Abstract
The National Agricultural Biotechnology Information Center (NABIC) reconstructed an AllergenPro database for allergenic proteins analysis and allergenicity prediction. The AllergenPro is an integrated web-based system providing information about allergen in foods, microorganisms, animals and plants. The allergen database has the three main features namely, (1) allergen list with epitopes, (2) searching of allergen using keyword, and (3) methods for allergenicity prediction. This updated AllergenPro outputs the search based allergen information through a user-friendly web interface, and users can run tools for allergenicity prediction using three different methods namely, (1) FAO/WHO, (2) motif-based and (3) epitope-based methods.
Availability
The database is available for free at http://nabic.rda.go.kr/allergen/
Keywords: allergenicity prediction, AllergenPro, allergen search
Background
Allergy has been one of the most common chronic health problems in the world, and other hypersensitivity reactions have become major causes of chronic suffering in human. With advances in genomic technologies, there is a rapid increase in allergy-related information and multiple tools have been developed for performing allergenicity prediction [1]. Allergen nomenclature website [2] is the official site for the systematic allergen nomenclature that is approved by the World Health Organization and International Union of Immunological Societies (WHO/IUIS) Allergen Nomenclature Sub-committee. Food Allergy Research and Resource Program [3] provide the food industry with credible information, expert opinions, tools, and services relating to allergenic foods. In order to assessment of potential allergenicity and patterns of cross-reactivity, AllerTool [4] provide a web server with essential tools for the assessment of predicted as well as published cross-reactivity patterns of allergens. AllergenOnline database [5] provides an allergen sequence entries and sequence searchable database intended for the identification of proteins that may present a potential risk of allergenic cross-reactivity.
Methodology
Data collection and development:
The allergen sequence, motif-allergen and epitopes data were collected from the IUIS allergen database, UniProtKB/Swiss-Prot and NCBI PubMed database. The allergen information were collected the 2,434 allergen sequence data without duplication, and were categorized the animals (i.e., worms, rodents, mammals, insects, mites, foods, others), microbes (i.e., bacteria, fungi), and plants (i.e., pollen, latex, foods, others) sections. The platform was developed using MYSQL and Oracle relational database management system. Using the collected allergen information, we have developed an allergen database with three main functions: (i) allergen list with structure and epitopes; (ii) searching of allergen by keyword and sequence information; and (iii) three computing methods for allergenicity prediction. In 2014, we have released a major update for allergenicity prediction to compare the three different methods such as the FAO/WHO, motif-based and epitope-based methods.
Implementation and features:
We have developed an integrated web database system which is comprised of allergenic proteins for food safety and homology search tools. The integrated allergen database has developed a web-based system that will provide information about allergen in microorganisms, foods, animals and plants. The database has three major parts and functions: an allergen list, allergen search and allergenicity prediction. The allergen list provides the information which selected allergen sequence data without duplication. The related information for allergen structure and epitopes were connected with UniProtKB/TrEMBL database, and users can search for specific-protein name by keywords, protein sequence and specific categories (Figure 1).
Figure 1.
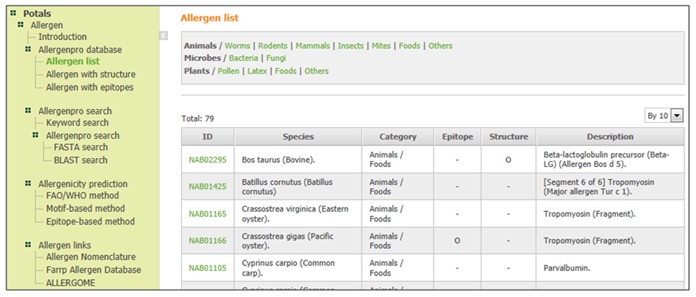
A snap shot of the database. This view table shows samples of search results such as the allergen information table by clicking a specific category.
In addition, we developed an allergenicity prediction tools to identify potential cross-reactivity with well-known allergens, and users can run tools for allergenicity prediction using three different methods such as the FAO/WHO, motif-based and epitope-based methods (Figure 2). The Food and Agriculture Organization of the United Nations (FAO) propose the method to analysis of sequence homology when the expressed protein is derived from a source with known allergenicity. According to the FAO/WHO method 7, AllergenPro provide the function to find the query protein for an 80 amino acids sliding window by a FASTA alignment program. Motif-based prediction method [6] provides the function to compare the two sequences with the specific profiles between the conserved motifs and the unique sequences. If specific protein is predicted to be allergenic, query result will be displayed to compare against a set of well-known allergenic motifs. Using the AllergenPro database, a user easily can predict the allergenic structure of query protein by changing related parameters and e-value. In browsing the result table, a user can access the individual characterization information tables by clicking a linked their respective hypertext.
Figure 2.
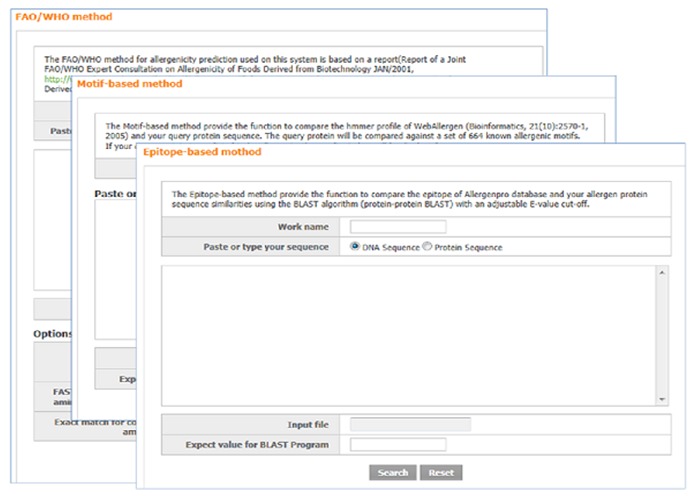
A snap shot shows individual searching table such as the FAO/WHO, motif-based and epitope-based methods.
Utility, caveats and future developments
The AllergenPro database provides allergen related information (motif structure and epitope of allergens) in microorganisms, foods, animals and plants. The database has three major utility features (dataset list, allergen search, and allergenicity prediction). The database provides options for (1) multiple allergen categories, (2) search of specific allergen, and (3) tools for allergenicity prediction using three different methods. The integrated allergen database provides a platform for allergen sequence search through a user-friendly web-interface. NABIC plans to provide this service to allergen research institutes and societies in the near future. We also plan to develop an integrated allergen database with web tools combining specific allergen datasets and tools for allergenicity prediction using various methods.
Acknowledgments
This study was conducted with support from the Research Program for Agricultural Science & Technology Development (Project No. PJ010112) of the National Academy of Agricultural Science, Rural Development Administration, Republic of Korea.
Footnotes
Citation:Kim et al, Bioinformation 10(6): 378-380 (2014)
References
- 1.Kim C, et al. Bioinformation. 2009;3:344. doi: 10.6026/97320630003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. http://www.allergen.org/
- 3. http://farrp.unl.edu/
- 4.Zhang ZH, et al. Bioinformatics. 2007;23:504. doi: 10.1093/bioinformatics/btl621. [DOI] [PubMed] [Google Scholar]
- 5. http://www.allergenonline.com/
- 6.Riaz T, et al. Bioinformatics. 2005;21:2570. doi: 10.1093/bioinformatics/bti356. [DOI] [PubMed] [Google Scholar]


