Abstract
Understanding the forces that shape patterns of genetic variation across the genome is a major aim in evolutionary genetics. An emerging insight from analyses of genome-wide polymorphism and divergence data is that selection on linked sites can have an important impact on neutral genetic variation. However, in contrast to Drosophila, which exhibits a signature of recurrent hitchhiking, many plant genomes studied so far seem to mainly be affected by background selection. Moreover, many plants do not exhibit classic signatures of linked selection, such as a correlation between recombination rate and neutral diversity. In this review, I discuss the impact of genome architecture and mating system on the expected signature of linked selection in plants and review empirical evidence for linked selection, with a focus on plant model systems. Finally, I discuss the implications of linked selection for inference of demographic history in plants.
Keywords: hitchhiking, genetic draft, recombination rate, genome size, FST, demographic inference
INTRODUCTION
Understanding the forces that shape genetic variation is of great general as well as applied interest. As a result of recent massive advances in sequencing technologies, we now have access to an unprecedented amount of genomic data (e.g. [1–5]). However, despite increasing data availability, many challenges remain when it comes to understanding what evolutionary forces dominate in shaping patterns of polymorphism across genomes.
Since the seminal work of Begun and Aquadro [6] it has been recognized that the interaction between selection and recombination, or linked selection, can have a profound impact on levels of genetic variation across the genome. This is true for different forms of selection: under a hitchhiking model, the increase in frequency of a beneficial mutation results in a local reduction of genetic variation as linked neutral variants are swept to fixation along with the beneficial mutation [7, 8] (Table 1). Under a background selection model, the continued removal of deleterious alleles by purifying selection also results in locally reduced variation at linked sites [9, 10] (Table 1). Finally, interference between linked selected variants reduces the efficacy of selection (Hill–Robertson interference; [11]). A common feature of most forms of linked selection is that they are expected to result in a characteristic signature of reduced levels of neutral variation in low-recombination regions of the genome that are more effected by selection at linked sites.
Table 1:
Glossary of linked selection
| Term | Explanation |
|---|---|
| Linked selection | When positive or purifying selection affects linked genetic variation. |
| Selective sweep | When positive selection on a beneficial allele leads to a rapid increase in its frequency. This process generally leads to reduced polymorphism at linked sites. |
| Background selection | When purifying selection on deleterious alleles leads to reduced diversity at linked sites. |
An emerging insight from analyses of genome-wide polymorphism and divergence data is that effects of linked selection may be much more pervasive than previously thought [12–14]. Indeed, it has been suggested that in some organisms, such as e.g. Drosophila simulans, most neutral sites in the genome have been affected by linked selection in the form of recurrent hitchhiking [13] (Box 1). However, while the role of linked selection is well established for Drosophila, the case is less clear when it comes to plants, which in many cases have not exhibited classical signatures of linked selection, such as a correlation between recombination rate and level of neutral diversity [15–17]. Here, I will discuss how plant genome architecture and mating system affect the signature of linked selection, review empirical evidence for linked selection, with a focus on plant model systems, and discuss the implications of linked selection for the estimation of demographic history in plants.
Box 1: Types of selective sweeps.
‘Soft sweeps’ occur when positive selection acts to increase the frequency of several equally beneficial alleles on different genetic backgrounds, in contrast to ‘hard sweeps’, which involve selective fixation of a new beneficial mutation. ‘Partial sweeps’ occur when selection acts on multiple loci that are involved in adaptation, but does not necessarily lead to fixation of beneficial alleles at any of them. Finally, ‘recurrent hitchhiking’ occurs when selective sweeps happen repeatedly over evolutionary time. Hard sweeps and recurrent hitchhiking often lead to a distinguishable signature of elevated divergence at sites under selection coupled with reduced silent diversity and skewed allele frequency distributions in the vicinity of those sites. In contrast, the signature of partial and soft sweeps can be considerably more difficult to detect.
THE IMPACT OF LINKED SELECTION ON PLANT GENOMIC VARIATION
Models of linked selection predict a positive correlation between recombination rates and levels of neutral diversity; however, this prediction only holds if the rate and intensity of selection are uniform across the genome. If the density of selected sites varies, a more general expectation is that neutral diversity will depend on the density of selected sites per recombinational map unit [10, 18, 19]. Thus, in contrast to neutral models, the specifics of genome architecture (e.g. density of genes and other functional elements, recombination rate variation, chromosome number and length) are important for the distribution of neutral variation under linked selection models.
A positive association between recombination rate and neutral polymorphism was observed early on in several plant species, including sea beets [20], Aegilops [21] and tomatoes [22]. However, the strength of the association between recombination rate and diversity was often considerably weaker than that seen in Drosophila (e.g. [23]). Moreover, in Arabidopsis thaliana, early studies found no correlation between recombination rate and neutral diversity [15, 24]. Likewise, an early study in Arabidopsis lyrata found no general reduction of non-coding polymorphism in low-recombination regions close to centromeres [16]. However, when recombination rates and gene densities are correlated, as they are in Arabidopsis [25], linked selection can result in a negative correlation between functional density and neutral diversity rather than the typical pattern of reduced diversity in low-recombination regions. Such a negative correlation between gene density and neutral diversity has indeed been observed in A. thaliana [15, 26]. A similar pattern was recently observed in Oryza rufipogon, where it was shown to be consistent with the action of background selection [17]. In the model legume Medicago truncatula, there is both a positive correlation between recombination rate and silent diversity, and a negative correlation between gene density and silent diversity [27]. In contrast, a negative correlation between gene density and neutral diversity has so far not been observed in A. lyrata [16], although this remains to be revisited using genome-wide data, now that the A. lyrata genome sequence is available [28].
When can we expect linked selection to result in a negative correlation between gene density and neutral diversity? This will depend on details of genome architecture, as well as on plant life history traits. In plants, a potentially important factor underlying variation in linked selection is variation in the mating system. Because self-fertilization (selfing) results in a lower degree of effective recombination, the extent of linkage disequilibrium is expected to be longer in selfers (Box 2), and linked selection is therefore expected to have an impact over larger genomic distances in highly, but not exclusively, selfing species [29]. However, aspects of genome architecture, such as correlations between the density of sites under selection and the recombination rate can obscure the signature of linked selection.
Box 2: Effects of mating system on effective recombination rates and linkage disequilibrium.
‘Linkage disequilibrium’ is defined as the non-random association of alleles among loci. Recombination breaks down allelic associations in double heterozygotes. Because self-fertilization reduces heterozygosity, recombination is less efficient at breaking up allelic associations in self-fertilizing species. Linkage disequilibrium can therefore be more extensive in self-fertilizers.
To investigate when a signature of linked selection would be expected to be evident in species with a genome architecture similar to those of A. thaliana and A. lyrata, I modelled the expected reduction of neutral diversity under the background selection model of Hudson and Kaplan [10], closely following the approach used in Rockman et al. [30] and Flowers et al. [17]. Briefly, the method of Hudson and Kaplan ([10]; Equation (15)) allows one to estimate the expected reduction of neutral diversity in discrete intervals across a chromosome, with the effect of background selection on neutral diversity expressed as a function of the recombination rate and the proportion of sites subject to deleterious mutations in linked intervals. The modification of Rockman et al. [30] incorporates the effects of partial selfing or other deviations from panmixia on effective recombination rates through a scaling factor, the index of panmixia (P). Calculations were performed over a grid of values of P and a combined parameter incorporating both the intensity of selection and the dominance coefficient (sh). The dominance coefficient is included because dominant mutations are expected to be more efficiently selected against than recessive alleles, especially when rare. The considered values of P were equally spaced on a log scale between 0.002 and 1, and those for sh were equally spaced on a log scale between 5 × 10−5 and 0.5.
Figures 1 and 2 show the expected effect of background selection on neutral diversity in A. thaliana and A. lyrata, respectively, over a range of outcrossing rates and varying strengths of selection, and with estimates of recombination rates and densities of selected sites based on empirical data. In these figures, the diploid genome-wide deleterious mutation rate U is assumed to be 0.33 and 0.32 for A. thaliana and A. lyrata, respectively. These values for U are based on estimates of on the proportion of sites under constraint of 0.177 for A. thaliana and 0.113 for A. lyrata [31], a genome size of 135 Mb for A. thaliana and 206.7 Mb for A. lyrata, and a mutation rate of 7.0 × 10−9 for both species [32]. Recombination rate estimates for A. thaliana are based on the P66 cross in Salomé et al. [33] and for A. lyrata they are based on Aalto et al. [34]. The proportion of sites subject to deleterious mutation in each interval is based on conserved regions identified in Haudry et al. [31].
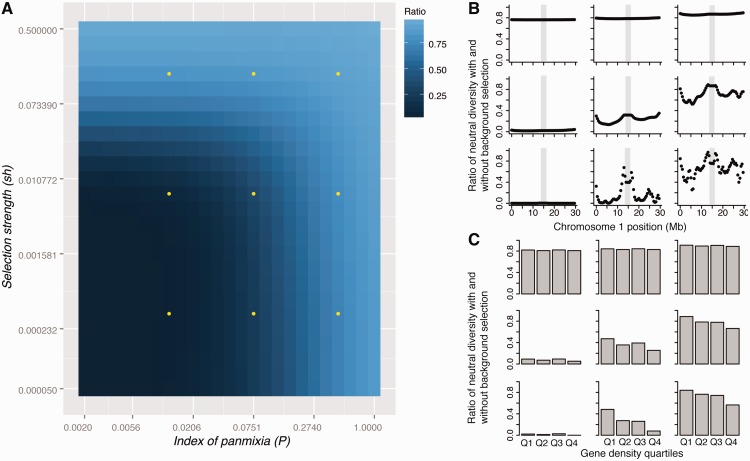
Figure 1:
Expected impact of background selection on neutral diversity in A. thaliana. (A) The predicted reduction in neutral diversity (ratio of neutral diversity with versus without background selection) is plotted over a grid of two parameters which measure the strength of selection (sh, a combined parameter incorporating the selection intensity and the dominance coefficient) and the deviation from panmixia (P). Dots indicate the parameter combinations plotted in panels (B) and (C). The three different values of P correspond to outcrossing rates of 0.06%, 3.9% and 29.9%, assuming all deviation from panmixia is a result of self-fertilization, and the sh values are 5 × 10−5, 3 × 10−3 and 0.1. (B) Relative proportions of neutral diversity across A. thaliana chromosome 1 for the nine parameter combinations indicated in A. Grey boxes mark the centromeric region on chromosome 1. (C) Conditions under which background selection is expected to lead to a negative correlation between gene density and neutral diversity. The predicted reduction of neutral diversity under background selection is shown for four quartiles of gene density, ranging from those with the lowest gene density (Q1) to those with the highest gene density (Q4).
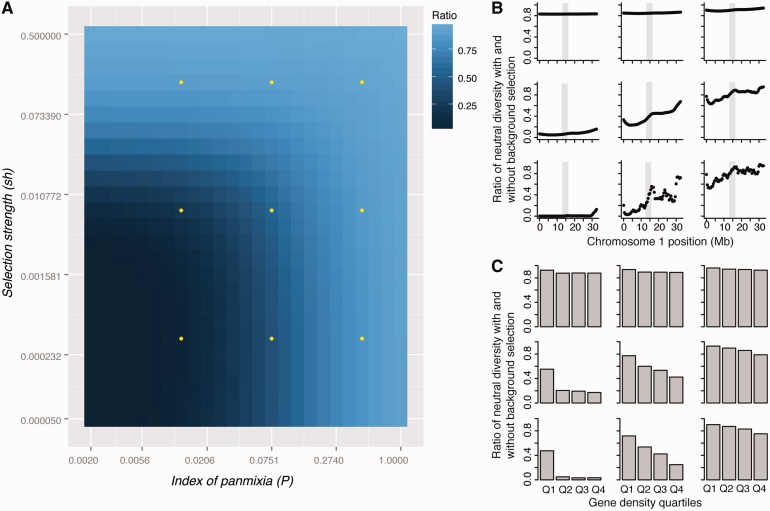
Figure 2:
Expected impact of background selection on neutral diversity in A. lyrata. (A) Predicted reduction in neutral diversity over the same grid of the index of panmixia and the compound selection parameter as in Figure 1A. (B) Relative proportions of neutral diversity across A. lyrata chromosome 1 for the nine parameter combinations indicated in A. Grey boxes mark the approximate location of the centromere. (C) Conditions under which background selection should lead to a negative correlation between gene density and neutral diversity. Expected reductions in neutral diversity under background selection are shown separately for four gene density quartiles labelled Q1 to Q4, ranging from lowest to highest gene density.
In both species, the genomic impact of background selection is expected to result in a negative correlation between gene density and neutral diversity, as long as there is not complete selfing (and/or other strong deviations from panmixia) and as long as purifying selection is not very strong (Figures 1 and 2). Furthermore, the model predicts higher levels of neutral diversity in pericentromeric regions, which harbor a lower density of sites subject to deleterious mutations. This is not strongly dependent on assumptions regarding U, as results are qualitatively similar for values of U of 0.15 and 0.60 (Supplementary Figures S1–S4).
In A. thaliana and A. lyrata, analyses of the distribution of fitness effects for non-synonymous mutations suggest that weak purifying selection is the predominant mode of selection and there is little evidence for high rates of adaptive non-synonymous fixations [35–37]. Furthermore, outcrossing rates have been estimated to be ∼3% in A. thaliana [38]. Under this level of outcrossing and with weak selection, a clear signature of background selection is a negative relationship between gene density and neutral diversity. This pattern has indeed been observed in A. thaliana [15]. Similar results have also been obtained for M. truncatula [39], suggesting that the negative correlation between gene density and silent diversity in this species [27] could also be explained by background selection. The model also predicts that the signature of background selection should be weaker in outcrossers, such as A. lyrata, in line with empirical observations [16]. However, elevated neutral diversity in low-recombination pericentromeric regions, which has been observed in A. lyrata [16], is consistent with the action of weak background selection (Figure 2). Qualitatively, therefore, population genetic patterns in A. thaliana and A. lyrata seem to fit a simple model of background selection quite well.
The patterns observed in many plant species so far seem to be consistent with a major role for weak purifying selection in shaping patterns of polymorphism, with linked selection mainly having an impact on the genomic distribution of neutral variation in selfing taxa. It is currently not clear why plants should be experiencing less recurrent hitchhiking than for instance Drosophila. Possible explanations include factors that reduce the efficacy of natural selection relative to drift, such as low effective population sizes, strong population structure[36, 40], or effects of mating system on adaptation [41, 42]. These effects have recently been reviewed thoroughly [43, 44] and hence will not be covered in more detail here. However, it should be noted that evidence for recurrent hitchhiking has recently been found in a plant species; the outcrossing species Capsella grandiflora [45]. This species has relatively weak population structure, a large effective population size and low levels of linkage disequilibrium, factors that are expected to render natural selection more effective [36]. To further elucidate when we can expect to observe recurrent hitchhiking in plants, genomic data for more species with a range of outcrossing rates and effective population sizes are required.
Another explanation for the relative dearth of evidence for recurrent hitchhiking in plants could be that forms of adaptation that do not necessarily lead to a signature of species-wide sweeps are common in plants. Local adaptation is one such form of selection that is not expected to lead to species-wide sweep signatures. There is increasing evidence for local adaptation in A. thaliana, both from reciprocal transplant studies [46] and from studies that have quantified fitness components in mapping populations grown in the field [47]. Evidence for local adaptation has also been found using genomic approaches that search for correlations between allele frequencies and environmental variables (e.g. [48, 49]) and by combining genomic analyses with common garden experiments [50] to identify locally adaptive alleles. However, the extent to which local adaptation affects patterns of polymorphism genome-wide is still an open question in A. thaliana as well as in most other plant species.
While recurrent hitchhiking may be rare in plants, this does not preclude an important role for selective sweeps in plant adaptation. For instance, a recent study that analysed genome sequences from 180 lines of A. thaliana from Sweden found many signatures of selective sweeps, including a massive sweep on chromosome 1 involving a 700-kb transposition [51]. There is also evidence for partial selective sweeps in A. thaliana [52], but the general importance of different forms of sweeps, such as partial and soft sweeps (Box 1) remains unclear.
Linked selection is also expected to have a major impact on levels of population differentiation. If there is diversifying selection, with positive selection driving alleles to high frequencies in some but not all populations under study, increased differentiation is expected at loci under selection, as well as at closely linked loci [53]. Similarly, background selection can lead to elevated FST, particularly in regions of low recombination, because it decreases the effective population size experienced by linked loci [54], although the locus under selection itself is expected to exhibit reduced FST. Thus, both forms of selection are expected to result in a negative correlation between recombination rate and FST [55]. Distinguishing between these hypotheses requires examining additional measures of differentiation that do not rely on within-population diversity; under background selection no elevation of absolute divergence is expected [53, 54]. This contrast was recently used to demonstrate that elevated values of FST on Silene latifolia Y-chromosomes are not a result of local adaptation, but instead caused by other processes reducing variability on Y-chromosomes [56]. Many studies have conducted genome scans in plants with the purpose of identifying candidate loci for local adaptation (reviewed in Strasburg et al. [57]), but these do not usually examine correlations between population differentiation and recombination rates, perhaps because estimates of recombination rates have previously not been available for many non-model species. With the recent rapid advances in genome sequencing and genotyping methods, this area seems ripe for further investigation.
EFFECTS OF PLANT GENOME SIZE VARIATION ON LINKED SELECTION
Plants vary over 1000-fold in genome size, due to polyploidy and variation in the content of repetitive elements [58]. This might have consequences for the impact of linked selection. In background selection models, a key parameter is the genome-wide deleterious mutation rate, U, a rough estimate of which can be obtained as a product of the mutation rate, the genome size, and the proportion of sites under constraint [59]. If plant genome-size variation affects U, it should also affect the impact of background selection. For instance, if polyploidization leads to relaxed selection on duplicate genes genome-wide, as theory predicts [60], background selection may be relaxed in polyploid genomes. Studies of paleopolyploid genomes, such as that of A. thaliana, suggest that duplicate gene loss is indeed the most frequent outcome of whole-genome duplication [61]. On the other hand, duplicate genes that are retained experience elevated levels of purifying selection [62]. Genome size increases due to expansion of repetitive elements may also be associated with reduced recombination rates, as the rate of crossovers is generally reduced in heterochromatic regions [58, 63]. Exploring the effects on linked selection when there are concomitant changes in genome size, recombination rates and levels of constraint will thus be important for interpretation of broad comparative genomic studies of the effects of linked selection in plants.
CONSEQUENCES OF LINKED SELECTION FOR DEMOGRAPHIC INFERENCE IN PLANTS
If linked selection is pervasive, patterns of variation at neutral sites linked to selected sites may largely reflect the rate and strength of selection (i.e. genetic draft; [64]), rather than demographic history. In a recent simulation study using realistic parameter estimates for human data, Messer and Petrov [65] demonstrated that linked selection can lead to significant skews in synonymous site frequency spectra, to the extent that demographic expansions were falsely inferred. The skew was exacerbated under higher levels of adaptive fixations, but was present at rates of adaptation as low as 0.1. The proportion of adaptive fixations at non-synonymous sites have been estimated to be higher than this in several plant species (e.g. Populus [66], C. grandiflora [36], and Helianthus [40, 67]). Indeed, a recent study of genomic patterns of variation in C. grandiflora found evidence for recurrent hitchhiking, as well as a skew towards rare alleles at synonymous sites [45], consistent with the results of Messer and Petrov [65]. While the exact effects of linked selection should be examined using simulations with realistic settings for the study species in question as well as with empirical data, these results suggest that care should be taken when choosing what sites for use for demographic inference. In the human literature, studies are already starting to appear that take this consideration seriously and consequently analyse demographic history using non-coding regions far from sites under selection (e.g. [68]). Such approaches may be more difficult in plants, as plant non-coding regions can be difficult to align reliably due to their dynamic nature and often high content of repetitive elements. However, with an improved understanding of the impact of linked selection in plant genomes, it is likely that considerations of the effects of linked selection will also become important for demographic inference in plants.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/.
Key Points.
Signatures of linked selection in plants often differ from those in animals, and taking account of variation in the density of selected sites is important.
There is accumulating evidence for an important role for background selection in many plants studied so far, whereas evidence for recurrent hitchhiking is scarce.
More work on the effects of plant genome size variation on linked selection is needed.
Linked selection can have a marked impact on inference of demographic history, and this should be considered when choosing sites for demographic inference in plants.
Acknowledgements
The author wishes to thank Peter Morrell, Thomas Kono and an anonymous reviewer for constructive comments that greatly helped improve this paper.
Biography
Tanja Slotte is an assistant professor in Ecological Genomics at Science for Life Laboratory, Stockholm University, Sweden. Her research is centred on plant evolutionary genomics, with a particular focus on the causes and consequences of plant mating system shifts.
FUNDING
This work was supported by the Swedish Research Council (grant no. A0550801).
References
- 1.Begun DJ, Holloway AK, Stevens K, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, Schneeberger K, Ossowski S, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–63. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 4.1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen EC, Gerke JP, Shapiro JA, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–90. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–20. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 7.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 8.Wiehe TH, Stephan W. Analysis of a genetic hitchhiking model, and its application to DNA polymorphism data from Drosophila melanogaster. Mol Biol Evol. 1993;10:842–54. doi: 10.1093/oxfordjournals.molbev.a040046. [DOI] [PubMed] [Google Scholar]
- 9.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson RR, Kaplan NL. Deleterious background selection with recombination. Genetics. 1995;141:1605–17. doi: 10.1093/genetics/141.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–94. [PubMed] [Google Scholar]
- 12.Hahn MW. Toward a selection theory of molecular evolution. Evolution. 2008;62:255–65. doi: 10.1111/j.1558-5646.2007.00308.x. [DOI] [PubMed] [Google Scholar]
- 13.Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cutter AD, Payseur BA. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet. 2013;14:262–74. doi: 10.1038/nrg3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nordborg M, Hu TT, Ishino Y, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright SI, Foxe JP, DeRose-Wilson L, et al. Testing for effects of recombination rate on nucleotide diversity in natural populations of Arabidopsis lyrata. Genetics. 2006;174:1421–30. doi: 10.1534/genetics.106.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flowers JM, Molina J, Rubinstein S, et al. Natural selection in gene-dense regions shapes the genomic pattern of polymorphism in wild and domesticated rice. Mol Biol Evol. 2012;29:675–87. doi: 10.1093/molbev/msr225. [DOI] [PubMed] [Google Scholar]
- 18.Barton NH. Linkage and the limits to natural selection. Genetics. 1995;140:821–41. doi: 10.1093/genetics/140.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordborg M, Charlesworth B, Charlesworth D. The effect of recombination on background selection. Genet Res. 1996;67:159–74. doi: 10.1017/s0016672300033619. [DOI] [PubMed] [Google Scholar]
- 20.Kraft T, Säll T, Magnusson-Rading I, et al. Positive correlation between recombination rates and levels of genetic variation in natural populations of sea beet (Beta vulgaris subsp. maritima) Genetics. 1998;150:1239–44. doi: 10.1093/genetics/150.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dvorák J, Luo M-C, Yang Z-L. Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics. 1998;148:423–34. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephan W, Langley CH. DNA polymorphism in Lycopersicon and crossing-over per physical length. Genetics. 1998;150:1585–93. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roselius K, Stephan W, Städler T. The relationship of nucleotide polymorphism, recombination rate and selection in wild tomato species. Genetics. 2005;171:753–63. doi: 10.1534/genetics.105.043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid KJ, Ramos-Onsins S, Ringys-Beckstein H, et al. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics. 2005;169:1601–15. doi: 10.1534/genetics.104.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 26.Gossmann TI, Woolfit M, Eyre-Walker A. Quantifying the variation in the effective population size within a genome. Genetics. 2011;189:1389–402. doi: 10.1534/genetics.111.132654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branca A, Paape TD, Zhou P, et al. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proc Natl Acad Sci USA. 2011;108:E864–70. doi: 10.1073/pnas.1104032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu TT, Pattyn P, Bakker EG, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–81. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cutter AD, Payseur BA. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol Biol Evol. 2003;14:262–74. doi: 10.1093/molbev/msg072. [DOI] [PubMed] [Google Scholar]
- 30.Rockman MV, Skrovanek SS, Kruglyak L. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science. 2010;330:372–6. doi: 10.1126/science.1194208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haudry A, Platts AE, Vello E, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45:891–8. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 32.Ossowski S, Schneeberger K, Lucas-Lledó JI, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–4. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salomé PA, Bomblies K, Fitz J, et al. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity. 2012;108:447–55. doi: 10.1038/hdy.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aalto EA, Koelewijn H-P, Savolainen O. Cytoplasmic male sterility contributes to hybrid incompatibility between subspecies of Arabidopsis lyrata. G3. 2013;3:1727–40. doi: 10.1534/g3.113.007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foxe JP, Dar V-U-N, Zheng H, et al. Selection on amino acid substitutions in Arabidopsis. Mol Biol Evol. 2008;25:1375–83. doi: 10.1093/molbev/msn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slotte T, Foxe JP, Hazzouri KM, et al. Genome-wide evidence for efficient positive and purifying selection in Capsella grandiflora, a plant species with a large effective population size. Mol Biol Evol. 2010;27:1813–21. doi: 10.1093/molbev/msq062. [DOI] [PubMed] [Google Scholar]
- 37.Slotte T, Bataillon T, Hansen TT, et al. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol Evol. 2011;3:1210–9. doi: 10.1093/gbe/evr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Platt A, Horton M, Huang YS, et al. The scale of population structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paape T, Bataillon T, Zhou P, et al. Selection, genome-wide fitness effects and evolutionary rates in the model legume Medicago truncatula. Mol Ecol. 2013;22:3525–38. doi: 10.1111/mec.12329. [DOI] [PubMed] [Google Scholar]
- 40.Gossmann TI, Song B-H, Windsor AJ, et al. Genome wide analyses reveal little evidence for adaptive evolution in many plant species. Mol Biol Evol. 2010;27:1822–32. doi: 10.1093/molbev/msq079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glémin S, Ronfort J. Adaptation and maladaptation in selfing and outcrossing species: new mutations versus standing variation. Evolution. 2013;67:225–40. doi: 10.1111/j.1558-5646.2012.01778.x. [DOI] [PubMed] [Google Scholar]
- 42.Wright SI, Kalisz S, Slotte T. Evolutionary consequences of self-fertilization in plants. Proc Biol Sci. 2013;280:20130133. doi: 10.1098/rspb.2013.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hough J, Williamson RJ, Wright SI. Patterns of selection in plant genomes. Annu Rev Ecol Evol Syst. 2013;44:31–49. [Google Scholar]
- 44.Hazzouri KM, Purugganan MD. Population genomics of plant species. In: Paterson AH, editor. Advances in Botanical Research. Vol. 69. Oxford, UK: Academic Press, Elsevier; 2014. pp. 311–34. [Google Scholar]
- 45.Williamson R, Josephs EB, Platts AE, et al. Evidence for widespread positive and negative selection in coding and conserved noncoding regions of Capsella grandiflora. bioRxiv. 2014 doi: 10.1371/journal.pgen.1004622. http://dx.doi.org/10.1101/002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ågren J, Schemske DW. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 2012;194:1112–22. doi: 10.1111/j.1469-8137.2012.04112.x. [DOI] [PubMed] [Google Scholar]
- 47.Ågren J, Oakley CG, McKay JK, et al. Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2013;110:21077–82. doi: 10.1073/pnas.1316773110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock AM, Brachi B, Faure N, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334:83–6. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 49.Lee C-R, Mitchell-Olds T. Environmental adaptation contributes to gene polymorphism across the Arabidopsis thaliana genome. Mol Biol Evol. 2012;29:3721–8. doi: 10.1093/molbev/mss174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fournier-Level A, Korte A, Cooper MD, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334:86–9. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 51.Long Q, Rabanal FA, Meng D, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45:884–90. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horton MW, Hancock AM, Huang YS, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44:212–6. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charlesworth B, Nordborg M. The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet Res. 1997;70:155–74. doi: 10.1017/s0016672397002954. [DOI] [PubMed] [Google Scholar]
- 54.Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 1998;15:538–43. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- 55.Keinan A, Reich D. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 2010;6:e1000886. doi: 10.1371/journal.pgen.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muir G, Bergero R, Charlesworth D, Filatov DA. Does local adaptation cause high population differentiation of Silene latifolia Y chromosomes? Evolution. 2011;65:3368–80. doi: 10.1111/j.1558-5646.2011.01410.x. [DOI] [PubMed] [Google Scholar]
- 57.Strasburg JL, Sherman NA, Wright KM, et al. What can patterns of differentiation across plant genomes tell us about adaptation and speciation? Philos Trans Roy Soc Lond B Biol Sci. 2012;367:364–73. doi: 10.1098/rstb.2011.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henderson IR. Control of meiotic recombination frequency in plant genomes. Curr Opin Plant Biol. 2012;15:556–61. doi: 10.1016/j.pbi.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Kondrashov AS, Crow JF. A molecular approach to estimating the human deleterious mutation rate. Hum Mutat. 1993;2:229–34. doi: 10.1002/humu.1380020312. [DOI] [PubMed] [Google Scholar]
- 60.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–37. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 61.Lynch M, Conery J. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 62.Guo H, Lee T-H, Wang X, Paterson AH. Function relaxation followed by diversifying selection after whole-genome duplication in flowering plants. Plant Physiol. 2013;162:769–78. doi: 10.1104/pp.112.213447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wijnker E, Velikkakam James G, Ding J, et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. Elife. 2013;2:e01426. doi: 10.7554/eLife.01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillespie JH. Genetic drift in an infinite population. The pseudohitchhiking model. Genetics. 2000;155:909–19. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Messer PW, Petrov DA. Frequent adaptation and the McDonald–Kreitman test. Proc Natl Acad Sci USA. 2013;21:8615–20. doi: 10.1073/pnas.1220835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ingvarsson PK. Natural selection on synonymous and nonsynonymous mutations shapes patterns of polymorphism in Populus tremula. Mol Biol Evol. 2010;27:650–60. doi: 10.1093/molbev/msp255. [DOI] [PubMed] [Google Scholar]
- 67.Strasburg JL, Kane NC, Raduski AR, et al. Effective population size is positively correlated with levels of adaptive divergence among annual sunflowers. Mol Biol Evol. 2011;28:1569–80. doi: 10.1093/molbev/msq270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gazave E, Ma L, Chang D, et al. Neutral genomic regions refine models of recent rapid human population growth. Proc Natl Acad Sci USA. 2014;111:757–62. doi: 10.1073/pnas.1310398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.