Abstract
Aims
Bone morphogenetic proteins (BMPs) are involved in embryonic and adult blood vessel formation in health and disease. Previous studies have shown that BMP endothelial cell precursor-derived regulator (BMPER) plays an important role in endothelial cell function and blood vessel formation. BMPER is a key regulator of BMP4 activity and a prerequisite for BMP pathway activation by BMP4 in endothelial cells. Here, we characterize the BMPER promoter and elucidate mechanisms of BMPER regulation.
Methods and results
To investigate transcriptional mechanisms of BMPER expression, the murine BMPER promoter was cloned and characterized. A series of 5′ deletions of the BMPER promoter revealed that the proximal promoter contains activating cis-elements. By overexpression or siRNA-based knockdown, we demonstrate that BMPER expression is activated by Krüppel-like factor (KLF) 15. As determined by gelshift analyses, KLF15 binds directly to a predicted KLF-binding element at −284 bp within the BMPER promoter. Co-expression experiments show that Sp1 acts as an antagonist for KLF15-induced promoter activation. Endothelin-1 was identified as a potent inhibitor of KLF15 and BMPER expression in endothelial cells, suggesting that KLF15 is a transducer of endothelin-1 activity on BMPER expression. The selective ETB endothelin receptor antagonist BQ788 abolished the downregulation of BMPER expression by endothelin-1.
Conclusion
Mechanistically, we found that KLF15 is a strong and direct activator of the BMPER expression. BMPER is downregulated by endothelin-1 in a dose-dependent fashion and in parallel to KLF15. As KLF15 deficiency is accompanied by a vascular phenotype and BMPER is necessary for proper blood vessel formation, we suggest a chain of events in which the effects of endothelin-1 on BMPER are mediated by KLF15.
Keywords: BMPER, Bone morphogenetic protein, Regulation, Krüppel-like factor 15, Endothelial cell
1. Introduction
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor-β (TGF-β) superfamily. Originally, they have been identified by their ability to induce ectopic bone formation and have been extensively studied during embryonic development, where they control axis formation and organogenesis. A growing body of evidence suggests that they serve also as important regulators in vascular development and disease.1 Important insights came from the discovery of mutations of the BMP receptors in patients with familial pulmonary artery hypertension or teleangiectasia, both being clinically important vascular diseases.2 BMP4 induces capillary sprouting of endothelial cells.3 Chronic infusion of BMP4 induces arterial hypertension and leads to endothelial dysfunction in mice.4 BMP2 and 4 exert pro-inflammatory effects on the endothelium.5–7 Recent studies suggest that BMPs are upregulated in athero-prone regions in blood vessels and may contribute to vascular calcification and the development of atherosclerotic plaques.8,9 Accordingly, loss of BMP pathway activity by genetic deletion of BMP receptors or BMP-dependent transcription factors (the Smads) results in a hypomorphic vascular phenotype during mouse embryogenesis.10–12 As unequivocally demonstrated by these data, a fine-tuned BMP activity is indeed necessary to maintain normal vasculature, and BMP4 contributes to unfavourable conditions of the vascular wall.
BMP endothelial cell precursor-derived regulator (BMPER) is a novel extracellular BMP modulator that we have identified earlier by its differential expression in embryonic endothelial cell precursors.13 In mouse and zebrafish, BMPER is expressed at sites and at the time of vasculogenesis consistent with a functional role for BMPER in vascular events. Indeed, BMPER controls the effect of BMP4 on critical endothelial cell functions such as sprouting and migration.14 In accordance with this endothelial cell sprouting and migration phenotype, lack of BMPER results in disarray of intersomitic blood vessels in developing zebrafish.15 Time course observations show that BMPER expression during mouse and zebrafish development is temporally and spatially controlled.13,15,16 Studies of the spatial expression of BMPER in relation to BMPs and other BMP modulators such as chordin suggest that BMPER expression is at least in part complementary to BMP4 expression.17 Taken together, BMPER is a key modulator of BMP signalling particularly in blood vessel formation, and its expression is tightly regulated.
Until now, the regulation of BMP activity has been studied at several levels including (i) protein–protein interactions of BMPs with extracellular modulators, (ii) BMP ligand binding to cell surface receptors, and (iii) intracellular signal transduction pathways. BMPs are transported within the extracellular space and thereby form concentration gradients. This process is modulated by the presence of extracellular BMP-binding proteins, such as chordin, which assist to transport BMPs over longer distances by protecting them from degradation. Thereby, chordin may contribute to the accumulation of BMPs at sites remote from their original site of expression and thus may also increase BMP activity at these sites. On the other hand, chordin is a direct BMP antagonist by interfering with BMP–BMP receptor interaction. In contrast, BMPER is not diffusible, and thus helps to accumulate BMP activity at the site of BMPER expression. In this scenario, the control of BMPER expression would be essential to regulate the local BMP activity and would reflect an additional level of BMP regulation.
2. Methods
2.1. Antibodies and reagents
Monoclonal anti-human BMPER antibody was purchased from R&D Systems (MAB1956); monoclonal anti-FLAG M2 antibody, recombinant endothelin-1 (E-7764), and selective ETB endothelin receptor antagonist BQ788 (B157) were purchased from Sigma-Aldrich.
2.2. Cell culture
Human umbilical vein endothelial cells (HUVECs) and bovine aortic endothelial cells (BAECs) were cultured in endothelial cell growth medium (Provitro) with 10% foetal bovine serum (FBS). C166 cells are mouse yolk sac-derived embryonic endothelial cells and were cultured as COS7 in DMEM (GIBCO) with 10% FBS.18 Cultures were kept at 37°C in a 5% CO2-humidified atmosphere. This project was approved by the local ethics committee of the University of Freiburg (208/04) and conforms with the principles outlined in the Declaration of Helsinki.
2.3. Promoter constructs
The 5′ flanking region of the BMPER gene was cloned by PCR using genomic DNA from C57/BL6 mice as template. A series of 5′ deleted fragments were cloned into promoterless pGL3-basic vector (Promega). A mutation of the Krüppel-like factor (KLF) 15-binding site was introduced into the p(-465)luc construct by a three-step PCR mutagenesis with selected sense and antisense primers. The sequences of all DNA constructs were confirmed by DNA sequencing.
2.4. Transfection and luciferase assay
BAEC or COS-7 cells were plated at a density of 1 × 105/well in a six-well dish. After 24 h, transient transfections were performed using Fugene 6 (Roche Molecular Biochemics) or Lipofectamine 2000 (Invitrogen) according to the instructions provided by the manufacturer. Cells were transfected using 2 µg of luciferase reporter vector and 0.5 µg of the pCMV-β-galactosidase (pCMV-β-Gal). For some experiments, expression plasmids for Sp/KLFs were co-transfected at this time. After 24 h, luciferase activity and β-galactosidase activity were measured. β-galactosidase activity was used to normalize for transfection efficiency. Data are shown as mean ± SD of triplicates of at least three independent experiments. To overexpress Foxo3A, HUVECs were plated at a density of 1×105 cells/well in a six-well dish and transfected with plasmids coding for pcDNA3, Foxo3A-wt, and constitutive active FoxO3A-A3 using PromoFectin (Promocell). After 48 h, HUVECs RNA was prepared and analysed by quantitative PCR (see Supplementary material online, Figure S1).
2.5. Semi-quantitative PCR
Total RNA was isolated from HUVECs using the Total RNA Mini Kit (Bio-Rad), and equal amounts were reverse-transcribed with iScript cDNA Synthesis Kit (Bio-Rad). Following primers were used: BMPER forward 5′-AGGACAGTGCTGCCCCAAATG-3′, BMPER reverse 5′-TACTGACACGTCCCCTGAAAG-3′, KLF15 forward 5′-CTTCCAGCCTACCCTGGAGGAG-3′, KLF15 reverse 5′-TTGGTGTACATCTTGCTGCAGCC-3′. hRPII forward 5′-GCACCACGTCCAATGACAT-3′ and hRPII reverse 5′-GTGCGGCTGCTTCCATAA-3′ served as internal control. The PCR amplification cycles consisted of an initial denaturation step at 94°C for 5 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 60–64.3°C for 30 s, and extension for 30 s. After a final extension of 5 min at 72°C, PCR products were analysed using an ethidium bromide-stained 2% agarose gel. Gel bands were semi-quantified using the Quantity One 1D Analysis Software Version 4.4 (Bio-Rad). All RT–PCR experiments were repeated at least three times and representative results are shown.
2.6. Western blot
HUVECs were stimulated for 72 h with 100 nM endothelin-1 in endothelial cell growth medium (Provitro) with 0.5% FBS. Cell lysates were resolved on a reducing polyacrylamide gel, plotted onto a nitrocellulose membrane (Amersham Bioscience), and blocked with 5% non-fat dry milk powder in PBS/Tris with 0.1% Tween 20 for 2 h at room temperature (20–22°C). The membrane was then incubated with primary antibody overnight at 4°C. After 1 h of incubation with the secondary antibody, proteins were visualized using ECL reagent (Amersham). All western blots were repeated at least three times and quantified data are shown.
2.7. siRNA transfection
siRNA for human KLF15 and a non-specific control siRNA were purchased from Invitrogen (KLF15siRNA1 sense: 5′-GCAUUUCUGCUUGCCCGAGUUUCCU-3′ and KLF15siRNA2 sense: 5′-UACCCUGGAGGAGAUUGAAGAGUUU-3′). HUVECs were plated 1 day before transfection at a density of 4 × 104/well in a 12-well dish in HUVEC medium with 10% FBS. HUVECs were transfected with siRNA using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocol. Cells were harvested for RNA. Knockdown efficiency was confirmed by RT–PCR. Gel bands were semi-quantified using the Quantity One 1D Analysis Software Version 4.4 (Bio-Rad).
2.8. Electrophoretic mobility shift assay
Mouse yolk sac-derived embryonic endothelial cells (C166) were used for binding analysis of endogenous KLF15 to the BMPER promoter as they express significant amounts of KLF15. For supershift (SS) analysis, KLF15-FLAG was overexpressed in COS7 cells. After 48 h, nuclear extracts were prepared with a nuclear extraction kit (Active Motif, Rixensart, Belgium). Gelshift analysis was performed with a 32P-labelled BMPER oligonucleotide (sequence: 5′-CGCTCAGCCACCCCTCCTCCCG-3′) containing the 5′- CACCC-3′ KLF-binding motif. The kinase reaction consists of 37 µL purified water, 1 µL BMPER oligonucleotide (25 ng/µL), 5 µL 10× kinase buffer, 5 µL of γ-32P desoxyadenosine triphosphate (Amersham), and 1.5 µL of T4 kinase and was incubated for 30 min at 37°C. The protein content of nuclear extracts was determined using the Bradford Assay (Bio-Rad). Equal amounts of protein were added to the labelled oligonucleotide. Afterwards, 18 µL of reaction mixture, containing 22.5 mM Hepes–KOH, pH 7.9, 2.6 mM MgCl2, 13.3% glycerol, 50 mM KCL, 0.125 mM EDTA, 0.5 mM dithiothreitol, 0.5 mg/mL poly(dI-dC), and subsequently 0.5 ng of 32P-labelled oligonucleotide, was added. The samples were incubated for 20 min at room temperature and then loaded on a 4% non-denaturating polyacrylamide gel running in 0.5 × Tris-borate-EDTA buffer. Gels were vacuum-dried for 40 min on a 3 mm chromatography filter (Whatman) and exposed to a radiograph film (Kodak Biomax MR).
2.9. Statistical analysis and quantification
Statistical analyses were performed using GraphPad Prism 4.0. Data are presented as mean ± SD, and comparisons were calculated by Student's t-test (two-sided, unpaired). Results were considered statistically significant when P < 0.05. Densitometric analysis of western blots and RT–PCR was performed using Quantity One 1-D Analysis Software Version 4.4 (Bio-Rad), and levels of significance were calculated by one-sample t-test.
3. Results
3.1. Characterization of the BMPER promoter
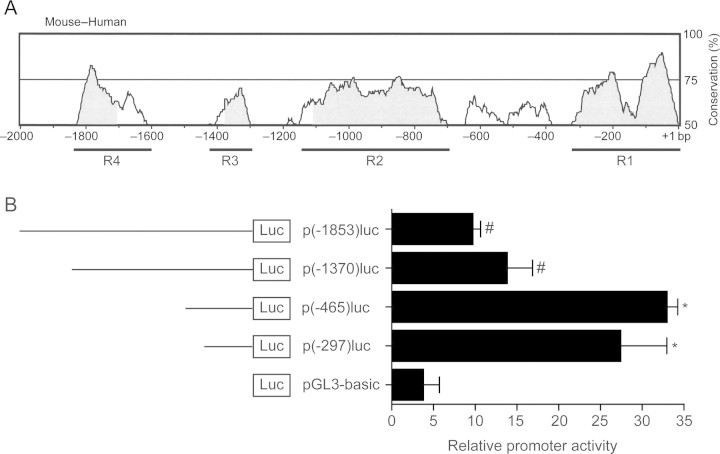
To study BMPER regulation, we dissected the murine BMPER promoter and searched for transcription factors which regulate BMPER promoter activity. Comparison of the 2000 base pairs upstream of the translational start site between human and mouse revealed four regions of particularly high degree of homology (R1–R4, Figure 1A). To characterize the role of these highly conserved regions, we generated deletion constructs of the BMPER promoter to drive luciferase as a reporter using the boundaries of these regions as deletion sites: p(-1853)luc, p(-1370)luc, p(-465)luc, p(-297)luc, and p(-97)luc. The BMPER promoter fragments were transfected into BAECs, and their transcriptional activity was measured (Figure 1B). Deletion of the promoter fragments from −1853 to −1370 bp as well as from −1370 to −465 bp resulted in an increase in promoter activity. This suggests that negative regulatory cis-elements are located within these regions (Figure 1B). In contrast, deletion of the proximal promoter led to a reduction of the promoter activity by 86%, pointing to activating cis-elements contained in the proximal promoter. In silico analysis indicated that the region from −297 to 1 bp contains two domains that are highly conserved between mouse and human (>75%). Furthermore, this promoter region is guanin- and cytosin-rich (74.5%) and contains predicted binding sites for members of the Sp1 and the KLF family of transcription factors (Figure 2A).
Figure 1.
The conserved domains of the 5′ flanking region of the BMPER gene contain positive and negative acting cis-elements. (A) Comparison of the human and mouse 5′ flanking region of the BMPER gene using mVISTA reveals four highly conserved regions (R1–R4) from −2000 bp to the translational start site. (B) Deletion analysis of the BMPER promoter in BAECs. Cells were transfected with the respective promoter construct, and luciferase activity was quantified. Values represent the mean ± SD of three independent experiments normalized to β-galactosidase (*P < 0.05 vs. pGL3-basic; #P < 0.05 vs. p(−465)luc).
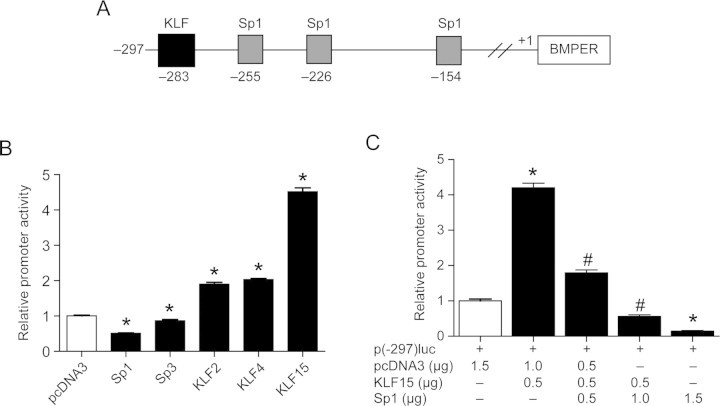
Figure 2.
Members of the SP/KLF family regulate BMPER expression. (A) Schematic representation of the Sp1- and KLF-binding sites within the region −297 to 1 bp of the translational start site of the BMPER gene as analysed by in silico prediction software (www.genomatix.de). (B) Overexpression of KLF15 induces BMPER promoter activity in COS-7 cells. The p(−297)luc promoter construct was co-transfected with pcDNA3 (empty vector) or the expression plasmids encoding for Sp1, Sp3, KLF2, KLF4, and KLF15, respectively. The results represent the mean ± SD of three independent experiments (*P < 0.05 vs. pcDNA3). (C) Overexpression of Sp1 reverses KLF15- induced BMPER promoter activity in a dose-dependent fashion. The promoter construct p(−297)luc was co-transfected with KLF15 and increasing amounts of Sp1. The relative promoter activity was represented as the fold increase compared with the control (pcDNA3). The results represent the mean ± SD of three independent experiments (*P < 0.05 vs. pcDNA3; #P < 0.05 vs. p(−297)luc+KLF15).
3.2. KLF15 regulates BMPER expression
On the basis of in silico prediction, we hypothesized that members of the Sp/KLF family of transcription factors may bind to the BMPER promoter and regulate BMPER expression (Figure 2A). Searching for a member of the Sp/KLF family with the ability to regulate the proximal BMPER promoter, we tested Sp1, Sp3, KLF2, KLF4, and KLF15 (Figure 2B). When the respective expression plasmids were transfected along with p(-297)luc, the strongest promoter induction was obtained by KLF15, followed by KLF2 and KLF4. Sp1 and Sp3 did not activate the BMPER promoter, and they even behaved as inhibitors (Figure 2B).
3.3. Sp1 attenuates KLF15-induced BMPER promoter activation
Next, we asked whether KLF15 interacts with Sp1 family members, resulting in the modification of BMPER promoter regulation, as it has been reported before for other promoters, such as AceCS2.19 To test this notion, KLF15 was transfected together with various amounts of Sp1, and the effect on promoter activity was compared. We found that KLF15-induced promoter activity is repressed by increasing amounts of Sp1, suggesting a functional antagonism between KLF15 and SP1 (Figure 2C).
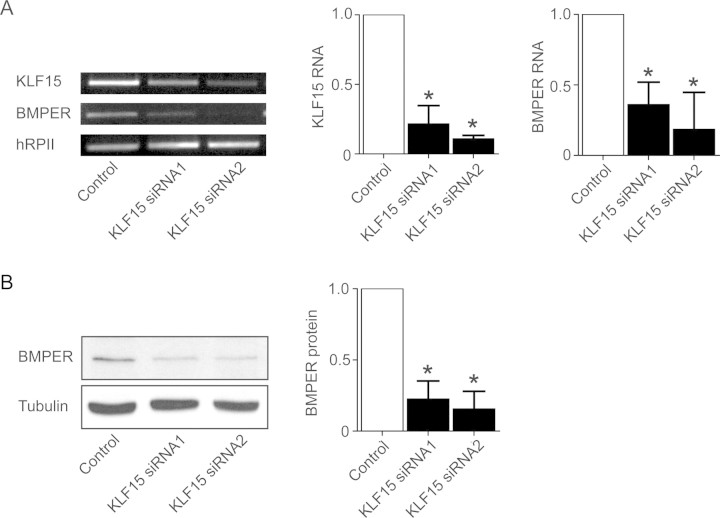
Along the same line of evidence, we found that the inhibition of KLF15 using specific siRNA reduced BMPER expression in endothelial cells as reflected by BMPER RNA (Figure 3A) and protein levels (Figure 3B). Taken together, these data suggest that KLF15 is a key activator of BMPER expression.
Figure 3.
Lack of KLF15 results in reduced BMPER RNA and protein expression in HUVECs. Specific silencing of KLF15 by siRNA in endothelial cells resulted in decreased BMPER RNA (A) and protein (B) expression. (A) Two different siRNAs for KLF15 or control siRNA were transfected in HUVECs. After 48 h, RNA was prepared and reverse-transcribed. RT–PCR was performed with specific primers for KLF15, BMPER, and loading control hRPII. Panels to the right demonstrate the quantification of three independent experiments, one of which is shown on the left (*P < 0.05 vs. control). (B) Lack of KLF15 results in reduced BMPER protein expression at 72 h post-transfection. β-Tubulin served as the loading control. The right panel demonstrates the quantification of three independent experiments, one of which is shown at the left (*P < 0.05 vs. control).
3.4. KLF15 binds the KLF-motif in the BMPER promoter
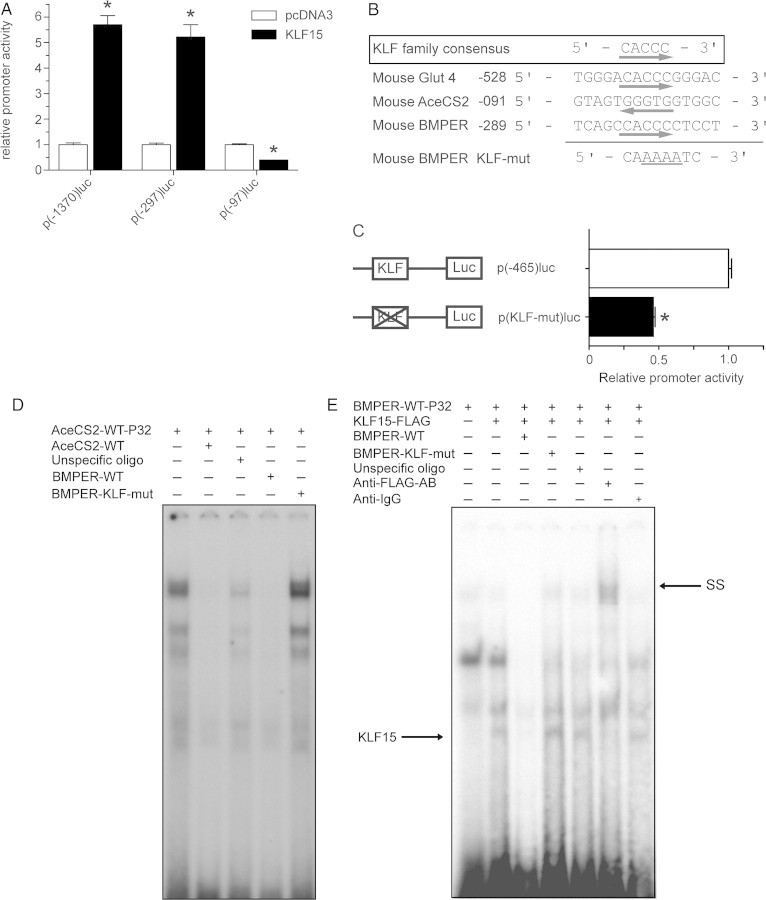
To narrow-down the region of KLF15 binding to the BMPER promoter, we investigated the ability of KLF15 to transactivate different promoter deletion fragments. Therefore, KLF15 was overexpressed in the presence of the various BMPER promoter reporter constructs, and luciferase activity was quantified (Figure 4A). As expected from our previous experiments, KLF15 strongly upregulated the reporter constructs containing the promoter fragments [p(-1370)luc (5.7-fold) and p(-297)luc (5.2-fold)]. But, a significant loss of KLF15 reactivity was detected between −297 and −97 bp upstream of the translational start site, suggesting the presence of a KLF15 response element in this region. Next, we analysed this 200 bp DNA fragment in more detail. A highly conserved KLF consensus-binding element 5′-CACCC-3′ was predicted by in silico analysis at −284 bp.19,20 Comparison of this sequence derived from the BMPER promoter with well-characterized KLF15 response elements from other promoters such as the GLUT4 and the AceCS2 promoter revealed a high degree of homology between the sequences (Figure 4B). To determine the functional relevance of this motif in the BMPER promoter, we inactivated the binding element by site-directed mutagenesis. The KLF-mutated promoter construct p(KLF-mut)luc showed only 45% promoter activity compared with the respective wildtype construct (Figure 4C). This result indicates that the −284 bp KLF-binding motif is essential for maximal promoter activity.
Figure 4.
(A–C) KLF15 binds to the positive-acting KLF motif in the BMPER promoter. (A) The inducibility of the BMPER promoter by KLF15 depends on the region from −297 to −97 bp. Three deletion constructs of the BMPER promoter were co-transfected with KLF15 or pcDNA3 (empty vector). At 24 h after transfection, cells were harvested for luciferase assay. The results represent the mean ± SD of three independent experiments (*P < 0.05 vs. pcDNA3). (B) Alignment of KLF motifs in the AceCS2 gene, the Glut4 gene along with the wildtype and the mutated motif in the BMPER gene. (C) The KLF motif in the BMPER promoter at −284 bp is essential for maximal transcriptional activity. The p(−465)luc and the mutated p(KLF-mut)luc constructs were transiently transfected in COS-7 cells. At 24 h post-transfection, cells were harvested for luciferase assay. The results represent the mean ± SD of three independent experiments. Mutation of the KLF15 motif in the BMPER promoter reduced promoter activity to 45% of wildtype activity (*P < 0.05 vs. p(465)luc). (D and E) KLF15 binds specifically to the KLF motif in the BMPER promoter. (D) Nuclear extracts from C166 cells were incubated with the 32P-labelled KLF15-binding-site-derived oligonucleotide from the AceCS2 promoter (lane 1). Cold competition was performed using the same but unlabelled oligonucleotide (lane 2) or wildtype KLF15-binding-site-derived oligonucleotide from the BMPER promoter (lane 4). Mutated KLF15-binding oligonucleotide from the BMPER promoter did not compete for DNA-protein complex formation (lane 3). Incubation with an unrelated control oligonucleotide had no effect (lane 5). (E) Nuclear extracts from COS7 cells transfected with pcDNA3 (lane 1) or the FLAG-tagged KLF15 expression vector were used in binding the 32P-labelled BMPER oligonucleotide (lane 2). Cold competition was performed with an excess of unlabelled BMPER oligonucleotide BMPER-WT (lane 3). Incubation with mutated BMPER oligonucleotide BMPER-KLF-mut (lane 4) or unrelated control oligonucleotide (lane 5) had no effect on the formation of the three bands. SS studies were performed using an anti-FLAG antibody (lane 6) or an unspecific IgG antibody as control (lane 7).
As we have identified the KLF15 response element within the BMPER promoter, we next set out to characterize the binding of KLF15 to the BMPER promoter in gelshift assays. Nuclear extracts of C166 cells, which express high levels of KLF15, were prepared and used for gelshift analysis. Incubation of the nuclear extracts with the radiolabelled AceCS2-oligonucleotide containing the known KLF15-binding site (AceCS2-wt-P32) resulted in the formation of three protein–DNA complexes (Figure 4D, lane 1). As expected, the three complexes could be effectively competed by the addition of excessive amounts of unlabelled AceCS2-wt oligonucleotide (Figure 4D, lane 2) or unlabelled BMPER-wt oligonucleotide (Figure 4D, lane 4). In contrast, competition with unlabelled oligonucleotide containing a mutated KLF-binding sequence (BMPER-KLF-mut, Figure 4D, lane 5) or a non-specific oligonucleotide (Figure 4D, lane 3) did not affect the formation of the protein–DNA complexes. These findings clearly demonstrate that the DNA–protein complexes that are formed by the KLF15-binding element within the AceCS2 promoter are formed by the newly described KLF15-binding element in the BMPER promoter. As expected, the mutated KLF15-binding element in the BMPER promoter is not able to bind the respective proteins. To prove direct binding of KLF15 to the BMPER promoter nuclear extracts of COS7 cells overexpressing KLF15-FLAG (Figure 4E, lanes 2–7) or pcDNA3 control (Figure 4E, lane 1) were incubated with the radiolabelled oligonucleotide containing the KLF15 response element (BMPER-WT-P32). Comparison of nuclear extracts from KLF15 overexpressing cells with control nuclear extracts identified a KLF15-specific band (Figure 4E, lanes 1 and 2). As expected, the KLF15-derived complex could be effectively competed by the addition of excessive amounts of unlabelled BMPER-WT oligo (lane 3). In contrast, competition with unlabelled oligonucleotide containing the KLF-mutated sequence (BMPER-KLF-mut) did not affect the KLF15 complex (lane 4). Similarly, the addition of unrelated non-specific oligonucleotide did not affect the KLF15 complex (lane 5). Incubation with an anti-FLAG antibody resulted in an SS of the KLF15 band, proving the specificity of this complex (lane 6). In contrast, the addition of a non-specific antibody had no effect (lane 7). Taken together, these findings demonstrate that KLF15 directly binds to the KLF-binding motif that we have identified in the BMPER promoter.
3.5. Endothelin-1 downregulates BMPER expression via the ETB receptor in endothelial cells
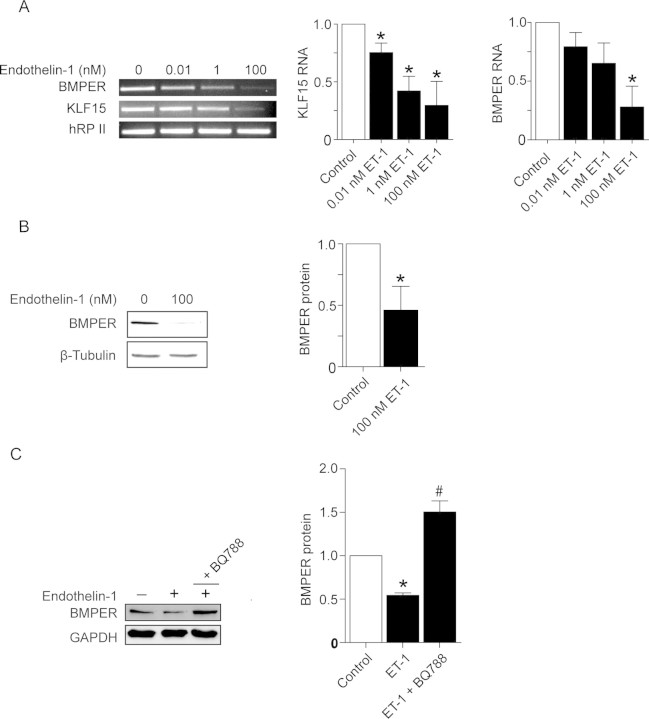
To identify regulators of BMPER, we tested known mediators of endothelial cell function and vascular homeostasis for their ability to regulate BMPER expression in vitro. Endothelin-1 was a potent, dose-dependent inhibitor of BMPER expression at the RNA and protein levels (Figure 5A and B). Incubation of HUVECs with endothelin-1 in the presence of the selective ETB endothelin receptor antagonist BQ788 abolished the endothelin-1 effect, suggesting that the ETB receptor transduces the downregulation of BMPER by endothelin-1 (Figure 5C). Thus, we identified endothelin-1 as the first cytokine shown to regulate BMPER and demonstrated that the ETB receptor transduces this effect.
Figure 5.
Endothelin-1 downregulates BMPER expression in endothelial cells. (A) Stimulation of HUVECs with the indicated concentrations of endothelin-1 for 48 h. RNA was prepared and reverse-transcribed for RT–PCR using specific primers for hBMPER, hKLF15, and hRPII as a control. One representative experiment out of three is shown. The panels to the right demonstrate the quantification of three independent experiments. (B) BMPER protein expression is reduced in HUVECs after stimulation with 100 nM endothelin-1. After 72 h stimulation with endothelin-1, cells were lysed, and western blot analysis for BMPER and β-tubulin (loading control) was performed. The right panel shows the quantification of three independent experiments (*P < 0.05 vs. control). (C) Co-incubation of selective ETB endothelin-1 receptor antagonist BQ788 blocks downregulation of BMPER expression by endothelin-1. After 72 h stimulation with endothelin-1 alone or in combination with BQ788 (2 µM), HUVECs were lysed and used for western blot analysis. GAPDH serves as loading control (*P < 0.05 vs. control).
3.6. Endothelin-1 represses KLF15 expression in HUVECs
We hypothesized that endothelin-1-induced inhibition of BMPER expression in endothelial cells may involve KLF15 because endothelin-1 has been shown to inhibit KLF15 expression in cardiomyocytes.21,22 To confirm this hypothesis, we incubated HUVECs with endothelin-1 and quantified KLF15 expression. Comparable with what was reported in cardiomyocytes, we found a decrease of KLF15 RNA levels upon endothelin-1 exposure in endothelial cells (Figure 5A). Thus, KLF15 is a downstream target and a potential mediator of endothelin-1. Taken all our data together, this suggests that KLF15 confers the effect of endothelin-1 on BMPER expression.
4. Discussion
We present several novel findings with regard to the regulation of BMPER in endothelial cells. First, within the BMPER promoter, repressive elements are located at distance from the translational start site, and activating elements are located in the proximal section. Second, KLF15 is a potent activator of BMPER expression and binds to a cis-acting KLF motif within the promoter (−284 bp) that is necessary for maximal promoter activity. Third, KLF15 and Sp1 functionally compete at the BMPER promoter. Fourth, we demonstrate that BMPER is reduced by endothelin-1 via the ETB receptor pathway. Fifth, endothelin-1 represses KLF15 in endothelial cells, providing a potential mechanism for BMPER regulation.
BMPER is a key modulator in BMP signalling particularly in blood vessel formation, and its expression is tightly regulated in a temporal and spatial fashion.13 Thus, the regulation of BMPER expression contributes to control BMP pathway activity. In the present work, we set out to study the regulation of BMPER in detail. In silico prediction revealed that the proximal promoter is highly conserved (>75%, Figure 1A), GC-rich (74.5%), and contains a binding site for KLF15 (Figure 2A). KLF15 is a member of the family of Krüppel-like transcription factors. Out of this family, so far KLF2, 4, and 6 have been implicated in physiological and pathological vascular conditions.23–26 Previous studies have identified KLF15 as a negative regulator of cardiac hypertrophy and fibrosis.21,27 Similar to BMPER, KLF15 is highly expressed in vascular structures.20 The deletion of KLF15 in mice revealed a vascular phenotype resulting from a thickened aortic media with signs for augmented inflammatory gene expression and foci of severe smooth muscle apoptosis, which further supports a role for KLF15 and BMPER in vascular disease. Here, we characterize KLF15 as an activator of endothelial BMPER expression (Figures 2D and 3).
Vascular homeostasis is regulated by cytokines and growth factors. Endothelin-1 is such a key vascular regulator. High concentrations of endothelin-1 cause oxidative stress and endothelial dysfunction, including increased expression of vascular inflammation markers (reviewed in 28). Moreover, endothelin-1 is involved in the pathogenesis of pulmonary hypertension.29 Endothelin receptor antagonists such as bosentan are in clinical use to lower pulmonary artery pressure. Interestingly, in cardiomyocytes, endothelin-1 downregulates KLF15 and upregulates BMP2, a BMP agonist that binds directly to BMPER.30
BMPs are involved in the lung and blood vessel formation. Loss of function mutations of the BMP pathway result in pulmonary hypertension.29 BMPER is a dose dependent regulator of BMP activity and is involved in BMP-dependent vascular diseases.14 We have not yet investigated the effect of BMPER on pulmonary artery hypertension, but the data obtained so far suggest that BMPER may have an influence on this disease. First, BMPER is strongly expressed in the lung,13 second, BMPER has activating effects on endothelial cells,14 and third, BMPER-deficient mice die from respiratory failure.31 In this paper, we link the endothelin-1 pathway with the BMP pathway by the downregulation of KLF15. Our data suggest a chain of events with KLF15 in the central position to confer the effect of endothelin-1 on BMPER expression.
Supporting in vivo evidence for the hypothesis that endothelin-1 and BMP signalling interact comes from the bone phenotypes in gain and loss of function models. In rats, endothelin-1 promotes bone formation, whereas the intake of an endothelin-1 receptor antagonist results in osteopenia.32 Along the same line of evidence, homozygous inactivation of endothelin-1 results in the impairment of bone and cartilage development with reduced mandibles and hypoplastic auricles.33 This phenotype is reminiscent to the bone phenotype observed in BMPER−/− mice.17,34
Taken together, we identify KLF15 as a direct transactivator of BMPER. Since BMPER is a key regulator of BMP signalling, it is crucial to understand how BMPER expression is regulated. In this context, it is of great physiological relevance that endothelin-1 downregulates BMPER as well as its activator KLF15, suggesting that KLF15 confers the endothelin-1 effect on BMPER expression.
Supplementary material
Supplementary Material is available at Cardiovascular Research online.
Funding
This work was supported by Deutsche Forschungsgemeinschaft SFB-TR23 (A1) to M.M.
Supplementary Material
Acknowledgements
We are indebted to Bianca Engert for her outstanding technical assistance. We thank M.K. Jain (Cleveland, OH, USA) for kindly sharing the KLF2, KLF4, and KLF15 expression plasmids; M. Potente (Frankfurt, Germany) for the FoxO3A-wt and FoxO3A-A3 plasmids; and G. Suske (Marburg, Germany) for providing us with Sp1 and Sp3 expression plasmids.
Conflict of interest: none declared.
References
- 1.Ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 2.Beppu H, Ichinose F, Kawai N, Jones RC, Yu PB, Zapol WM, et al. BMPR-II heterozygous mice have mild pulmonary hypertension and an impaired pulmonary vascular remodeling response to prolonged hypoxia. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1241–L1247. doi: 10.1152/ajplung.00239.2004. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Heinke J, Vargas A, Winnik S, Krauss T, Bode C, et al. ERK signaling is a central regulator for BMP-4 dependent capillary sprouting. Cardiovasc Res. 2007;76:390–399. doi: 10.1016/j.cardiores.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Miriyala S, Gongora Nieto MC, Mingone C, Smith D, Dikalov S, Harrison DG, et al. Bone morphogenic protein-4 induces hypertension in mice: role of noggin, vascular NADPH oxidases, and impaired vasorelaxation. Circulation. 2006;113:2818–2825. doi: 10.1161/CIRCULATIONAHA.106.611822. [DOI] [PubMed] [Google Scholar]
- 5.Csiszar A, Ahmad M, Smith KE, Labinskyy N, Gao Q, Kaley G, et al. Bone morphogenetic protein-2 induces proinflammatory endothelial phenotype. Am J Pathol. 2006;168:629–638. doi: 10.2353/ajpath.2006.050284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorescu GP, Sykes M, Weiss D, Platt MO, Saha A, Hwang J, et al. Bone morphogenic protein 4 produced in endothelial cells by oscillatory shear stress stimulates an inflammatory response. J Biol Chem. 2003;278:31128–31135. doi: 10.1074/jbc.M300703200. [DOI] [PubMed] [Google Scholar]
- 7.Sorescu GP, Song H, Tressel SL, Hwang J, Dikalov S, Smith DA, et al. Bone morphogenic protein 4 produced in endothelial cells by oscillatory shear stress induces monocyte adhesion by stimulating reactive oxygen species production from a nox1-based NADPH oxidase. Circ Res. 2004;95:773–779. doi: 10.1161/01.RES.0000145728.22878.45. [DOI] [PubMed] [Google Scholar]
- 8.Bostrom K, Watson KE, Horn S, Wortham C, Herman IM, Demer LL. Bone morphogenetic protein expression in human atherosclerotic lesions. J Clin Invest. 1993;91:1800–1809. doi: 10.1172/JCI116391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhore CR, Cleutjens JP, Lutgens E, Cleutjens KB, Geusens PP, Kitslaar PJ, et al. Differential expression of bone matrix regulatory proteins in human atherosclerotic plaques. Arterioscler Thromb Vasc Biol. 2001;21:1998–2003. doi: 10.1161/hq1201.100229. [DOI] [PubMed] [Google Scholar]
- 10.Beppu H, Kawabata M, Hamamoto T, Chytil A, Minowa O, Noda T, et al. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev Biol. 2000;221:249–258. doi: 10.1006/dbio.2000.9670. [DOI] [PubMed] [Google Scholar]
- 11.Lechleider RJ, Ryan JL, Garrett L, Eng C, Deng C, Wynshaw-Boris A, et al. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Dev Biol. 2001;240:157–167. doi: 10.1006/dbio.2001.0469. [DOI] [PubMed] [Google Scholar]
- 12.Tremblay KD, Dunn NR, Robertson EJ. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development. 2001;128:3609–3621. doi: 10.1242/dev.128.18.3609. [DOI] [PubMed] [Google Scholar]
- 13.Moser M, Binder O, Wu Y, Aitsebaomo J, Ren R, Bode C, et al. BMPER, a novel endothelial cell precursor-derived protein, antagonizes bone morphogenetic protein signaling and endothelial cell differentiation. Mol Cell Biol. 2003;23:5664–5679. doi: 10.1128/MCB.23.16.5664-5679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinke J, Wehofsits L, Zhou Q, Zoeller C, Baar KM, Helbing T, et al. BMPER is an endothelial cell regulator and controls bone morphogenetic protein-4-dependent angiogenesis. Circ Res. 2008;103:804–812. doi: 10.1161/CIRCRESAHA.108.178434. [DOI] [PubMed] [Google Scholar]
- 15.Moser M, Yu Q, Bode C, Xiong J-W, Patterson C. BMPER is a conserved regulator of hematopoietic and vascular development in zebrafish. J Mol Cell Cardiol. 2007;43:243–253. doi: 10.1016/j.yjmcc.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffinier C, Ketpura N, Tran U, Geissert D, De Robertis EM. Mouse Crossveinless-2 is the vertebrate homolog of a Drosophila extracellular regulator of BMP signaling. Gene Expr Patterns. 2002;2:189–194. doi: 10.1016/s0925-4773(02)00420-3. [DOI] [PubMed] [Google Scholar]
- 17.Zakin L, Metzinger CA, Chang EY, Coffinier C, De Robertis EM. Development of the vertebral morphogenetic field in the mouse: interactions between Crossveinless-2 and twisted gastrulation. Dev Biol. 2008;323:6–18. doi: 10.1016/j.ydbio.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang SJ, Greer P, Auerbach R. Isolation and propagation of yolk-sac-derived endothelial cells from a hypervascular transgenic mouse expressing a gain-of-function fps/fes proto-oncogene. In vitro Cell Dev Biol. 1996;32:292–299. doi: 10.1007/BF02723062. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, et al. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem. 2004;279:16954–16962. doi: 10.1074/jbc.M312079200. [DOI] [PubMed] [Google Scholar]
- 20.Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, et al. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322–34328. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 21.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci USA. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullingford TE, Butler MJ, Marshall AK, Tham El L, Sugden PH, Clerk A. Differential regulation of Kruppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta. 2008;1783:1229–1236. doi: 10.1016/j.bbamcr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–1141. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 24.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 25.Kuo CT, Veselits ML, Barton KP, Lu MM, Clendenin C, Leiden JM. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 27.Wang B, Haldar SM, Lu Y, Ibrahim OA, Fisch S, Gray S, et al. The Kruppel-like factor KLF15 inhibits connective tissue growth factor (CTGF) expression in cardiac fibroblasts. J Mol Cell Cardiol. 2008;45:193–197. doi: 10.1016/j.yjmcc.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bohm F, Pernow J. The importance of endothelin-1 for vascular dysfunction in cardiovascular disease. Cardiovasc Res. 2007;76:8–18. doi: 10.1016/j.cardiores.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Cody RJ, Haas GJ, Binkley PF, Capers Q, Kelley R. Plasma endothelin correlates with the extent of pulmonary hypertension in patients with chronic congestive heart failure. Circulation. 1992;85:504–509. doi: 10.1161/01.cir.85.2.504. [DOI] [PubMed] [Google Scholar]
- 30.Cullingford TE, Markou T, Fuller SJ, Giraldo A, Pikkarainen S, Zoumpoulidou G, et al. Temporal regulation of expression of immediate early and second phase transcripts by endothelin-1 in cardiomyocytes. Genome Biol. 2008;9:R32. doi: 10.1186/gb-2008-9-2-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley R, Ren R, Pi X, Wu Y, Moreno I, Willis M, et al. A concentration-dependent endocytic trap and sink mechanism converts BMPER from an activator to an inhibitor of BMP signaling. J Cell Biol. 2009;184:597–609. doi: 10.1083/jcb.200808064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukahara H, Hori C, Hiraoka M, Yamamoto K, Ishii Y, Mayumi M. Endothelin subtype A receptor antagonist induces osteopenia in growing rats. Metabolism. 1998;47:1403–1407. doi: 10.1016/s0026-0495(98)90313-4. [DOI] [PubMed] [Google Scholar]
- 33.Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- 34.Ikeya M, Kawada M, Kiyonari H, Sasai N, Nakao K, Furuta Y, et al. Essential pro-BMP roles of Crossveinless 2 in mouse organogenesis. Development. 2006;133:4463–4473. doi: 10.1242/dev.02647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.