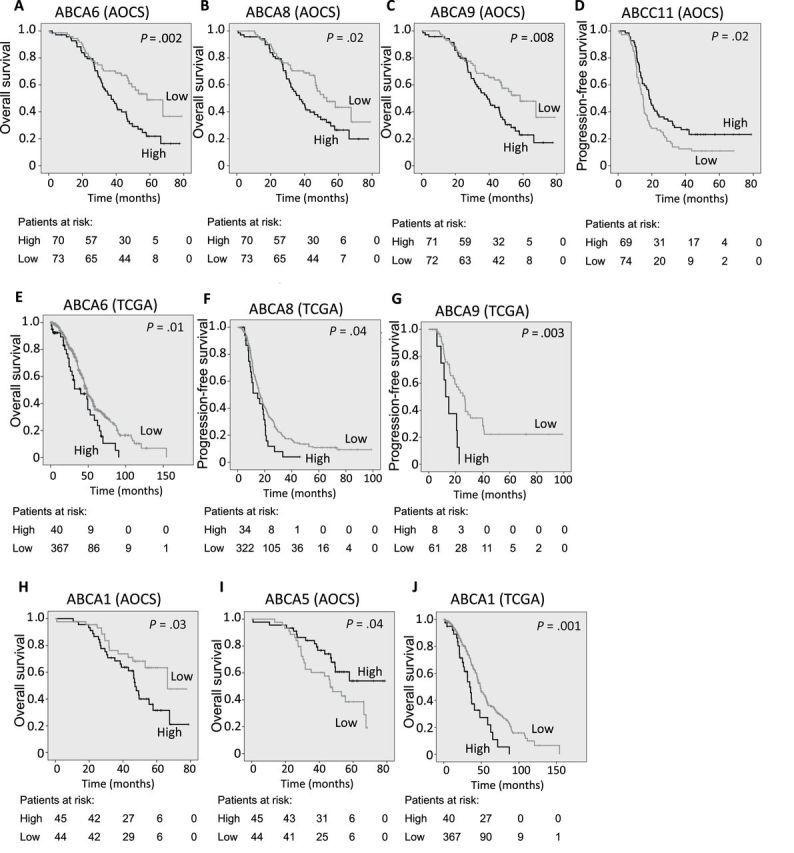
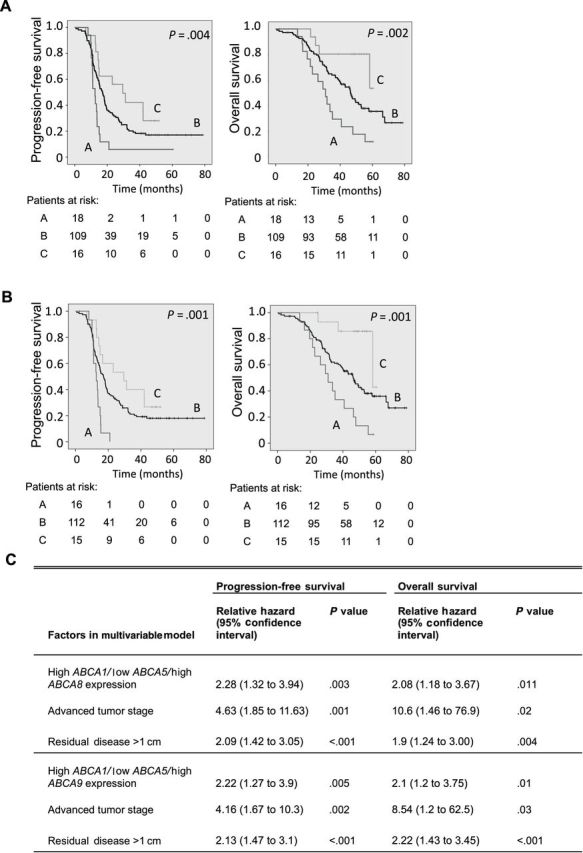
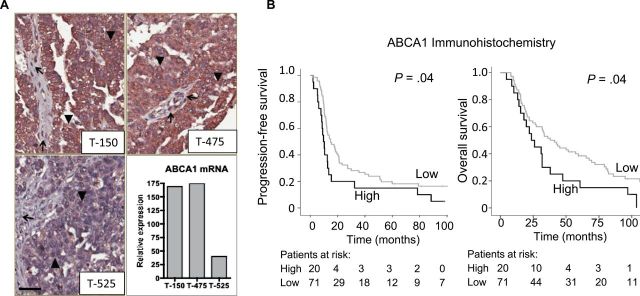
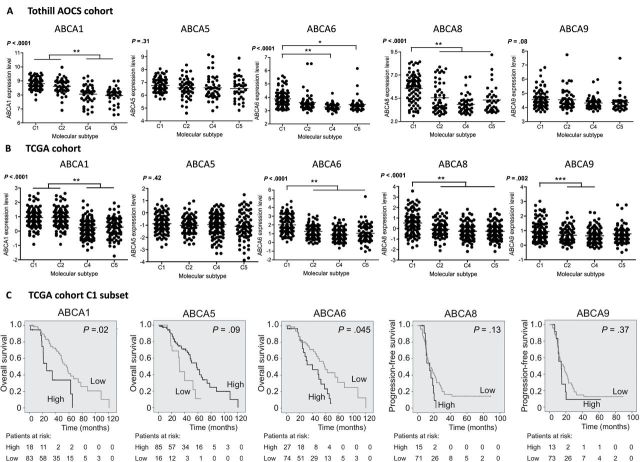
Ellen L Hedditch
Ellen L Hedditch
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,*,
Bo Gao
Bo Gao
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,*,
Amanda J Russell
Amanda J Russell
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,*,
Yi Lu
Yi Lu
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Catherine Emmanuel
Catherine Emmanuel
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Jonathan Beesley
Jonathan Beesley
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Sharon E Johnatty
Sharon E Johnatty
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Xiaoqing Chen
Xiaoqing Chen
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Paul Harnett
Paul Harnett
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Joshy George
Joshy George
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1;
Australian Ovarian Cancer Study Group1,
Rebekka T Williams
Rebekka T Williams
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Claudia Flemming
Claudia Flemming
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Diether Lambrechts
Diether Lambrechts
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Evelyn Despierre
Evelyn Despierre
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Sandrina Lambrechts
Sandrina Lambrechts
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Ignace Vergote
Ignace Vergote
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Beth Karlan
Beth Karlan
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Jenny Lester
Jenny Lester
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Sandra Orsulic
Sandra Orsulic
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Christine Walsh
Christine Walsh
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Peter Fasching
Peter Fasching
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Matthias W Beckmann
Matthias W Beckmann
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Arif B Ekici
Arif B Ekici
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Alexander Hein
Alexander Hein
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Keitaro Matsuo
Keitaro Matsuo
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Satoyo Hosono
Satoyo Hosono
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Toru Nakanishi
Toru Nakanishi
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Yasushi Yatabe
Yasushi Yatabe
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Tanja Pejovic
Tanja Pejovic
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Yukie Bean
Yukie Bean
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Florian Heitz
Florian Heitz
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Philipp Harter
Philipp Harter
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Andreas du Bois
Andreas du Bois
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Ira Schwaab
Ira Schwaab
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Estrid Hogdall
Estrid Hogdall
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Susan K Kjaer
Susan K Kjaer
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Allan Jensen
Allan Jensen
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Claus Hogdall
Claus Hogdall
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Lene Lundvall
Lene Lundvall
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Svend Aage Engelholm
Svend Aage Engelholm
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Bob Brown
Bob Brown
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
James Flanagan
James Flanagan
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Michelle D Metcalf
Michelle D Metcalf
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Nadeem Siddiqui
Nadeem Siddiqui
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Thomas Sellers
Thomas Sellers
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Brooke Fridley
Brooke Fridley
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Julie Cunningham
Julie Cunningham
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Joellen Schildkraut
Joellen Schildkraut
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Ed Iversen
Ed Iversen
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Rachel P Weber
Rachel P Weber
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Andrew Berchuck
Andrew Berchuck
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Ellen Goode
Ellen Goode
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
David D Bowtell
David D Bowtell
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Georgia Chenevix-Trench
Georgia Chenevix-Trench
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Anna deFazio
Anna deFazio
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Murray D Norris
Murray D Norris
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Stuart MacGregor
Stuart MacGregor
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,
Michelle Haber
Michelle Haber
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,*,
Michelle J Henderson
Michelle J Henderson
1Affiliations of authors:Children’s Cancer Institute Australia, Randwick, Australia (ELH, AJR, RTW, CF, MDN, MH, MJH); Department of Gynaecological Oncology and Westmead Institute for Cancer Research (BG, CE, AOCSG, AdF), Crown Princess Mary Cancer Centre and Westmead Institute for Cancer Research (PH) University of Sydney at the Westmead Millennium Institute, Westmead Hospital, Sydney, Australia; Queensland Institute of Medical Research, Brisbane, Australia (YL, JB, SEJ, XC, AOCSG, GC-T, SM); Peter MacCallum Cancer Centre, Melbourne, Australia (JG, AOCSG, DDB); Vesalius Research Center, VIB, Leuven, Belgium (DL); Department of Oncology, University of Leuven and University Hospitals Leuven, Leuven, Belgium (ED, SL, IV); Women’s Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA (BK, JL, SO, CW); Department of Gynecology and Obstetrics, University Hospital Erlangen, Friedrich-Alexander University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen-Nuremberg, Erlangen, Germany (PF, MWB, AH); University of California at Los Angeles, David Geffen School of Medicine, Department of Medicine, Division of Hematology and Oncology, David Geffen School of Medicine, University of California at Los Angeles, Los Angeles, CA (PF); Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, Aichi, Japan (ABE, KM, SH); Department of Gynecology (TN), and Department of Pathology and Molecular Diagnostics (YY), Aichi Cancer Center Central Hospital, Nagoya, Aichi, Japan; Department of Obstetrics and Gynecology (TP, YB), and Knight Cancer Institute (TP, YB), Oregon Health & Science University, Portland, OR; Institute of Human Genetics, Friedrich-Alexander-University Erlangen-Nuremberg, Comprehensive Cancer Center Erlangen Nuremberg, Erlangen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Kliniken Essen-Mitte/ Evang. Huyssens-Stiftung/Knappschaft GmbH, Essen, Germany (FH); Department of Gynecology and Gynecologic Oncology, Dr. Horst Schmidt Kliniken Wiesbaden, Wiesbaden, Germany (FH); Institut für Humangenetik, Wiesbaden, Germany (PH, AdB, IS); Danish Cancer Society Research Center, Unit of Virus, Lifestyle and Genes, Copenhagen, Denmark (EH, SKK, AJ); Molecular Unit, Department of Pathology, Herlev Hospital, University of Copenhagen, Copenhagen, Denmark (EH); Department of Obstetrics and Gynecology (SKK), Gynecology Clinic (CH, LL), and Department of Oncology (SAE), Rigshospitalet, University of Copenhagen, Copenhagen, Denmark; Department Surgery & Cancer, Imperial College London, London, UK (BB, JF, MDM); North Glasgow University Hospitals NHS Trust, Stobhill Hospitall, Glasgow, UK (NS); Department of Cancer Epidemiology, Moffitt Cancer Center, Tampa, FL (TS); Department of Health Science Research, Division of Epidemiology (BF, EG), and Department of Laboratory Medicine and Pathology (JC), Mayo Clinic, Rochester, MN; Department of Community and Family Medicine (JS, RPW), and Department of Obstetrics and Gynecology (AB), Duke University Medical Center, Durham, NC; Cancer Prevention, Detection & Control Research Program, Duke Cancer Institute, Durham, NC (JS); Department of Statistical Science, Duke University, Durham, NC (EI).
1,*