is coincident with the high levels of RNA granules in the brain and germ cells (3, 5, 57). Knock-out of NFAR1/2 leads to increased apoptosis, especially in cortical neurons and skeletal muscle (56). In skeletal muscle, NFAR1/2 bind to mRNAs for myogenic regulators such as MyoD and p21WAF1/CIP1 and stabilize these mRNAs (56). It is notable that p21WAF1/CIP1 mRNA also binds to the RNA granule component HuR (58, 59) and is recruited into RNA granules to be stabilized (60). These reports suggest an association between NFAR2 function in RNA granules and skeletal muscle development and survival. Increased neuronal cell death and respiratory failure in NFAR1/2 knock-out mice suggest that NFAR1/2 also play important roles in neurons (56). Similar phenotypes have been reported in knock-out mice of other RNA granule components, including RNG105/caprin1 and G3BP (23, 24). Elucidating the roles of NFAR1/2 in neuronal RNA granules as well as analyzing the genetic and functional interactions of NFAR1/2 with RNG105/caprin1, G3BP, and other RNA granule components in knock-out and mutant mice will be a topic for future studies.
NFAR2, particularly in its phosphorylated form, promoted large RNA granules. Although differences between large and small RNA granules are not well studied, large RNA granules are implicated in slower dissolution of mRNAs and other components from the granules (61, 62). Large RNA granules trigger eIF2α phosphorylation through PKR, suggesting a positive feedback regulation of granule assembly in large RNA granules (22). Large RNA granules are also induced by neurodegenerative disease-associated mutations in RNA granule components such as TDP-43, FUS/TLS, hnRNPA2B1, and hnRNPA1 (6–8). These proteins contain prion-like LC sequence domains responsible for RNA granule assembly (6–9), which has also been found in NFAR2. It is suggested that defective nuclear localization and cytoplasmic accumulation of disease-associated mutant TDP-43 and FUS are key events in neurodegenerative pathogenesis (6, 7), which is reminiscent of our present result that increased cytoplasmic retention of phosphorylated NFAR2 was linked to the formation of large RNA granules. The structural and functional mechanisms of RNA granule assembly regulated by these RNA granule components provide a common basis for the generation of pathological large RNA granules in neurodegenerative disease.
Acknowledgments
We thank C. Matsuda for technical assistance. We also thank T. Mori and the Functional Genomics Facility of National Institute for Basic Biology for the mass spectrometry analysis.
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Uehara Memorial Foundation (to N. S.).
- FUS
- fused in sarcoma
- mRNP
- messenger ribonucleoprotein
- NF
- nuclear factor
- NFAR
- nuclear factor associated with dsRNA
- PKR
- double-stranded RNA-activated kinase
- IP
- immunoprecipitation
- poly(I-C)
- polyinosinic:polycytidylic acid
- TLS
- translocated in sarcoma
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- LC
- low complexity
- SG
- stress granule
- G3BP
- RasGAP SH3 domain-binding protein
- ILF
- interleukin enhancer binding factor
- DZF domain
- zinc-finger nucleic acid binding domain
- GQSY domain
- domain enriched with Gly, Gln, Ser, and Tyr residues
- mRFP1
- monomeric red fluorescent protein 1
- TA
- phosphodeficient mutant
- TD
- phosphomimetic mutant
- DNA-PK
- DNA-dependent protein kinase
- RFP
- red fluorescent protein.
REFERENCES
- 1. Anderson P., Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kindler S., Wang H., Richter D., Tiedge H. (2005) RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21, 223–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kiebler M. A., Bassell G. J. (2006) Neuronal RNA granules: movers and makers. Neuron 51, 685–690 [DOI] [PubMed] [Google Scholar]
- 4. Sossin W. S., DesGroseillers L. (2006) Intracellular trafficking of RNA in neurons. Traffic 7, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 5. Martin K. C., Ephrussi A. (2009) mRNA localization: gene expression in the spatial dimension. Cell 136, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bentmann E., Haass C., Dormann D. (2013) Stress granules in neurodegeneration– lessons learnt from TDP-43 and FUS. FEBS J. 280, 4348–4370 [DOI] [PubMed] [Google Scholar]
- 7. Ling S. C., Polymenidou M., Cleveland D. W. (2013) Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., Pinkus J. L., Greenberg S. A., Trojanowski J. Q., Traynor B. J., Smith B. N., Topp S., Gkazi A. S., Miller J., Shaw C. E., Kottlors M., Kirschner J., Pestronk A., Li Y. R., Ford A. F., Gitler A. D., Benatar M., King O. D., Kimonis V. E., Ross E. D., Weihl C. C., Shorter J., Taylor J. P. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L. (2012) Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anderson P., Kedersha N. (2008) Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33, 141–150 [DOI] [PubMed] [Google Scholar]
- 11. Buchan J. R., Parker R. (2009) Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bramham C. R., Wells D. G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 13. Costa-Mattioli M., Sossin W. S., Klann E., Sonenberg N. (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang D. O., Martin K. C., Zukin R. S. (2010) Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 33, 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moser J. J., Fritzler M. J. (2010) Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 42, 828–843 [DOI] [PubMed] [Google Scholar]
- 16. El Fatimy R., Tremblay S., Dury A. Y., Solomon S., De Koninck P., Schrader J. W., Khandjian E. W. (2012) Fragile X mental retardation protein interacts with the RNA-binding protein Caprin1 in neuronal RiboNucleoProtein complexes. PLoS One 7, e39338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shiina N., Shinkura K., Tokunaga M. (2005) A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 25, 4420–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Solomon S., Xu Y., Wang B., David M. D., Schubert P., Kennedy D., Schrader J. W. (2007) Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2α, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell Biol. 27, 2324–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katsafanas G. C., Moss B. (2004) Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 279, 52210–52217 [DOI] [PubMed] [Google Scholar]
- 20. Shiina N., Tokunaga M. (2010) RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J. Biol. Chem. 285, 24260–24269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J. M., Bertrand E., Tazi J. (2003) The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22. Reineke L. C., Dougherty J. D., Pierre P., Lloyd R. E. (2012) Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell 23, 3499–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shiina N., Yamaguchi K., Tokunaga M. (2010) RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. J. Neurosci. 30, 12816–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zekri L., Chebli K., Tourrière H., Nielsen F. C., Hansen T. V., Rami A., Tazi J. (2005) Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol. Cell Biol. 25, 8703–8716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Duchange N., Pidoux J., Camus E., Sauvaget D. (2000) Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene 261, 345–353 [DOI] [PubMed] [Google Scholar]
- 26. Saunders L. R., Jurecic V., Barber G. N. (2001) The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics 71, 256–259 [DOI] [PubMed] [Google Scholar]
- 27. Corthésy B., Kao P. N. (1994) Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 269, 20682–20690 [PubMed] [Google Scholar]
- 28. Orford R. L., Robinson C., Haydon J. M., Patient R. K., Guille M. J. (1998) The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double-stranded-RNA-binding subunit. Mol. Cell Biol. 18, 5557–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shim J., Lim H., Yates J. R., Karin M. (2002) Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10, 1331–1344 [DOI] [PubMed] [Google Scholar]
- 30. Kuwano Y., Kim H. H., Abdelmohsen K., Pullmann R., Jr., Martindale J. L., Yang X., Gorospe M. (2008) MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 28, 4562–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Larcher J. C., Gasmi L., Viranaïicken W., Eddé B., Bernard R., Ginzburg I., Denoulet P. (2004) Ilf3 and NF90 associate with the axonal targeting element of Tau mRNA. FASEB J. 18, 1761–1763 [DOI] [PubMed] [Google Scholar]
- 32. Pfeifer I., Elsby R., Fernandez M., Faria P. A., Nussenzveig D. R., Lossos I. S., Fontoura B. M., Martin W. D., Barber G. N. (2008) NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl. Acad. Sci. U.S.A. 105, 4173–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harashima A., Guettouche T., Barber G. N. (2010) Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 24, 2640–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wolkowicz U. M., Cook A. G. (2012) NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res. 40, 9356–9368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reichman T. W., Parrott A. M., Fierro-Monti I., Caron D. J., Kao P. N., Lee C. G., Li H., Mathews M. B. (2003) Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 332, 85–98 [DOI] [PubMed] [Google Scholar]
- 36. Shiina N., Tsukita S. (1999) Mutations at phosphorylation sites of Xenopus microtubule-associated protein 4 affect its microtubule-binding ability and chromosome movement during mitosis. Mol. Biol. Cell 10, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 38. Xue B., Dunbrack R. L., Williams R. W., Dunker A. K., Uversky V. N. (2010) PONDR-Fit: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 1804, 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A. K. (2005) Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins 61, Suppl 7, 176–182 [DOI] [PubMed] [Google Scholar]
- 40. Mészáros B., Simon I., Dosztányi Z. (2009) Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 5, e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Disfani F. M., Hsu W. L., Mizianty M. J., Oldfield C. J., Xue B., Dunker A. K., Uversky V. N., Kurgan L. (2012) MoRFpred, a computational tool for sequence-based prediction and characterization of disorder-to-order transitioning binding sites in proteins. Bioinformatics 28, i75–i83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guan D., Altan-Bonnet N., Parrott A. M., Arrigo C. J., Li Q., Khaleduzzaman M., Li H., Lee C. G., Pe'ery T., Mathews M. B. (2008) Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol. Cell Biol. 28, 4629–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uversky V. N. (2002) Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11, 739–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S. L. (2012) Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 45. Patel C. V., Handy I., Goldsmith T., Patel R. C. (2000) PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275, 37993–37998 [DOI] [PubMed] [Google Scholar]
- 46. Daher A., Laraki G., Singh M., Melendez-Peña C. E., Bannwarth S., Peters A. H., Meurs E. F., Braun R. E., Patel R. C., Gatignol A. (2009) TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol. Cell Biol. 29, 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J. J., Anderson P., Kaufman R. J. (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933 [DOI] [PubMed] [Google Scholar]
- 48. Höck J., Weinmann L., Ender C., Rudelü S., Kremmer E., Raabe M., Urlaub H., Meister G. (2007) Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 8, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uversky V. N. (2010) Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D2 concept. Expert Rev. Proteomics 7, 543–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kedersha N., Ivanov P., Anderson P. (2013) Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ting N. S., Kao P. N., Chan D. W., Lintott L. G., Lees-Miller S. P. (1998) DNA-dependent protein kinase interacts with antigen receptor response element binding proteins NF90 and NF45. J. Biol. Chem. 273, 2136–2145 [DOI] [PubMed] [Google Scholar]
- 53. Arimoto K., Fukuda H., Imajoh-Ohmi S., Saito H., Takekawa M. (2008) Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 10, 1324–1332 [DOI] [PubMed] [Google Scholar]
- 54. Kim W. J., Back S. H., Kim V., Ryu I., Jang S. K. (2005) Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell Biol. 25, 2450–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kotani T., Yasuda K., Ota R., Yamashita M. (2013) Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J. Cell Biol. 202, 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shi L., Zhao G., Qiu D., Godfrey W. R., Vogel H., Rando T. A., Hu H., Kao P. N. (2005) NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 280, 18981–18989 [DOI] [PubMed] [Google Scholar]
- 57. Voronina E., Seydoux G., Sassone-Corsi P., Nagamori I. (2011) RNA granules in germ cells. Cold Spring Harbor Perspect. Biol. 3, a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Figueroa A., Cuadrado A., Fan J., Atasoy U., Muscat G. E., Muñoz-Canoves P., Gorospe M., Muñoz A. (2003) Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol. Cell Biol. 23, 4991–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mazroui R., Di Marco S., Kaufman R. J., Gallouzi I. E. (2007) Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 18, 2603–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Buchan J. R., Yoon J. H., Parker R. (2011) Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang J., Okabe K., Tani T., Funatsu T. (2011) Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J. Cell Sci. 124, 4087–4095 [DOI] [PubMed] [Google Scholar]