Abstract
Aerial green algae of Trentepohliaceae can form conspicuous free-living colonies, be parasites of plants or photobionts of lichen-forming ascomycetes. So far, their diversity in temperate regions is still poorly known as it has been mostly studied by phenotypic approaches only. We present new insights in the phylogenetic relationships of lichenized representatives from temperate and Mediterranean parts of Europe by analysis of 18S rRNA and rbcL gene fragments, and nuclear ITS sequence data. For this purpose we isolated the trentepohlialean photobionts from lichens representing different genera. Algal cultures from lichenized and free-living Trentepohliaceae were used to design new primers for amplification of the marker loci. We constructed a phylogenetic hypothesis to reveal the phylogenetic placements of lichenized lineages with 18S rRNA and rbcL sequences. ITS variation among the clades was substantial and did not allow including them in the general phylogenetic assessment, yet ITS appears to be a promising marker for DNA-barcoding approaches. Specific algae were found in particular lichen but the overall diversity of photobionts was limited. The multilocus tree does not support the current morphological classification of genera in Trentepohliaceae, suggesting that morphology is more variable than previously thought in this group of algae.
Electronic supplementary material
The online version of this article (doi:10.1007/s13199-014-0285-z) contains supplementary material, which is available to authorized users.
Keywords: ITS region, Lichen symbioses, Photobionts, Phylogeny, Temperate regions, Trentepohliaceae
Introduction
The Trentepohliaceae are a widespread family of aero-terrestrial green algae which differs from other green algae in terms of their reproductive structures, phragmoplast-mediated cytokinesis, the lack of pyrenoids in the chloroplast and other characters (Rindi et al. 2009). In particular, the phragmoplasts are otherwise only known from the Charophyceae and from land plants (Chapman et al. 2001). Trentepohlia colonies and of their allied genera are frequent on rocks, buildings, tree barks, leaves, stems, and fruits (Printz 1939; Chapman 1984; López-Bautista et al. 2002; Chapman and Waters 2004; López-Bautista et al. 2006; Gaylarde et al. 2006; Nelson 2008). Among the aero-terrestrial algae, the Trentepohliaceae are particularly well-adapted to habitats with high levels of air humidity. These habitats are also frequently colonized by species of lichen-forming fungi which form symbioses with trentepohlialean algae. Nelsen et al. (2011) estimated that approximately 23 % of all lichen-forming fungi are associated with trentepohlialean photobionts. The highest species diversity of Trentepohliaceae is so far reported from tropical to subtropical regions (Thompson and Wujek 1997; Chapman 1984; Chapman and Waters 2004; López-Bautista et al. 2002; Rindi et al. 2010), but it is still unclear how many of these species are also symbionts of lichens.
The free-living representatives of the type genus Trentepohlia consists of small shrubby or cushion-like thalli composed of branched filaments. As the cells produce a vast amount of carotenoid pigments in light-exposed locations, the colonies are recognizable from a distance by their orange color. In contrast, lichenized forms of Trentepohliaceae do not develop well-formed filaments as the fungal hyphae control the photobionts, but instead form only short concatenations of cells (Honegger 1998; Chapman and Waters 2004). In lichen thalli, the trentepohlialean photobionts are usually sheltered below an upper fungal layer which protects the algal cells against detrimental UV radiation. Consequently, in the lichenized state, the production of carotenoids is reduced and the green chloroplasts become more distinct. As a result of these alterations, and because sexual stages are usually suppressed, the identification of Trentepohliaceae on the basis of morphological features is difficult.
Molecular analyses have facilitated the classification of lichenized green algae. So far, this has been mostly achieved with coccal photobionts of the family Trebouxiophyceae (e.g. Cordeiro et al. 2005; del Campo et al. 2010; Bock et al. 2011; Ruprecht et al. 2012). Few studies of trentepohlialean photobionts have focused on analysis of the fungal selectivity. So far, partial small subunit rRNA genes and the large subunit of the ribulose-bisphosphate carboxylase gene (rbcL) have been used for phylogenetic placement of free-living trentepohlialean algae and in Viridiplantae, especially in the class Ulvophyceae (López-Bautista and Chapman 2003; López-Bautista et al. 2006; Rindi et al. 2009). Moreover, the relationships within the order Trentepohliales and also between the genera Trentepohlia and Printzina have been analyzed using these two loci. The phylogenetic trees generated with the 18S rRNA gene were poorly resolved at the genus and species level. However, combined analysis with the rbcL gene resulted in higher support values for various branches (Rindi et al. 2009). Despite these results, the assignment of named trentepohlialean species to terminal branches remains problematic. These findings were taken as evidence for the insufficiency of the current classification within Trentepohliaceae. Nelsen et al. (2011) discussed a similar situation in their initial molecular study of Trentepohliaceae associated with lichen-forming fungi and used the rbcL gene. Their study mainly included tropical lichens and their topology of the phylogenetic hypothesis is not well supported for a majority of the clades. Before it is possible to complete a revision of classification and have a better knowledge of relationships in general, the phylogeny of Trentepohliaceae requires further support with the inclusion of additional markers.
At a low taxonomic level the ITS region of the ribosomal gene cluster has been used in diverse organismal groups. It was therefore our plan to include this locus in studies of Trentepohliaceae. In the context of a larger survey of trentepohlialean lichen symbionts, we were interested in the diversity of trentepohlialean photobionts in lichens from temperate and Mediterranean regions in Europe. Initially, we used published algal-specific-primers and found that the published primers by Nelsen et al. (2011) can amplify both trentepohlialean and trebouxiophycean algae. This represented a problem of a background signal when epithalline algae are abundant in thallus material used for total DNA extractions. Our goal was therefore also to design Trentepohliaceae-specific primers that avoid contamination problems. Using phylogenetic analyses, we then assessed the relationships of trentepohlialean photobionts in selected lichens collected from temperate regions and with free-living representatives.
Material and methods
Sampling
Thirty-eight lichens, representing different genera, were collected from various sites in temperate and Mediterranean habitats (Table 1). All voucher specimens, except for Dimerella pineti (deposited at the University of Graz, Institute of Plant Science, herbarium GZU 5650), are in the herbarium of E. Stocker-Wörgötter, which is publicly accessible through the Department of Organismic Biology at the University of Salzburg.
Table 1.
Photobiont strain descriptions of different lichen samples used in this study for phylogenetic analyses
| Lichen-taxa | Trentepohliacea-taxa | Location | GenBank accession numbers | ||
|---|---|---|---|---|---|
| SSU | ITS | rbcL | |||
| Acrocordia gemmata (156) | Trentepohlia sp. | United Kingdom, Scotland, Loch Carron and West Monar, Attadale (N57°23′45.80″ W5°27′1.76″) | JQ618000 | JQ617977 | JQ617931 |
| Arthonia cinnabarina (137) | Printzina cf. lagenifera | United Kingdom, Scotland, Isle of Skye, Kinloch & Kyleakin (N57°10′55.10″ W 5°48′9.77″) | JQ618002 | JQ617961 | JQ617944 |
| Arthonia cinnabarina (12) | Printzina lagenifera | France, Forêt du Cranou (N48°18′57.00″ W 4° 5′42.00″) | - | JQ617952 | JQ617932 |
| Arthonia radiata (155) | Trentepohlia sp. | United Kingdom, Scotland, Loch Carron and West Monar, Attadale (N57°23′45.80″ W5°27′1.76″) | JQ617998 | JQ617979 | JQ617929 |
| Arthothelium ruanum (22) | Printzina cf. lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13° 6′00.52″) | JQ618017 | JQ617960 | JQ617914 |
| Cystocoleus ebeneus (1) | Trentepohlia sp. | Austria, Salzburg, Krimmler waterfalls (N 47°12′41.25″ E12°10′6.13″) | JQ617982 | JQ617945 | JQ617917 |
| Cystocoleus ebeneus (2) | Trentepohlia sp. | Austria, Salzburg, Krimmler waterfalls (N 47°12′41.25″ E12°10′6.13″) | JQ617983 | JQ617946 | - |
| Dimerella pineti (65) | Printzina sp. | Austria, Styria, Grazer uplands, Dürrgraben (N47°07′15″ E015°28′05″) | JQ618011 | JQ617950 | - |
| Graphis scripta (13) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JF727812 | JF727811 | JF727813 |
| Graphis scripta (14) | Printzina lagenifera | Italy, Friaul, Sella Nevea Pass after pass altitude (N46°23′31.13″ E13°28′29.21″) | JF727815 | JF727814 | JF727816 |
| Graphis scripta (19) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618004 | JQ617959 | JQ617940 |
| Graphis scripta (20) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618005 | JQ617956 | JQ617938 |
| Graphis scripta (21) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618008 | JQ617958 | JQ617943 |
| Graphis scripta (27) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618007 | JQ617957 | JQ617939 |
| Graphis scripta (28) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618012 | JQ617951 | JQ617915 |
| Graphis scripta (29) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618009 | JQ617953 | JQ617937 |
| Graphis scripta (30) | Printzina lagenifera | Austria, Salzburg, Bluntautal (N47°33′59.76″ E13°6′00.52″) | JQ618006 | JQ617955 | JQ617941 |
| Graphis scripta (143) | Printzina lagenifera | United Kingdom, Scotland, Isle of Skye, Kinloch & Kyleakin (N57°10′55.10″ W5°48′9.77″) | JQ618003 | JQ617954 | JQ617942 |
| Gyalecta jenensis (25) | Trentepohlia aurea | Austria, Salzburg, Paß Lueg near Golling (N47°34′50.23″ E13°11′6.43″) | JQ618016 | JQ617948 | JQ617916 |
| Mycoporum sparsellum (136) | Trentepohlia sp. | United Kingdom, Scotland, Isle of Skye, Kinloch & Kyleakin (N57°10′55.10″ W5°48′9.77′) | JQ617997 | JQ617978 | JQ617930 |
| Opegrapha atra (149) | Trentepohlia sp. | United Kingdom, Scotland, Kyle of Lochalsh, Duirinish (N 57°19′9.00″ W5°40′13.06″) | JQ618014 | JQ617963 | JQ617933 |
| Opegrapha atra (151) | Trentepohlia sp. | United Kingdom, Scotland, Kyle of Lochalsh, Duirinish (N 57°19′9.00″ W5°40′13.06″) | JQ618013 | JQ617947 | JQ617934 |
| Pyrenula laevigata (26) | Printzina sp. | Austria, St. Bartholomä, Königssee (N47°32′45.46″ E12°58′11.61″) | JQ618010 | JQ617949 | JQ617913 |
| Pyrenula laevigata (135) | Printzina sp. | United Kingdom, Scotland, Isle of Skye, Kinloch & Kyleakin (N57°10′55.10″ W5°48′9.77″) | JQ618015 | JQ617962 | JQ617935 |
| Roccella decipiens (7) | Trentepohlia sp. | Spain, Gran Canaria, Jardin Botánico Canario Viera y Clavijo (N28°3′55.73″ W15°27′36.60″) | JQ617992 | JQ617967 | JQ617920 |
| Roccella galapagoensis (8) | Trentepohlia sp. | Spain, Gran Canaria, Jardin Botánico Canario Viera y Clavijo (N28°3′55.73″ W5°27′36.60″) | JQ617991 | JQ617966 | JQ617921 |
| Roccella linearis (5) | Trentepohlia sp. | Spain, Gran Canaria, Andés Verdes (N27°50′20.11″ W15°33′45.77″) | JQ617987 | JQ617968 | JQ617924 |
| Roccella lirellina (10) | Trentepohlia sp. | Spain, Gran Canaria, Jardin Botánico Canario Viera y Clavijo (N28°3′55.73″ W15°27′36.60″) | JQ617984 | JQ617964 | JQ617918 |
| Roccella maderensis (5) | Trentepohlia sp. | Portugal, Azoren, Fortress of São João Baptista (N38°39′5.19″ W27°13′36.71″) | JQ617996 | JQ617971 | JQ617925 |
| Roccella cf. montagnei (9) | Trentepohlia sp. | Spain, Gran Canaria, Jardin Botánico Canario Viera y Clavijo (N28°3′55.73″ W15°27′36.60″) | JQ617994 | JQ617976 | JQ617923 |
| Roccella phycopsis (7) | Trentepohlia sp. | Malta, Ggantija Tempel Gozo (N36°2′50.08″ E14°16′8.77″) | JQ617988 | JQ617975 | - |
| Roccella phycopsis (6) | Trentepohlia sp. | Spain, Gran Canaria, Andés Verdes (N27°50′20.11″ W15°33′45.77″) | JQ617986 | JQ617969 | JQ617922 |
| Roccella tinctoria (2) | Trentepohlia sp. | Spain, Teneriffa, Puertito de los Silos (N28°22′29.19″ W16°48′30.98″) | JQ617993 | JQ617972 | JQ617927 |
| Roccella tinctoria (4) | Trentepohlia sp. | Spain, Teneriffa, Puertito de los Silos (N28°22′29.19″ W16°48′30.98″) | JQ617985 | JQ617970 | - |
| Roccella tinctoria (6) | Trentepohlia sp. | Portugal, Azoren, São Sebastião (N38°39′46.53″ W27° 5′35.97″) | JQ617989 | JQ617974 | JQ617926 |
| Roccella tinctoria (11) | Trentepohlia sp. | Spain, Gran Canaria, Jardin Botánico Canario Viera y Clavijo (N28°3′55.73″ W15°27′36.60″) | JQ617990 | JQ617965 | JQ617919 |
| Roccella tuberculata (8) | Trentepohlia sp. | Portugal, Azoren, Fortress of São João Baptista (N38°39′5.19″ W27°13′36.71″) | JQ617995 | JQ617973 | - |
| Thelotrema lepadinum (144) | Trentepohlia sp. | United Kingdom, Scotland, Isle of Skye, Kinloch & Kyleakin (N57°10′55.10″ W5°48′9.77″) | JQ617999 | JQ617980 | JQ617928 |
Culture experiments
Trentepohlialean photobionts of selected lichens (see Online Resource 1) were isolated according to the “Yamamoto-method” (Yamamoto 1990) with the modifications specified in Stocker-Wörgötter (2002). Fragments of lichen thalli (of 2–3 mm size) were washed in sterile bi-distilled water for 15 min, then one drop of Tween 80 was added to the water and the fragments were washed again for 10 min. After the transfer to fresh bi-distilled water, the fragments were washed for another 20 min by agitating in water using a magnetic stirrer. The fragments were then gently homogenized in several drops of sterile water with an autoclaved mortar and pestle. This suspension was filtered first through a sieve with 500 μm mesh-size and then through a sieve with 150 μm mesh-size. Individual lichen pieces (around 150 μm in size) were picked by a sterile bamboo stick and transferred to test tubes with slanted solid nutrient media under a stereomicroscope.
The isolates were grown on three different nutrient media for 5–6 months under a changing light dark regime of 14 h at 20 °C and 10 h at 15 C° with a light intensity of 100 μE m−2 s−1. We used modified Bold’s Basal Medium (BBM; Deason and Bold 1960), Woods Hole MBL (WHM; Nichols 1973), and optimal Haematococcus medium (OHM; Fábregas et al. 2000). After the Trentepohliaceae-colonies reached a size of approximately 5 mm in diameter, some algal filaments were removed with a sterile inoculation needle for subculture in sterilized 50 ml-Erlenmeyer flasks with liquid modified BBM, WHM or OHM, and plugged with sterile cotton. Flask necks were covered with aluminium foil to avoid contaminations. The isolates were further incubated for several months in a culture chamber under the same conditions as mentioned and specified above.
Molecular methods
Trentepohlialean filaments of the subcultures were transferred to Eppendorf tubes using a sterile inoculation needle in a clean bench and afterwards dried using the speed-vac UNIVAPO 150 ECH (UniEquip, Planegg, Germany). The dried filaments were frozen with liquid nitrogen to facilitate the grinding procedure with the grinding mill MM301 (Retsch GmbH, Haan, Germany). DNA was extracted using a modified CTAB-method (Doyle and Doyle 1987). The modifications included the addition of 400 μl CTAB buffer, which contained 1 % Polyvinylpyrrolidone (PVP-40; Sigma Aldrich, Vienna, Austria) instead of 2-mercaptoethanol, and 2 μl RNase A (10 mg/ml; Fermentas, Vienna, Austria) directly onto the grinded material. Additionally, the samples were centrifuged at 13,300 rpm for 5 min after the preheating procedure and the chloroform isoamyl alcohol (CI) extraction, the CI extraction was carried out twice and step 7 of the original CTAB-method was omitted. Furthermore, the pellet was washed twice with 500 μl 70 % ethanol at 12,000 rpm for 2 min, afterwards dried at 50 °C for 5 min and dissolved in appropriate volume of double-distilled water.
The PCR was initially accomplished with universal primers and parameters as used in Rindi et al. (2009) for the 18S rRNA and rbcL genes. For amplification of the ITS region, we initially used the primers ITS5 (White et al. 1990) and ITS4 (White et al. 1990). PCR-products were cleaned with QIAquick PCR product purification Kit (Qiagen, Hilden, Germany) and sequenced by Macrogen Inc. (Korea). The obtained sequences were classified using NCBI Blast searches, and then aligned with sequences retrieved from the NCBI GenBank, representing both diverse green algal groups (e.g. Chlorophyceae, Trebouxiophyceae, Trentepohliaceae, etc.) and lichenized fungi (e.g. Arthoniaceae, Graphidaceae, etc.) with Geneious (Drummond et al. 2011). After the inspection by eye, the alignments of each of the three markers (18S rRNA and rbcL genes, nuclear ITS) were used to find appropriate regions for the design of Trentepohliaceae-specific primers.
To apply and test the newly designed primers, total DNA was extracted from thallus fragments by the modified CTAB-method as mentioned above, and using the Plant Mini DNA-isolation Kit (Qiagen). In addition, DNA was also extracted of four samples of cultured, free-living Trentepohliaceae from the culture collection of algae in Göttingen, Germany (strains SAG 73.90, SAG 25.83, SAG 20.94 and SAG 483–1), of the cultured free-living Trentepohlia iolithus strain ASIB505 obtained from the culture collection of Prof. Dr. Gärtner (Innsbruck, Austria) and single colonies of free-living Printzina cf. lagenifera collected from Almbachklamm, Salzburg, Austria (Acc.no. JX675739). The amplification of the 18S rRNA gene was performed with various combinations of primers at an optimal annealing temperature of 57 °C (see Table 2). Nested PCR was performed occasionally, when starting material was scant. The ITS region were amplified with similar conditions as used for the 18S-amplification, but with an annealing temperature at 56 °C.
Table 2.
Primer information containing primer name, amplified gene region, primer sequence, source of primers and PCR-product size
| Combination | Primer name | Gene | Primer sequence | Source of primers | Product size (bp) |
|---|---|---|---|---|---|
| 1 | Tre18S_N1_for | 18S rRNA | 5′CCC GAC CTT CGG TGA ATC3′ | this lab | ~ 820 |
| CHtrente1.rev | 18S rRNA | 3′CCA CCT CCG ATC CCT AGT5′ | this lab | ||
| 2 (nested) | Tre18S_N2_for | 18S rRNA | 5′TAG GGT AGT GGC CTA CCG3′ | this lab | ~ 700 |
| CHtrente0.rev | 18S rRNA | 3′GTC GAG ACT ACG ACG GT5′ | this lab | ||
| 3 | Tre18S_N5a_for | 18S rRNA | 5′TAG CAT GGG ATG ACA CGA TAG GA3′ | this lab | ~ 760 |
| CHtrente2.rev | 18S rRNA | 3′ACA AAG CTC TAG CCC CAT CA5′ | this lab | ||
| 4 (nested) | Tre18S_N5_for | 18S rRNA | 5′GGA TGA CAC GAT AGG ACT TCG3′ | this lab | ~ 750 |
| CHtrente2.rev | 18S rRNA | 3′ACA AAG CTC TAG CCC CAT CA5′ | this lab | ||
| 5 | CHtrente1.for | ITS | 5′ACT AGG GAT CGG AGG TGG3′ | this lab | ~ 700 |
| ITS4 | ITS | 3′TCC TCC GCT TAT TGA TAT GC5′ | White et al. (1990) | ||
| 6 | CHtrente2.for | ITS | 5′TGA TGG GGC TAG AGC TTT GT3′ | this lab | ~ 900 |
| ITS4 | ITS | 3′TCC TCC GCT TAT TGA TAT GC5′ | White et al. (1990) |
For rbcL-amplification, PCR was performed first with TrerbcL_mos_for (5′ GAA GCW ATT CCR GGA GAA G 3′) and TrerbcL_mos_rev (3′ CAT CCA TTC TTG AGW AAA GAA TAC 5′), and for semi-nested PCR with the internal reverse primer TrerbcL_sel_rev (3′ GAT AGT CGT GCA TRA YAA TTG G 5′); all cycles using a 50 °C annealing temperature.
Sequence alignment and phylogenetic analyses
The DNA sequences of the lichenized Trentepohliaceae and the free-living trentepohlialean algae were aligned using the program Geneious. We used two representatives of the order Ulvales as outgroup of the 18S rRNA and rbcL genes. Nucleotide sequences of all samples used in this study have been submitted to NCBI GenBank (Accession numbers of the photobionts see Table 1).
The alignments in Fasta-format were converted into Nexus-files by the web-portal Alter alignment (Glez-Peña et al. 2010). Each dataset were analyzed with the program jModelTest 2.1.1 using Akaike Information Criterion (AIC) scores (Darriba et al. 2012; Guindon and Gascuel 2003) to find GTR + G + I as the optimal substitution model for the phylogenetic analyses. The analyses of each single dataset were performed by Cipres Science Gateway (Miller et al. 2010) with MrBayes v.3.1.2. (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). Analyses of individual ITS-datasets for clades R with Ld1, clade Gs, clade GPD, clades Ta with Td, and clades Ce with Ld2, employed different substitution models and settings, which are described in Online Resource 1. For the analysis of the rbcL-sequence alignment, parameters of the GTR + G + I model were adjusted for each codon position. For each of the analyses, 3 000 000 generations were generated, every 1000th tree was sampled and the initial 750 trees were discarded as burn-in. The combined analysis of the marker regions 18S rRNA and rbcL contained the same 52 sequences (see Online Resource 2) as used for the separate analyses, and was computed in BEAST 1.7.4 (Drummond and Rambaut 2007) with the appropriate settings of each locus specified for the separate calculations. Three parallel analyses were run for 6 000 000 chains, which were performed under a strict clock, and every 1000th tree was sampled. The three obtained log-files were controlled with the program Tracer v1.5 (Rambaut and Drummond 2007). The tree-files with branch lengths in units of substitutions of each run were combined with a burn-in of 1200 to one common tree-file using LogCombiner1.7.4 in BEAST. Finally, this file was used to summarize the sampled trees to a maximum clade credibility tree with the setting to reflect the posterior median node heights for the clades by TreeAnnotator1.7.4 in BEAST.
The datasets were also analyzed by maximum parsimony with PAUP* (Swofford 2003). The combined Paup-analysis of 18S rRNA and rbcL genes was computed under heuristic search, 100 random addition sequence replicates with one trees held at each step, tree bisection reconnection (TBR) and no more than five trees per replicate (treescore ≥1) were saved. Bootstrapping (Felsenstein 1985) was performed with 1,000 replicates. All trees were illustrated using the program FigTree v1.3.1 (Rambaut 2006–2009).
Results
By using several loci we wanted to increase the phylogenetic resolution of Trentepohliales. So far, this has been achieved with concatenated 18S rRNA and rbcL sequences, which increased the support of clades which did not resolve well in separate analyses of individual loci (see Online Resource 3). It was not possible to include the newly generated ITS sequences in the analysis of the whole dataset due to the variation being too high for reliable alignments and also the sample size differed between the data sets. We therefore decided to use ITS to check for congruence with the 18S-rbcL phylogeny and for the resolution of terminal branches (see below).
The 18S-rbcL phylogeny (Fig. 1) reveals several major clades with high support values (MPB 96 %/1.00 PP and MPB 99 %/1.00 PP). Clade R is closely related to strains representing Printzina bossae, Trentepohlia annulata (1.00 PP), and T. iolithus (MPB 75 %/1.00 PP). The sister clade to this group is clade Ld1 (MPB 100 %/1.00), which is not fully supported by the separate analysis of the ITS-dataset, according to which T. annulata groups together with clade Ld1. An incongruence between the overall and ITS- phylogeny can also be found in clade Gs. The photobiont of Pyrenula laevigata (135) JQ617962 is not included in the Graphis scripta-group of the ITS-tree calculated with the alignment of all ITS-sequences (data not presented); in contrast, this strain shows a close relationship to the trentepohlialean alga lichenized with Arthothelium ruanum (22) JQ617960. The free-living Printzina lagenifera forms a sister species to the monophyletic clade Gs, on the basis of the combined analysis. Clade GPD is composed of the photobionts from Dimerella pineti (65), Graphis scripta (28), Pyrenula laevigata (26), and the free-living alga P. cf. lagenifera (MPB 90 %/1.00) and is closely related to the photobiont of Arthothelium ruanum (22), the free-living Trentepohlia arborum and clade Gs. The remaining clades (OaA, Td, Ta, CeT and Ld2) of the maximum clade credibility tree consist of the same lichenized algal clusters, as shown in the Bayesian analysis using the rbcL-dataset. Clade Td represents a close relationship to Ta with a high posterior probability of 0.99, as well as clade CeT to clade Ld2 (1.00 PP). Despite the support of the CeT-Ld2 relation, the separate ITS-consensus tree reveals only a high supported relation within the algal strains of Cystocoleus ebeneus No.1 and 2 (1.00 PP), and uncertain associations from the photobionts of Mycoporum sparsellum (136) and Acrocordia gemmata (156). Trentepohlia umbrina (free-living) forms an individual branch which agrees with the 18S-rbcL-analysis.
Fig. 1.
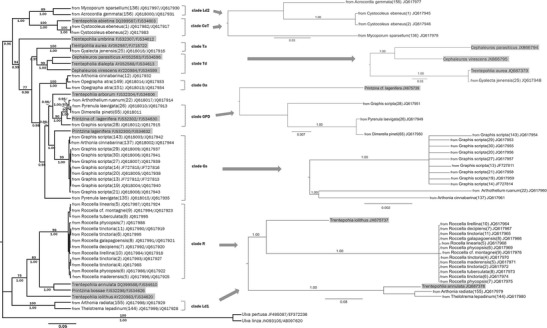
Mid-point rooted maximum clade credibility tree of lichenized and free-living Trentepohliaceae calculated with the concatenated datasets of marker regions 18S rRNA and rbcL using Bayesian MCMC analysis implemented in BEAST. Branches with bootstrap support (MPB) ≥70 % and posterior probabilities (PP) ≥0.95, which reflect the posterior median node heights for the clades, were considered as strongly supported. MPB values were illustrated above the branches, whereas PP values were shown below. The tree was rooted using two outgroup sequences of the genus Ulva. Single consensus trees with posterior probability values (PP) ≥0.95 of different clades using the ITS rDNA are shown on the right side of the combined tree. The bars specify the substitutions per site. The trentepohlialean algae in free-living stage are grey-shaded and were used as reference for the identification of the photobiont strains
Discussion
Previous phylogenetic analyses of trentepohlialean algae revealed their position to be in the Ulvophyceae in Chlorophyta (López-Bautista and Chapman 2003; Leliaert et al. 2012). However, more detailed molecular analyses within the order Trentepohliales showed that the taxonomy was not clearly settled, especially in the case of the genera Printzina and Trentepohlia (Rindi et al. 2009). As expected, our initial separate, single-locus analyses of lichenized and free-living Trentepohliaceae were poorly supported at internal branches, and did not resolve relationships within the family. The 18S-rbcL tree received higher support, which agrees with the results of Rindi et al. (2009) for free-living Trentepohliaceae. They recommended the addition of another molecular dataset, such as ITS or D1-D2 28S rRNA, for a better resolution within some problematic taxa, including Printzina lagenifera and Trentepohlia arborum (Rindi et al. 2009). Therefore, it appeared reasonable to include ITS-sequences in the analyses because this locus was informative at a lower taxonomic level in other green algal orders (Coleman and Mai 1997; Blaha et al. 2006; Van Der Strate et al. 2008). ITS, in particular the ITS2 region, was also considered to be promising for the DNA-barcoding of green algae, besides the suggested chloroplast genes rbcL and tufA, the LSU of rDNA and the mitochondrial COI gene (Leliaert et al. 2009; Grube and Muggia 2010; Hall et al. 2010; Saunders and Kucera 2010; Škaloud and Peksa 2010; Fucikova et al. 2011; Saunders and McDevit 2012). Our analyses actually support the use of ITS for barcoding in Trentepohliaceae. Variation between species appears to be rather high while there is limited variation within the branches we consider as species. In lichens, DNA barcoding approaches could be promising for an assessment of algal selectivity and uniformity of photobionts in individual lichen thalli.
All phylogenetic analyses in this study showed that Roccella species (clade R) only associate with a single and distinct trentepohlialean species. The selectivity of this species might correlate with the coastal habitat of the genus Roccella. The collection sites comprise various islands which were distantly located about 3,700 km from one another. The roccelloid lichens so far studied contain an undescribed trentepohlialean species as photobiont, which has not yet been found in association with other lichenized ascomycetes. A more detailed determination of this photobiont was not possible without morphological examination of cultured material. It remains to be shown if this algal species is also characterized by a higher salt tolerance compared with the other cultured Trentepohliaceae. Printzina bossae, Trentepohlia annulata, and T. iolithus are sister species of the algal partner from the studied Roccella species, but apparently not conspecific with it. The phylogenetically related strains of clade R- photobionts prefer completely different habitats to Roccella. Thus, Printzina bossae, was collected from the bark of unidentified trees located on an island of Barro Colorado and from a public park in Gamboa (Panama) (Rindi et al. 2008). Other specimens were collected from the bark of an oil palm (French Guiana), bark of Cryptomeria sp. (Azores), as well as the bark of an unidentified tree in Florida (U.S.A) (Rindi and López-Bautista 2008; Nelsen et al. 2011).
The free-living alga Trentepohlia annulata, also related to clade R, has been reported from vastly distant localities such as Trebitsch (Central Europe) and French Guiana (South America), although the affiliation to T. annulata of the specimen from French Guiana has not been 100 % confirmed (Rindi et al. 2009). The T. annulata, found in Trebitsch, was growing on the roots and on a cross-section of a conifer trunk (Prat 1914). This algal species was also identified in the apothecia and as an epiphyte on the lichen Micarea misella (Voytsekhovich et al. 2011), which normally associates with coccoid algal photobionts. Finally, the cultured strain T. annulata No. 20.94 of the algal collection SAG originated from Czechoslovakia.
The cosmopolitan Trentepohlia iolithus, also related to clade R, forms red coatings on rocky cliffs of the British Isles, tree bark, periodically submerged rocks along calcareous streams (John 2002) and also grow on limestone used to face buildings, old concrete and cement walls in unpolluted urban areas (Rindi and Guiry 2002; Rindi et al. 2003). The algal symbiotic partners of Arthonia radiata and Thelotrema lepadinum (clade Ld1) were specified as unknown Trentepohlia-species based on relationships with three free-living Trentepohliaceae grouped in clade R and with the NCBI Blast results.
The maximum clade credibility tree computed in BEAST clearly revealed (support values of MPB 100 % and PP 1.00) that the lichenized fungus Cystocoleus ebeneus associates with trentepohlialean representatives closely related to Trentepohlia abietina (clade CeT). Free-living T. abietina has so far been reported from temperate and tropical regions throughout the world (Wildeman 1900; Jose and Chowdary 1980; Tracanna 1989; Ettl and Gärtner 1995; John 2002; Rindi et al. 2005; Rindi et al. 2006). In the free-living stage, this species is restricted to bark as a substrate where it forms distinct yellow-orange patches (Rindi et al. 2005; Rindi et al. 2008). The photobionts of Acrocordia gemmata and Mycoporum sparsellum (clade Ld2) formed a sister clade to the Trentepohlia abietina-strains and are as yet unnamed trentepohlialean lineages.
According to sequence data, the photobiont strains associated with ascomycetes of the lichen species Graphis scripta (clade Gs) were identified as Printzina lagenifera, and this is confirmed based on morphological observations of cultured strains (Hametner et al. 2014). This mycobiont does not seem to be specific for a single trentepohlialean clade and has the ability to switch photobionts. For example, specimen Graphis scripta (28), which was collected at the same locality in Austria as the other specimens from Austria, was found to associate with Printzina cf. lagenifera. This alga is also found as the photobiont of Arthothelium ruanum, Dimerella pineti and Pyrenula laevigata (clade GPD). Nelsen et al. (2011) also discovered two specimens of the genus Graphis in symbiosis with the alga P. cf. lagenifera.
The lichen Arthonia cinnabarina is another example of a species that can switch algae. It can associate with the algal Printzina lagenifera (clade Gs), as well as another lineage of the genus Printzina which was found in Arthonia cinnabarina (12), and which we assigned to this genus on the basis of the NCBI Blast searches and concatenated phylogenetic analyses with all three loci (data not presented). Based on the 18S-rbcL analysis of this study, the photobiont of A. cinnabarina (12) clustered together with algal strains of the lichen Opegrapha atra which were determined as Trentepohlia sp. (clade Oa). This grouping appeared due to the fact that only the rbcL-sequence of the Arthonia-cinnabarina- photobiont was involved as a substantial factor in the combined 18S-rbcL analysis.
The free-living P. lagenifera seems not to be very specific for substrata (e.g. occurring on the barks of trees, cement, limestone, etc.) and has a very wide distribution in both temperate and tropical regions (Rindi and Guiry 2002; Rindi et al. 2006; Rindi et al. 2008). All lichen species in our study were collected in Europe, which led to the conclusion that in this region Printzina lagenifera is a common algal species and tends to be an adequate photobiont for several lichens.
Finally, we found that Trentepohlia aurea (Fig. 2a and b) is the typical photobiont of Gyalecta jenensis (clade Ta). This has previously been suspected as free-living stages of this species are usually found adjacent to the thalli or growing out from the upper surface.
Fig. 2.
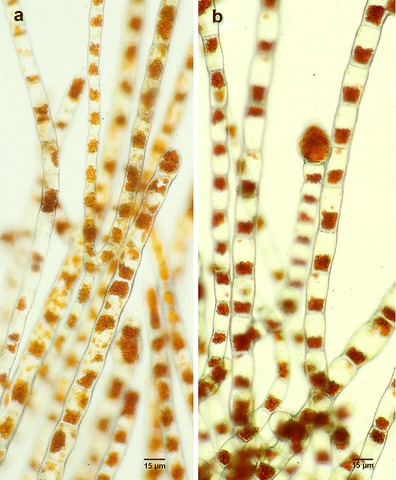
Isolated trentepohlialean photobiont from the lichen Gyalecta jenensis compared to the algal strain Trentepohlia aurea SAG483-1 (free-living) originated from the culture collection of Göttingen. a Algal filaments of photobiont T. aurea grown in liquid Woods Hole MBL medium; b Algal filaments of free-living T. aurea grown on solid Woods Hole MBL medium
This study included different lichen genera and collection sites in temperate Europe and revealed that the species diversity of Trentepohliaceae in lichens from this region appears to be limited. Low photobiont variation was also shown previously for trentepohlialean lichens from Japan using culture experiments (Nakano 1988). On the other hand, it appears that at least some bark-inhabiting lichens may switch their trentepohlialean photobionts (Nelsen et al. 2011). This parallels the situation in lichens with coccoid green algae, where selectivity patterns have been found to vary in different lineages (Yahr et al. 2006). The classification of genera belonging to Trentepohlia, based on phenotypic characters, appears to be complicated by morphological polymorphism and it needs to be clarified as to whether widely distributed conspecific strains actually belong to the same lineage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 16 kb)
(DOCX 145 kb)
(DOCX 936 kb)
Acknowledgments
The study was generously supported by the Austrian Science Foundation, FWF grant No. 20887 to EST-W. We thank Marcela Cáceres (Brazil) and Andre Aptroot (Netherlands) for collecting free-living algal specimens. Special thanks are due to Georg Gärtner (Innsbruck, Austria) for submitting trentepohlialean filaments in culture. We are also grateful to Georg Brunauer and Matthias Affenzeller (Salzburg, Austria) for technical support.
References
- Blaha J, Baloch E, Grube M. High photobiont diversity associated with the euryoecious lichen-forming ascomycete Lecanora rupicola (Lecanoraceae, Ascomycota) Biol J Linn Soc. 2006;88:283–293. doi: 10.1111/j.1095-8312.2006.00640.x. [DOI] [Google Scholar]
- Bock C, Krienitz L, Pröschold T. Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea. 2011;11:293–312. [Google Scholar]
- Chapman RL. An assessment of the current state of our knowledge of the Trentepohliaceae. In: Irvine D, John D, editors. Systematics of the green algae. London: Academic Press; 1984. pp. 223–250. [Google Scholar]
- Chapman RL, Waters DA (2004) Lichenization of the Trentepohliales-complex algae and odd relationship. In: Seckbach J (ed) Symbiosis, Volume 4. Kluwer Academic Publishers, Netherlands, pp 361–369
- Chapman RL, Borkhsenious O, Brown RC, Henk MC, Waters DA. Phragmoplast-mediated cytokinesis in Trentepohlia: results of TEM and immunofluorescence cytochemistry. Int J Syst Evol Microbiol. 2001;51:759–765. doi: 10.1099/00207713-51-3-759. [DOI] [PubMed] [Google Scholar]
- Coleman AW, Mai JC. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J Mol Evol. 1997;45:168–177. doi: 10.1007/PL00006217. [DOI] [PubMed] [Google Scholar]
- Cordeiro LMC, Reis RA, Stocker-Wörgötter E, Cruz LM, Grube M, Jacomin M. Molecular studies of photobionts of selected lichens from the coastal vegetation of Brazil. FEMS Microbiol Ecol. 2005;54:381–390. doi: 10.1016/j.femsec.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deason TR, Bold HC. Phycological studies. I. Exploratory studies of Texas soil algae. Austin: University of Texas Publications; 1960. [Google Scholar]
- del Campo EM, Casano LM, Gasulla F, Barreno E. Suitability of chloroplast LSU rDNA and its diverse group I introns for species recognition and phylogenetic analyses of lichen-forming Trebouxia algae. Mol Phylogenet Evol. 2010;54:437–444. doi: 10.1016/j.ympev.2009.10.024. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011) Geneious v5.4. http://www.geneious.com [DOI] [PMC free article] [PubMed]
- Ettl H, Gärtner G. Syllabus der Boden-Luft- und Flechtenalgen. New York: Gustav Fischer Verlag, Stuttgart, Jena; 1995. [Google Scholar]
- Fábregas J, Dominguez A, Regueiro M, Maseda A, Otero A. Optimization of culture medium for the continuous cultivation of the microalga Haematococcus pluvialis. Appl Microbiol Biotechnol. 2000;53:530–535. doi: 10.1007/s002530051652. [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed]
- Fucikova K, Rada JC, Lukesova A, Lewis LA. Cryptic diversity within the genus Pseudomuriella Hanagata (Chlorophyta, Chlorophyceae, Sphaeropleales) assessed using four barcode markers. Nova Hedwigia. 2011;93:29–46. doi: 10.1127/0029-5035/2011/0093-0029. [DOI] [Google Scholar]
- Gaylarde P, Englert G, Ortega-Morales O, Gaylarde C. Lichen-like colonies of pure Trentepohlia of limestone monuments. Int Biodeterioration Biodegrad. 2006;58(3–4):119–123. doi: 10.1016/j.ibiod.2006.05.005. [DOI] [Google Scholar]
- Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D (2010) ALTER: program-oriented format conversion of DNA and protein alignments. Nucleic Acids Res. doi:10.1093/nar/gkq321 [DOI] [PMC free article] [PubMed]
- Grube M, Muggia L. Identifying algal symbionts in lichen symbioses. In: Nimis PL, Vignes Lebbe R, editors. Tools for identifying biodiversity. Trieste: Progress and Problems. EUT; 2010. pp. 295–299. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall JD, Fucikova K, Lo C, Lewis LA, Karol KG. An assessment of proposed DNA barcodes in freshwater green algae. Cryptogamie Algol. 2010;31:529–555. [Google Scholar]
- Hametner C, Stocker-Wörgötter E, Rindi F, Grube M. Phylogenetic position and morphology of lichenized Trentepohliales (Ulvophyceae, Chlorophyta) from selected species of Graphidaceae. Phycol Res. 2014 [Google Scholar]
- Honegger R. The lichen symbiosis—what is so spectacular about it? Lichenol. 1998;30:193–212. doi: 10.1017/S002428299200015X. [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- John DM (2002) Order Trentepohliales. In: John DM, Whitton BA, Brook AJ (eds) The freshwater Algal Flora of the British isles: An identification Guide to Freshwater and terrestrial Algae, No. 478. Cambridge University Press, Australia, Spain, South Africa, U.S.A.
- Jose G, Chowdary YBK. New records of Trentepohliales from India. Nova Hedwigia. 1980;32:725–731. [Google Scholar]
- Leliaert F, Verbruggen H, Wysor B, De Clerck O. DNA taxonomy in morphologically plastic taxa: algorithmic species delimitation in the Boodlea complex (Chlorophyta: Cladophorales) Mol Phylogenet Evol. 2009;53:122–133. doi: 10.1016/j.ympev.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- López-Bautista JM, Chapman RL. Phylogenetic affinities of the Trentepohliales inferred from small-subunit rDNA. Int J Syst Evol Microbiol. 2003;53:2099–106. doi: 10.1099/ijs.0.02256-0. [DOI] [PubMed] [Google Scholar]
- López-Bautista JM, Waters DA, Chapman RL (2002) The Trentepohliales revisited. Constancea 83. http://ucjeps.berkeley.edu/constancea/83/lopez_etal/trentepohliales.html
- López-Bautista JM, Rindi F, Guiry M. Molecular systematics of the subaerial green algal order Trentepohliales: an assessement based on morphological and molecular data. Int J Syst Evol Microbiol. 2006;56:1709–1715. doi: 10.1099/ijs.0.63990-0. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. “Creating the CIPRES Science Gateway for inference of large phylogenetic trees”. New Orleans: In Proceedings of the Gateway Computing Environments Workshop (GCE); 2010. pp. 1–8. [Google Scholar]
- Nakano T. Phycobionts of some Japanese species of the Graphidaceae. Lichenol. 1988;20:353–360. doi: 10.1017/S0024282988000453. [DOI] [Google Scholar]
- Nelsen MP, Rivas Plata E, Andrew CJ, Lücking R, Lumbsch HT. Phylogenetic diversity of trentepohlialean algae associated with lichen-forming fungi. J Phycol. 2011;47:282–290. doi: 10.1111/j.1529-8817.2011.00962.x. [DOI] [PubMed] [Google Scholar]
- Nelson SC. Cephaleuros species, the plant-parasitic green algae. Plant Dis. 2008;43:1–6. [Google Scholar]
- Nichols HW. Growth media-freshwater. In: Stein JR, editor. Handbook of phycological methods, culture methods and growth measurements, No. Cambridge: Cambridge University Press; 1973. pp. 7–25. [Google Scholar]
- Prat S. Trentepohlia annulata brand in Mähren. Plant Syst Evol. 1914;64:420–421. doi: 10.1007/BF01642289. [DOI] [Google Scholar]
- Printz H. Vorarbeiten zu einer monographie der trentepohliaceae. Nyt Mag f Naturvid. 1939;80:137–216. [Google Scholar]
- Rambaut A (2006–2009) FigTree: tree figure drawing tool. Version 1.3.1.
- Rambaut A, Drummond AJ (2007) Tracer v1.4. http://beast.bio.ed.ac.uk/Tracer
- Rindi F, Guiry MD. Diversity, life history and ecology of Trentepohlia and Printzina (Trentepohliales, Chlorophyta) in urban habitats in Western Ireland. J Phycol. 2002;38:39–54. doi: 10.1046/j.1529-8817.2002.01193.x. [DOI] [Google Scholar]
- Rindi F, López-Bautista JM. Diversity and ecology of Trentepohliales (Ulvophyceae, Chlorophyta) in French Guiana. Cryptogamie Algol. 2008;29:13–43. [Google Scholar]
- Rindi F, Guiry MD, Critchley AT, Ar Gall E. The distribution of some species of Trentepohliaceae (Trentepohliales, Chlorophyta) in France. Cryptogamie Algol. 2003;24:133–144. [Google Scholar]
- Rindi F, Sherwood AR, Guiry MD. Taxonomy and distribution of Trentepohlia and Prinzina (Trentepohliales, Chlorophyta) in the Hawaiian Islands. Phycologia. 2005;44:270–284. doi: 10.2216/0031-8884(2005)44[270:TADOTA]2.0.CO;2. [DOI] [Google Scholar]
- Rindi F, Guiry MD, López-Bautista JM. New records of Trentepohliales (Ulvophyceae, Chlorophyta) from Africa. Nova Hedwigia. 2006;83:431–449. doi: 10.1127/0029-5035/2006/0083-0431. [DOI] [Google Scholar]
- Rindi F, Lam DW, López-Bautista JM. Trentepohliales (Ulvophyceae, Chlorophyta) from Panama. Nova Hedwigia. 2008;87:421–444. doi: 10.1127/0029-5035/2008/0087-0421. [DOI] [Google Scholar]
- Rindi F, Lam DW, López-Bautista JM. Phylogenetic relationships and species circumscription in Trentepohlia and Printzina (Trentepohliales, Chlorophyta) Mol Phylogenet Evol. 2009;52:329–339. doi: 10.1016/j.ympev.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Rindi F, Allali HA, Lam DW, López-Bautista JM. An overview of the biodiversity and biogeography of terrestrial green algae. In: Rescigno V, Maletta S, editors. Biodiversity hotspots. New York: Nova Science Publishers Inc.; 2010. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruprecht U, Brunauer G, Printzen C. Genetic diversity of photobionts in antarctic lecideoid lichens from an ecological point of view. Lichenol. 2012;44:661–678. doi: 10.1017/S0024282912000291. [DOI] [Google Scholar]
- Saunders GW, Kucera H. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie Algol. 2010;31:487–528. [Google Scholar]
- Saunders GW, McDevit DC. Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms. Methods Mol Biol. 2012;858:207–222. doi: 10.1007/978-1-61779-591-6_10. [DOI] [PubMed] [Google Scholar]
- Škaloud P, Peksa O. Evolutionary inferences based on ITS rDNA and actin sequences reveal extensive diversity of the common lichen alga Asterochloris (Trebouxiophyceae, Chlorophyta) Mol Phylogenet Evol. 2010;54:36–46. doi: 10.1016/j.ympev.2009.09.035. [DOI] [PubMed] [Google Scholar]
- Stocker-Wörgötter E. Resynthesis of photosymbiodemes. In: Kramer I, Beckett RP, Varma AK, editors. Protocols in lichenology: culturing, biochemistry, ecophysiology and use in biomonitoring. Berlin: Springer Lab Manual; 2002. pp. 47–60. [Google Scholar]
- Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4.0b10. Sinauer Associates
- Thompson RH, Wujek D. Trentepohliales: Cephaleuros, Phycopeltis, and Stomatochroon. morphology, taxanomy, and ecology. New Hampshire: Science Publishers; 1997. [Google Scholar]
- Tracanna BC. Trentepohliales (chlorophyta): el género Trentepohlia Martius para las provincias de Salta y Tucumán (Argentina) Lilloa. 1989;37:47–59. [Google Scholar]
- Van Der Strate HJ, Boele-Bos SA, Olsen JL, Van De Zande L, Stam WT. Phylogeographic studies in the tropical seaweed Cladophoropsis membranacea (Chlorophyta, Ulvophyceae) reveal a cryptic species complex. J Phycol. 2008;38:572–582. doi: 10.1046/j.1529-8817.2002.01170.x. [DOI] [Google Scholar]
- Voytsekhovich A, Dymytrova L, Nadyeina O. Photobiont composition of some taxa of the genera Micarea and Placynthiella (Lecanoramycetes, lichenized Ascomycota) from Ukraine. Folia Cryptogam Est. 2011;48:135–148. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Wildeman E. Les algues de la flore de Buitenzorg. Leiden: E.G. Brill; 1900. [Google Scholar]
- Yahr R, Vylgalys R, DePriest PT. Geographic variation in algal partners of Cladonia subtenuis (Cladoniaceae) highlights the dynamic nature of a lichen symbiosis. New Phytol. 2006;171:847–860. doi: 10.1111/j.1469-8137.2006.01792.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. Studies of cell aggregates and the production of natural pigments in plant cell culture. Osaka: Nippon Paint Publication; 1990. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 16 kb)
(DOCX 145 kb)
(DOCX 936 kb)


