Summary
We are just starting to understand the functional importance of the non-coding genomes in development and disease. This article will review the emerging effort that investigates the functions of small ncRNAs, long ncRNAs and retrotransposons in the development of cancer.
Abstract
Neoplastic transformation is caused by accumulation of genetic and epigenetic alterations that ultimately convert normal cells into tumor cells with uncontrolled proliferation and survival, unlimited replicative potential and invasive growth [Hanahan,D. et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674]. Although the majority of the cancer studies have focused on the functions of protein-coding genes, emerging evidence has started to reveal the importance of the vast non-coding genome, which constitutes more than 98% of the human genome. A number of non-coding RNAs (ncRNAs) derived from the ‘dark matter’ of the human genome exhibit cancer-specific differential expression and/or genomic alterations, and it is increasingly clear that ncRNAs, including small ncRNAs and long ncRNAs (lncRNAs), play an important role in cancer development by regulating protein-coding gene expression through diverse mechanisms. In addition to ncRNAs, nearly half of the mammalian genomes consist of transposable elements, particularly retrotransposons. Once depicted as selfish genomic parasites that propagate at the expense of host fitness, retrotransposon elements could also confer regulatory complexity to the host genomes during development and disease. Reactivation of retrotransposons in cancer, while capable of causing insertional mutagenesis and genome rearrangements to promote oncogenesis, could also alter host gene expression networks to favor tumor development. Taken together, the functional significance of non-coding genome in tumorigenesis has been previously underestimated, and diverse transcripts derived from the non-coding genome could act as integral functional components of the oncogene and tumor suppressor network.
Introduction
One striking observation from the Human Genome Project is the existence of only ~25 000 protein-coding genes, a surprisingly low number that does not seem to scale with human developmental and pathological complexity. The genomic regions with protein-coding capacity only account for 1.5% of the human genome; and instead, a vast proportion of non-coding genomes in mammals are clearly correlated with the extent of their genomic complexity in evolution (1). The mammalian non-coding genomes include sequences encoding introns, cis-regulatory elements, non-coding RNAs (ncRNAs), and most abundantly, repetitive elements (2,3). Contrary to the conventional wisdom that the non-coding sequences have little functional importance, emerging evidence has revealed cell type- and context-dependent transcriptional activity within these non-coding genomic regions, and further highlighted their important biological functions in development and disease (4,5).
Recent advance in sequencing technology has revolutionized the functional characterization of the non-coding genome. Numerous ncRNAs and transposable elements in the mammalian genome exceed protein-coding genes in numbers and in functional complexity. Emerging evidence has demonstrated that the genetic and epigenetic alteration of protein-coding genes cannot constitute the entire molecular basis underlying the pathogenesis of tumor development. It is increasingly clear that the functional importance of the non-coding genome in cancer biology, particularly that of ncRNAs and transposable elements, has been largely overlooked until recently. ncRNAs do not possess protein-coding capacity, and can be further divided into small ncRNAs (<200 nt) and long ncRNAs (>200 nt), purely based on the length of the molecules. The majority of ncRNAs that impact tumorigenesis act to regulate gene expression through a diverse range of molecular mechanisms. Transposable elements, on the other hand, contain abundant retrotransposons and DNA transposons, with retrotransposons starting to emerge with a potential role in promoting tumorigenesis. Here, we will review the recent advance in cancer biology to reveal the functional importance outside the protein-coding genome.
MicroRNAs as integral components of oncogene and tumor suppressor network
Small ncRNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi interacting RNAs, refer to a class of ncRNAs that are less than 200 nt in length. As the best functionally characterized small ncRNAs, miRNAs regulate gene expression through posttranscriptional repression (6). Nascent miRNA transcripts (pri-miRNAs) are processed sequentially by two ribonuclease III enzymes, Drosha and Dicer, to yield mature miRNAs (7,8). Upon maturation, one strand of the miRNA duplex is selectively incorporated into the RNA-induced silencing complex, subsequently mediating the posttranscriptional gene silencing of specific mRNA targets through imperfect complementarity (9). The specificity of the miRNA-mRNA binding is often, although not exclusively, achieved by a perfect base-pairing at the miRNA ‘seed’ region—the 2–7 nucleotides at the miRNA 5′ end (10). miRNA-mediated posttranscriptional silencing can occur through degradation of target mRNAs (11,12), and/or inhibition of protein synthesis at the initiation stage (13) (Figure 1). Owing to their small size and imperfect base-pairing with the targets, miRNAs have the capacity to regulate many target mRNAs, and therefore act as global regulators for gene expression.
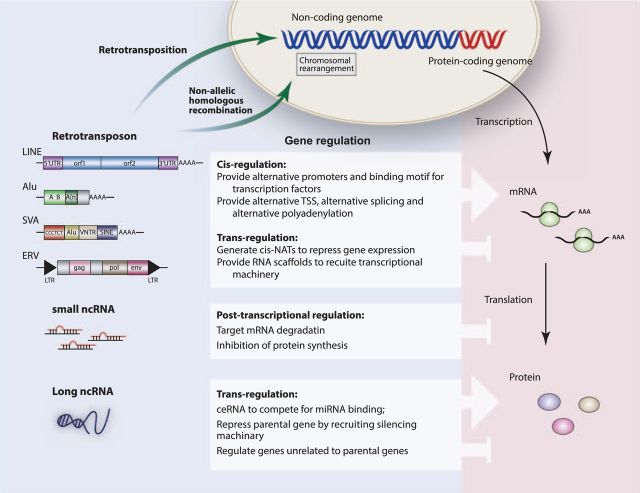
Figure 1.
A summary of diverse transcripts derived from the non-coding genome and their potential roles in cancer development.
The connection between miRNAs and cancer was first suggested by their frequent genomic alteration and dysregulated expression in various human tumors (14). Subsequently, the first oncogenic miRNA, mir17-92, have been characterize with potent oncogenic activity both in mouse models and in cell culture studies (15). To date, many miRNAs have been identified to promote or suppress oncogenesis in mouse tumor models, cell culture systems and clinical studies, regulating nearly all essential cellular processes during tumorigenesis. Consistently, both specific miRNAs and components of the global miRNA biogenesis machinery undergo genetic and epigenetic alterations in a variety of human cancers (16,17). Aberrant alteration of miRNA levels and activities often lead to aberrant dosage of their target genes, which often dictates aberrant functional readouts of multiple molecular pathways during tumorigenesis. In addition, a single miRNA could regulate a specific oncogene or tumor suppressor pathway by repressing multiple components, and ultimately provide selective advantages at different stages of tumor development. Taken together, miRNAs are integral components of the oncogene and tumor suppressor network. Their functional significance during cancer development makes these small RNAs great candidates for novel diagnostic markers and therapeutical targets. In fact, the miRNA mimics to miR-34a, a p53-regulated miRNA with potent tumor suppressor effects upon overexpression, has reached Phase I studies in clinics for treating liver cancer. The functions of miRNAs in cancer development is a subject of extensive discussion in a few recent reviews (17,18), and will not be the focus of this article.
A potential role for retrotransposons in tumor development
Nearly half of the mammalian genome consists of transposable elements, which are divided into DNA transposons and retrotransposons. DNA transposons undergo excision and reintegration through a self-encoded transposase enzyme, using a ‘cut-and-paste’ mechanism that does not increase their copy number. In contrast, retrotransposons propagate by a ‘copy-and-paste’ mechanism, utilizing an RNA intermediate that is reverse-transcribed into DNA before integration into a new genomic locus. Thus, retrotransposons can quickly accumulate within host genomes, and greatly outnumber other species of transposable elements (2). The conventional wisdom depicts retrotransposons as selfish genomic parasites that remain strictly silenced in somatic tissues. However, retrotransposons occasionally escape transcriptional silencing under specific developmental and pathological contexts; the resulting aberrant propagation can compromise host fitness, at least in part due to mutagenic events and increased genomic instability. Furthermore, increasing evidence also suggests that at least a subset of retrotransposon elements could act both in cis and in trans to alter the structure and expression of adjacent protein-coding genes (19,20). Thus, reactivation of retrotransposons during tumor development could contribute to oncogenesis through multiple mechanisms (Figure 1).
In mammals, retrotransposons constitute ~40% of the genome, and can be further classified as long-terminal repeat (LTR) retrotransposons or non-LTR retrotransposons. LTR retrotransposons, also called endogenous retroviruses (ERVs) in mammals, represent evolutionary remnants of ancient retroviral invasions into the host genome. ERVs consist of two LTRs flanking an internal sequence that contains gag, pol and env genes encoding the core viral proteins. ERVs are further grouped into three classes on the basis of homology to exogenous retroviruses: Class I (including MuLV and VL30), Class II (including IAP and ERVK) and Class III (including ERVL and MaLR (21)). Although most ERVs are heavily mutated and/or truncated, a small number of retrotransposon elements in mammalian genomes resemble intact exogenous retroviruses in overall structure and retrotransposition capacity. Interestingly, the human ERVs (HERVs) are present as molecular fossils, the majority of which have no retrotransposition activity as a result of accumulated mutations and truncations (2,22). Nevertheless, HERVs still confer strong gene regulatory effects, possibly contributing to malignant transformation of a variety of cell types (see below). Non-LTR retrotransposons mainly comprise long interspersed elements (LINEs) and short interspersed elements (SINEs). Hominid genomes also contain SVAs, elements that each consists of a SINE region, a Variable number of tandem repeats and an Alu-like sequence. In contrast to the inactive human ERVs, a small number of intact LINE-1, Alu and SVA elements still remain active in retrotransposition (23–25), and de novo integration events caused by all three retrotransposon classes have been described in a variety of human tumors (26–28).
Expression regulation of retrotransposons in cancer
As remnants of integrations of exogenous retroviruses and self-propagation during evolution, retrotransposons are thought to be mostly silenced by host genome surveillance through various mechanisms, including DNA hypermethylation (29,30), histone modification, polycomb complex interaction (31), Piwi-interacting RNA and endo-siRNA-mediated silencing (32,33). Nevertheless, a surge of specific retrotransposon expression is observed during specific stage of embryogenesis and germ line development, possibly through multiple intricate machineries that precisely regulate such processes (31,34–38).
The delicate retrotransposon suppression network is likely dysregulated in cancer cells, as a result of genetic and epigenetic alterations. Cancer genomes are overall hypomethylated in the repeat-rich heterochromatin regions, a pattern that could lead to the transcriptional activation of these repeats (39,40). In addition, aberrant retrotransposon expression in cancer cells could be induced by stress stimuli, including, but not limited to, metabolic stress, unfavorable tumor microenvironment and genotoxic agents. Stress-induced activation of retrotransposons has been observed in many organisms. Heat shock and cycloheximide treatment can rapidly increase Alu and SINE levels (41); benzo(a)pyrene (BaP), a ubiquitous environmental carcinogen, can upregulate L1 RNA levels and increase L1 retrotransposition activity (42), by decreasing DNA methylation and increasing H3K4 trimethylation (H4K4Me3) and H3K4 acetylation (H3K9Ac) at LINE-1 promoters (43). Given these observations, it is likely that genetic and epigenetic alterations in specific cancer types induce aberrant retrotransposon expression that has profound functional impact on tumor development.
Retrotransposition of reactivated non-LTR retrotransposons in human cancer
In the human genome, only a small number of intact non-LTR retrotransposons, including LINE, Alu and SVA, have the ability for retrotransposition. The human genome has nearly 500 000 LINE elements and 1 500 000 SINE elements. Most tumor-specific retrotransposition events identified so far are attributed to the aberrant activity of LINE-1 (L1) retrotransposon class, due to its abundance in the genome, as well as its intact and autonomous retrotransposition machinery. L1 retrotransposition leads to insertional mutagenesis events that are potentially detrimental to the host genome (23). The full length of the human L1 retrotransposon is ~6kb, including a 5′ untranslated region (5′ UTR) with a RNA polymerase II promoter, two open reading frames (ORFs), and a 3′ UTR that contains a polyadenylation signal and a polyA tail. ORF2 encodes a ~150-kDa protein that has reverse transcriptase and endonuclease activities crucial for LINE1 retrotransposition (44,45), whereas the protein encoded by ORF1 forms a trimer that serves as a RNA chaperone (46,47). The intact L1 can directly retrotranspose into genomic DNA through a mechanism called target-primed reverse transcription, during which the ORF2p generates a single-strand endonucleolytic nick at target genomic loci. The liberated 3′-OH then is used as a primer by the L1 reverse transcriptase to initiate cDNA synthesis using the L1 mRNA as a template. The second strand of targeted DNA then is cleaved and used to prime second-strand synthesis. The signatures of this L1 integration include 5′ truncation, shortening of oligo(dA) at the 3′ end, and target site duplications of 7–20 base pairs in length (48). L1s can also insert into transcribed genes, in either the sense or antisense orientation, and retrotranspose sequences derived from their 3′ flanks to new genomic locations (49). By doing so, L1 has the potential to serve as a vehicle to mobilize fragments of protein-coding genes into other genomic loci to create new genes, or to alter the expression of existing genes (50,51). These L1 retrotransposition events provide a mechanism for insertional mutagenesis in somatic cells, possibly yielding various pathological conditions, including cancer (Figure 1).
Of the 500 000 copies of LINE elements in the human genome, the majority of L1 elements have lost their retrotransposition activity by truncation, inversion, mutation or recombination (52,53), leaving fewer than 100 copies competent for retrotransposition (2). It has been long suspected that L1 retrotransposition is involved in cancer development, yet the repetitive and diverse nature of L1 elements poses a technical difficulty in identifying cancer-specific integration sites. Historically, the identification of de novo L1 insertion was performed by analyzing new L1 insertion sites near well-defined tumor suppressors or oncogenes using classic molecular biology approaches. For example, new L1 integration has been observed by Southern analysis in the last exon of the tumor suppressor adenomatous polyposis coli in colorectal cancers, which disrupt intact adenomatous polyposis coli function to control cell growth (26). Another classic example of L1 insertion in cancer was observed in canine transmissible venereal tumor, where L1 insertion occurs (at high frequency) 5′ to the first exon of the proto-oncogene c-myc, driving high c-Myc expression and predisposing cells to malignant transformation (54–56).
Recent advances in high-throughput sequencing technology have revolutionized our ability to analyze L1 activity during tumor development (57). The first effort to identify novel somatic L1 insertions on a genome-wide scale was performed in human lung tumors using L1-Ta junction PCR followed by pyrosequencing. In that study, tumor-specific somatic L1 insertions are observed to occur at high frequency in lung cancer cells, but not in adjacent healthy tissues (58). The prevalence of L1 insertion was further confirmed in colorectal cancers using whole-genome paired-end sequencing (27,28). Here, a third of tumor-specific L1 insertions lead to the disruption of annotated protein-coding genes, some of which have implicated tumor suppressor functions with reoccurring mutations. Furthermore, the oncogenic role of somatic L1 insertion also is demonstrated in a subset of hepatocellular carcinomas (59), where either inherited or somatic de novo L1 integrations possibly yield a strong oncogenic effect. In this study, a germ line L1 insertion occurs in the tumor suppressor MCC (mutated in clolorecal cancers) gene, which ablated MCC expression and activates oncogenic β-Catenin/Wnt signaling. In addition, a tumor-specific L1 inserted into the enhancer region of tumorigenicity 18 (ST18), a candidate oncogene in hepatocellular carcinoma, thus aberrantly activating ST18 by interrupting its negative feedback loop (59). The exact molecular mechanism governing the cancer-specificity of L1 reactivation remains largely unclear; in addition to target-primed reverse transcription, multiple mechanisms have been proposed by which L1 mobilize to alter cancer genome.
To date, de novo L1 insertions have been found in a number of human tumor types, including colon cancer, lung cancer, ovarian cancer, prostate cancer and hepatocellular carcinoma (59). In these studies, aberrant L1 retrotransposition activity varies among tumor types and among individual patients; and it is yet to be determined whether the occurrence of L1 retrotransposition has any prognostic value. Interestingly, de novo L1 insertions in somatic tissues may also occur in normal development. Although still controversial, researchers have reported a high level of somatic LINE-1 retrotransposition in human neuronal progenitor cells, which is likely to alter the transcriptome dynamics in mature neurons and contribute to their plasticity(60). Taken together, aberrant L1 insertion in somatic tissue could constitute a novel mechanism for mutagenesis of the cancer genome. The findings described above could have profound impact on our understanding of the pathogenesis of cancer.
Alu and SVA, two non-LTR retrotransposon classes, also are capable of retrotransposition, although their retrotransposition frequency is much lower than that of L1s in human cancer (61). Unlike the intact L1 elements, which encode all machineries required for retrotransposition, both Alu and SVA elements hijack the activated L1 ORF2p protein in trans to achieve retrotransposition (62). Alu elements constitute the most abundant class of transposable elements in human, with ~1 000 000 copies interspersed throughout the genome. Alu elements are derived from the small cytoplasmic 7SL RNA, without any protein-coding capacity. The transcription of Alu elements is initiated by a RNA polymerase III-binding promoter (63), yet their low-fidelity transcription generates many mutations during the expansion of Alu elements in the genome. Most of the intact mobile Alu elements belong to a young Alu family, AluY, whose de novo insertions are mostly confined to the non-coding genome (64), but occasionally disrupting protein-coding genes to compromise the integrity of the tumor suppressor network (65). SVA is a human-specific retrotransposon comprising ~7000 copies in the genome. Although de novo SVA insertions have been associated with some human diseases (such as X-linked agammaglobulinaemia and Fukuyama-type congenital muscular dystrophy) (66,67), cancer-causing SVA insertions are yet to be identified and validated.
Retrotransposons mediate chromosomal rearrangement
The presence of abundant and highly homologous retrotransposon elements in the human genome can mediate non-allelic homologous recombination (NAHR). NAHR is initiated by double-strand breaks, followed by homologous recombination between two highly similar DNA fragments (usually >1kb apart). One of the single-stranded DNA tails formed at the break site invades the non-allelic homologous DNA duplex, forming a displacement-loop, which then is extended by DNA synthesis. The 3′ single-stranded DNA tail then is captured and forms a double Holliday junction. Depending on position, orientation and resolution of this junction, NAHR between two identical or highly homologous Alu elements can result in deletion, inversion, duplication or translocation. Since cancer cells tolerate high level of DSBs by compromising checkpoint machinery, evading apoptosis and overexpressing DSB repair proteins, retrotransposon-mediated NAHR could contribute to frequent chromosomal rearrangement in cancer cells (Figure 1), possibly leading to pathogenic copy number variations (68,69).
Unlike L1 elements that are enriched in intergenic regions, Alu elements are preferentially enriched in gene-rich regions (70,71). The chromosomal rearrangements mediated by Alu–Alu homologous recombination can have direct functional impacts on protein-coding genes. For example, BRCA1 and BRCA2 are important DNA repair proteins, mutations of which are associated with inherited breast/ovarian cancer. Interestingly, high densities of repetitive elements occur in both loci, with BRCA1 consisting of 42% Alu sequences and BRCA2 containing 20% Alu and 27% LINE sequences (72–74). Thus, both BRCA1 and BRCA2 genes are susceptible to NAHR-mediated mutagenesis, as demonstrated in patients with hereditary breast/ovarian cancer. In recent studies, an 89-kb deletion encompassing BRCA1 exons 7–11 and a 23-kb deletion containing BRCA1 exons 11–15 were both found to be flanked by two highly homologous Alu elements (75). Not surprisingly, L1-mediated NAHR has not been identified in cancer, possibly due to lower L1 recombination frequency and/or a negative selection pressure imposed by deletion of the much larger L1 element.
Similarly, active ERVs also have the potential to mediate chromosomal rearrangements. The human ERV type K (HERV-K) is the best-characterized ERV associated with increased genomic variation. Members of the HERV-K superfamily are considered to be the most recent and active members of human ERVs. There are ~550 HERV-K loci and 6400 HERV-K derived solo LTRs in the human genome (76). Although evidence indicates the occurrence of HERV-K-mediated chromosomal rearrangements during human genome evolution (77,78), it is unclear whether the divergence and low copy number of HERV-K would permit NAHR in the cancer genome.
Retrotransposons confer regulation of adjacent genes.
While L1 and Alu retrotransposition occur in a tumor type-specific manner and possibly generates mutagenic events to promote tumor progression, transcriptional regulation conferred by aberrantly retrotransposon derepression could also constitute a mechanism dictating an aberrant transcriptional program that favor tumor development. In 1950s, when Barbara McClintock first discovered transposable elements, she speculated that such sequences could act as mobile ‘controlling elements’ that regulate host gene transcription and alter phenotypes. Indeed, a subset of mammalian retrotransposons resides at the vicinity of protein-coding genes, and could have the capacity to alter the structure and expression of adjacent host genes upon their escape of the transcriptional silencing (79).
Retrotransposons, particularly LTR retrotransposons (ERVs) and their evolutionary remnants, impact the neighboring genes through a variety of mechanisms. First, ERV reactivation, either as a solo-LTR or as an intact ERV element, yields a strong viral LTR promoter that could act to enhance the expression level of adjacent genes on the same strand (80). Additionally, ERVs could reside at gene promoters, enhancers or silence/insulator regions, and provide binding motifs for important oncogenic and tumor suppressor transcription factors to confer a unique transcriptional regulation on the adjacent genes (81). Second, an intronic retrotransposon, upon its reactivation, could lead to alternative transcription start site usage, alternative splicing or alternative polyadenylation, thus generating a unique isoform of the adjacent gene (79,82,83). Third, antisense transcripts could be derived from retrotransposons that overlap with adjacent genes in the opposite orientation, and subsequently disrupt the expression of the neighboring genes with sequence complementarity (84). Finally, retrotransposon transcript could act as an RNA scaffold in trans to recruit epigenetic machineries to confer transcriptional regulation on nearby genes (20). Taken together, reactivation retrotransposons, even those without retrotransposition capacity, could confer strong regulation on adjacent genes (Figure 1).
Consistent with this idea, 18.1 and 31.4% of transcription start sites map within transposable elements in the mouse and human genomes, respectively. However, retrotransposon-initiated transcription varies considerably among tissues and cell types, with one of the strongest representations observed in embryonic tissues (85). This result is consistent with widespread ERV derepression during normal embryonic development, particularly in preimplantation embryos (86). One of the best-characterized examples is the derepression of MERVL retrotransposons in two-cell-stage mouse embryos. The MERVL-mediated regulation on adjacent protein-coding genes constitutes the molecular basis for the unique developmental potential of two-cell embryos (19). Given the gene regulatory effects caused by retrotransposon derepression in normal embryonic development, it is plausible that cancer cells, particularly cancer stem cells, could harbor the same retrotransposon-initiated transcription activity that occurs in embryonic cell types. Thus, retrotransposon derepression could yield altered expression and structure of the adjacent genes, possibly constituting at least one mechanism for the frequent alternative transcription start site usage observed in various cancers (87–90).
Intronic retrotransposons can alter the intron–exon distribution of the RNA transcripts by enforcing alternative splicing, intron retention, exonization and/or premature polyadenylation. Almost 90% of multiexon human genes undergo alternative splicing and generate multiple splice variants during development and cell differentiation (91–93). So far, most relevant studies have focused on the role of SINEs in mediating alternative splicing. Both the sense and antisense strands of Alu contain a number of potential splice sites; therefore, the pre-mRNAs containing Alu sequence will be recognized by the splicing machinery and lead to exonization of a Alu fragment (82). The abundance of Alu in gene-rich regions provides a repertoire of alternative splicing sites. In fact, Alu exonization is such a widespread phenomenon that in the human brain as many as 50% transcripts contain Alu sequence (94). In addition, adenosine-to-inosine (A-to-I) RNA editing by ADAR protein on intramolecular pair of inverted Alu repeats can create or eliminate splicing signals on pre-mRNAs. As 90% of A-to-I editing occurs on intronic Alu sequences, these Alu-containing transcripts can be subject to aberrant splicing (95). Similarly, there is evidence suggesting that ERVs also can disrupt transcription by regulating splicing. By sequence prediction, human ERVs such as ETn, HERV-W and HERV9 show strong internal exonization by providing cryptic splice donor/splice acceptor sequences or polyadenylation signal in an orientation-sensitive manner (96). Cancer cells have been shown to display a change in transcript splicing pattern from normal cells (97,98). Many critical genes involved in cell proliferation and DNA damage have cancer-specific variants resulting in dysfunctional or even antagonizing proteins. Dysregulation of the splicing machinery and RNA editing have been reported in human cancers (99,100); these defects might target retrotransposons-containing pre-mRNAs, thereby generating oncogenic splicing variants.
Another mechanism through which retrotransposons could regulate adjacent gene expression is through the generation of antisense transcripts. When retrotransposons and corresponding adjacent genes are located on opposite strands, the retrotransposon-associated transcription can generate cis-natural antisense transcripts (cis-NATs). Indeed, transcriptome analysis has demonstrated that there are extensive occurrences of antisense transcription of retrotransposon cis-NATs (86). The functions of these antisense transcripts are still puzzling. However, some ERV families exhibit antisense bias in regions close to genes, suggesting some degree of selection for this antisense orientation (96,101). The antisense ERV transcription, as seen with ERV9, has been observed to strongly associate with the transcriptional disruption of adjacent genes (102,103). A LINE-1 derived antisense, LCT13 has been show to direct target metastasis-suppressor gene TFPI-2 and suppress its expression and impact on tumor progression (104). In some case, these cis-NATs can be further processed into miRNAs (105) or RNAs complementary to the adjacent gene to mediate this posttranscriptional repression (106).
Whether retrotranspositions contribute to or are simply consequence of the massive genomic changes that occur throughout cancer genomes is not yet clear. Assessment of the phenotypic effect of an insertion will require evaluation of the selective advantage imparted to the populations of cells that possess specific novel insertions. If tumors cells that contain new L1 insertions can undergo colony expansion, these insertions would be enriched, generating sufficient reads to be detected in high-throughput sequencing. Given the clonal nature of cancer evolution, a single clone that possesses metastatic capability or drug resistance will be further selected during tumor progression (107). Therefore, opportunities to capture the endogenous retrotransposition events are likely to be higher in the tumor metastases and drug-resistant samples. Current studies of endogenous retrotransposons is impeded by limitations in accurate alignment and mapping of repetitive sequences, and further hindered by a lack of functional assays. The development of more sophisticated tools for genome sequencing and computational analysis will be critical for the progression of these critical studies.
A potential role of long ncRNAs in cancer development
The functional importance of long ncRNAs (lncRNAs) in development has been well demonstrated for decades. A number of classic lncRNAs act as the key regulators for multiple essential developmental processes, such as X-inactivation, dosage compensation and imprinting. Currently, lncRNAs are defined as ncRNAs greater than 200 nucleotides in length, and encompass a board spectrum of different RNA classes, including enhancer intergenic RNAs (formerly lincRNAs), RNAs (eRNAs), circular RNAs, pseudogenes and sense and antisense RNAs overlapping other protein-coding or non-coding transcripts (108). This classification of lncRNAs by length defines an ncRNA class different from small ncRNAs. lncRNAs have diverse structural features, expression patterns and functional readout. Next-generation sequencing projects, such as FANTOM (Functional Annotation of Mammalian cDNA), have revealed the abundance and the complexity of numerous ncRNAs across human genome, which exceed the protein-coding genes in numbers and complexity (109).
We are only starting to understand the realm of ncRNA biology and the diverse mechanisms through which they regulate development and disease. Contrary to protein-coding genes and miRNAs that are largely evolutionarily conserved, lncRNAs often exhibit much weaker conservation, and even lncRNAs with strong biological phenotypes, such as Air and Xist, are poorly conserved, suggesting that evolutionary selection on lncRNAs largely act to preserve the RNA structure rather than the primary sequence (110). Despite the diverse structure features, expression patterns and mechanisms of action, lncRNA functions often converge on gene regulation. lncRNAs could target transcription factors, basal transcription machinery and even DNA to mediate transcriptional regulation. lncRNAs also mediate posttranscriptional regulation, at least in part by incomplete base-pairing with complementary mRNA to regulate pre-mRNA processing, splicing, transport, translation and degradation. Finally, lncRNAs could act as integral components of chromatin complexes, mediating epigenetic regulation via recruiting and directing chromatin modifying complexes to the target loci (Figure 1).
Expression studies have identified considerable lncRNA species with cancer-specific alterations in various tumor types at distinct stages of cancer progression. lncRNA MALAT1 is aberrantly upregulated during metastasis of non-small cell lung cancer, and acts as an early prognostic marker for poor survival (111). LncRNAs HOTAIR and HULC also exhibit strong expression level specifically in cancer. In addition, specific lncRNAs are regulated transcriptionally by key oncogene and tumor suppressor pathways, and likely to act as integral components of such signaling network (112). For example, lincp21 is a bona fide p53 transcriptional target, whose upregulation mediate a portion of p53 downstream effects (113).
Despite the ample examples of altered lncRNA expression in cancer, genomic analyses and functional studies has yet to generate a comprehensive understanding on lncRNA functions in cancer. Nevertheless, the importance of specific lncRNAs in tumor development has started to emerge. For example, the classic lncRNA Xist has a potent tumor suppressor effect. The deletion of Xist in the blood compartment leads to a highly aggressive myeloproliferative neoplasm and myelodysplastic syndrome in mice due to aberrant X reactivation and multiple autosomal changes (114). In addition, some protein-coding gene loci, including well-defined oncogenes and tumor suppressors, generate antisense transcripts, which in turn cause DNA replication and mitotic anomalies, genome instability and dysregulation of the hematopoiesis pathway (115). Cancer cells also employ antisense RNAs to repress the transcription of tumor suppressor genes by epigenetic mechanisms. One of the best examples is an antisense transcript derived from the tumor suppressor locus, p15, which enhances the heterochromatin and DNA methylation of p15 to transcriptionally silence this important tumor suppressor gene (116).
Among the diverse lncRNAs, pseudogenes represent an ncRNA class that has long been speculated to impact on tumor development. Pseudogenes are defective relatives of parental protein-coding genes, such that the pseudogenes have lost protein-coding capacity due to accumulation of mutations. There are two types of pseudogenes: non-processed pseudogenes (believed to have originated by gene duplication) and processed pseudogenes (believed to have arises by retrotransposition) (117–119). In addition to the 26 000 well-annotated protein-coding genes, the human genome is estimated to contain more than 17 000 pseudogenes, two-thirds of which are of the processed type (120). Originating from mRNA transcripts, processed pseudogenes do not contain introns and typically are located on different chromosomes from that of the gene of origin. Although reverse transcriptase activity of L1 has been implicated to act in trans in generating processed pseudogenes, such pseudogenes do not necessarily associate with L1 sequences. Given the aberrant L1 activity frequently observed in cancer cells (121), it is likely that de novo generation of somatic pseudogenes arise from aberrant retrotransposon activity during tumorigenesis. Consistently, novel pseudogenes have been identified from various human cancers using next-generation sequencing.
Despite the diversity of pseudogenes in the human genome, little is known about the function of these non-coding transcripts. Emerging evidence suggests that pseudogenes are not merely ‘junk DNA’; instead, pseudogenes could exert physiological and pathological functions through distinct mechanisms. First, pseudogene transcripts are proposed to function as competitive endogenous RNAs competing with the parental gene for miRNAs or RNA-binding proteins (122,123). Although this decoy mechanism is still under debate, it could constitute a mechanism through which pseudogenes regulate the expression and function of their parental genes, by allowing the parent loci to bypass miRNA-mediated posttranscriptional-silencing or RNA-binding-protein-mediated gene regulation. For example, PTENP1, a pseudogene of the tumor suppressor PTEN, is frequently downregulated in various cancer types. PTENP1 serves as a decoy for PTEN-targeting miRNAs to maintain a high-level expression for PTEN, and PTENP1 downregulation/deletion leads to reduced PTEN level (124). By analogy, pseudogenes of key oncogenes and tumor suppressors are likely to be integral components of the oncogene and tumor suppressor pathways. Second, antisense transcripts derived from the pseudogene loci can be recruited to the parental genomic loci for transcription repression. For instance, the antisense RNA derived from OCT4-pg5, an OCT4 pseudogene can recruit silencing complex to Oct4 promoter region and repress its transcription (125). Finally, pseudogene transcripts also could function as long ncRNAs and affect the expression of genes unrelated to the parental genes. The best example is lncRNA Xist, a major effector of X-inactivation, which is a pseudogene of functional-unrelated Lnx3 protein-coding gene (126). Taken together, these data suggest that pseudogenes might confer unexpected gene regulatory activity, potentially acting as integral components of the oncogene and tumor suppressor network.
Conclusion
Once regarded as ‘junk DNA’, the functional importance of the non-coding genome is increasingly recognized in human development and disease. Both small ncRNAs and lncRNAs are integral components of the oncogene and tumor suppressor network. Their genetic and epigenetic alterations have profound functional impacts on the expression of specific protein-coding and non-coding genes through diverse mechanisms. In addition, retrotransposons could be aberrantly expressed and/or activated during tumor development. Being the most abundant elements in the human genome, a subset of reactivated retrotransposons could mediate insertional mutagenesis, genome recombination and gene conversion and, more interestingly, regulate the structure and expression of adjacent genes. The key challenge we now face is to pinpoint, with gene-specific resolution, the specific non-coding loci that have disease-causing functions. With the recent advances in sequencing technology, we are finally able to identify specific transcripts from the non-coding genome specific to cancer cells, and to define the retrotransposon-mediated regulation of adjacent genes. In the near future, the battlefield of the war against cancer is likely to hinge upon our thorough understanding about this ‘dark matter’ of our genome.
Acknowledgments
We thank members of the He lab for their helpful discussion. L.H. acknowledges the support of an RO1 grant from NCI (R01 CA139067); a new faculty award from California Institute of Regenerative Medicine (CIRM, RN2-00923-1); A research grant from Tobacco-Related Disease Research Program (21RT-0133), and a research scholar award from American Cancer Society (ACS, 123339-RSG-12-265-01-RMC).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- ERV
endogenous retrovirus
- LINE
long interspersed element
- LTR
long-terminal repeat
- miRNA
microRNA
- NAHR
non-allelic homologous recombination
- ncRNA
non-coding RNA
- ORF
open reading frame
- SINE
short interspersed element.
References
- 1. Batista P.J., et al. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell, 152, 1298–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cordaux R., et al. (2009). The impact of retrotransposons on human genome evolution. Nat. Rev. Genet., 10, 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goodier J.L., et al. (2008). Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell, 135, 23–35 [DOI] [PubMed] [Google Scholar]
- 4. Licatalosi D.D., et al. (2010). RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet., 11, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rinn J.L., et al. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem., 81, 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He L., et al. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet., 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 7. Lee Y., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature, 425, 415–419 [DOI] [PubMed] [Google Scholar]
- 8. Lee Y.S., et al. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell, 117, 69–81 [DOI] [PubMed] [Google Scholar]
- 9. Hutvágner G., et al. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060 [DOI] [PubMed] [Google Scholar]
- 10. Bartel D.P. (2009). MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tuschl T., et al. (1999). Targeted mRNA degradation by double-stranded RNA in vitro . Genes Dev., 13, 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammond S.M., et al. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature, 404, 293–296 [DOI] [PubMed] [Google Scholar]
- 13. Pillai R.S., et al. (2005). Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science, 309, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 14. Zhang L., et al. (2006). microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA, 103, 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He L., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature, 435, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macfarlane L.-A., et al. (2010). MicroRNA: biogenesis, function and role in cancer. Curr. Genomics, 11, 537–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esquela-Kerscher A., et al. (2006). Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer, 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 18. Mendell J.T., et al. (2012). MicroRNAs in stress signaling and human disease. Cell, 148, 1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Macfarlan T.S., et al. (2012). Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature, 487, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu X., et al. (2014). The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat. Struct. Mol. Biol., 21, 423–425 [DOI] [PubMed] [Google Scholar]
- 21. Stocking C., et al. (2008). Murine endogenous retroviruses. Cell. Mol. Life Sci., 65, 3383–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lander E.S., et al. (2001). Initial sequencing and analysis of the human genome. Nature, 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 23. Beck C.R., et al. (2011). LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet., 12, 187–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang C.R., et al. (2010). Mobile interspersed repeats are major structural variants in the human genome. Cell, 141, 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang H., et al. (2005). SVA elements: a hominid-specific retroposon family. J. Mol. Biol., 354, 994–1007 [DOI] [PubMed] [Google Scholar]
- 26. Miki Y., et al. (1992). Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res., 52, 643–645 [PubMed] [Google Scholar]
- 27. Solyom S., et al. (2012). Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res., 22, 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee E., et al. (2012). Landscape of somatic retrotransposition in human cancers. Science, 337, 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bock C., et al. (2006). CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet., 2, e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das R., et al. (2006). Computational prediction of methylation status in human genomic sequences. Proc. Natl Acad. Sci. USA, 103, 10713–10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maksakova I.A., et al. (2008). Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell. Mol. Life Sci., 65, 3329–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aravin A.A., et al. (2007). Developmentally regulated piRNA clusters implicate MILI in transposon control. Science, 316, 744–747 [DOI] [PubMed] [Google Scholar]
- 33. Ciaudo C., et al. (2013). RNAi-dependent and independent control of LINE1 accumulation and mobility in mouse embryonic stem cells. PLoS Genet., 9, e1003791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34. Brunmeir R., et al. (2010). Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet., 6, e1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunican D.S., et al. (2013). Lsh regulates LTR retrotransposon repression independently of Dnmt3b function. Genome Biol., 14, R146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fadloun A., et al. (2013). Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol., 20, 332–338 [DOI] [PubMed] [Google Scholar]
- 37. Kuramochi-Miyagawa S., et al. (2008). DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev., 22, 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reichmann J., et al. (2012). Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS Comput. Biol., 8, e1002486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Estécio M.R., et al. (2010). Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res., 20, 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robertson K.D. (2005). DNA methylation and human disease. Nat. Rev. Genet., 6, 597–610 [DOI] [PubMed] [Google Scholar]
- 41. Liu W.M., et al. (1995). Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res., 23, 1758–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stribinskis V., et al. (2006). Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res., 66, 2616–2620 [DOI] [PubMed] [Google Scholar]
- 43. Teneng I., et al. (2011). Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics, 6, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feng Q., et al. (1996). Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell, 87, 905–916 [DOI] [PubMed] [Google Scholar]
- 45. Mathias S.L., et al. (1991). Reverse transcriptase encoded by a human transposable element. Science, 254, 1808–1810 [DOI] [PubMed] [Google Scholar]
- 46. Khazina E., et al. (2011). Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol., 18, 1006–1014 [DOI] [PubMed] [Google Scholar]
- 47. Kolosha V.O., et al. (1997). In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc. Natl Acad. Sci. USA. 94, 10155–10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szak S., et al. (2002). Molecular archeology of L1 insertions in the human genome. Genome Biol., 3, research0052.1–research0052.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Moran J.V., et al. (1999). Exon shuffling by L1 retrotransposition. Science, 283, 1530–1534 [DOI] [PubMed] [Google Scholar]
- 50. Rangwala S.H., et al. (2009). Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol., 10, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Speek M. (2001). Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol., 21, 1973–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brouha B., et al. (2003). Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA, 100, 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sassaman D.M., et al. (1997). Many human L1 elements are capable of retrotransposition. Nat. Genet., 16, 37–43 [DOI] [PubMed] [Google Scholar]
- 54. Amariglio E.N., et al. (1991). Identity of rearranged LINE/c-MYC junction sequences specific for the canine transmissible venereal tumor. Proc. Natl Acad. Sci. USA, 88, 8136–8139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Katzir N., et al. (1985). “Retroposon” insertion into the cellular oncogene c-myc in canine transmissible venereal tumor. Proc. Natl Acad. Sci. USA, 82, 1054–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Murgia C., et al. (2006). Clonal origin and evolution of a transmissible cancer. Cell, 126, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ewing A.D., et al. (2010). High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iskow R.C., et al. (2010). Natural mutagenesis of human genomes by endogenous retrotransposons. Cell, 141, 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shukla R., et al. (2013). Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell, 153, 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Thomas C.A., et al. (2012) LINE-1 retrotransposition in the nervous system. Annu. Rev. Cell Dev. Biol., 28, 555–573 [DOI] [PubMed] [Google Scholar]
- 61. Deininger P.L., et al. (1999). Alu repeats and human disease. Mol. Genet. Metab., 67, 183–193 [DOI] [PubMed] [Google Scholar]
- 62. Batzer M.A., et al. (2002). Alu repeats and human genomic diversity. Nat. Rev. Genet., 3, 370–379 [DOI] [PubMed] [Google Scholar]
- 63. Deininger P.L., et al. (2002). Mammalian retroelements. Genome Res., 12, 1455–1465 [DOI] [PubMed] [Google Scholar]
- 64. Bennett E.A., et al. (2008). Active Alu retrotransposons in the human genome. Genome Res., 18, 1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang W., et al. (2011). Alu distribution and mutation types of cancer genes. BMC Genomics, 12, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Watanabe M., et al. (2005). Founder SVA retrotransposal insertion in Fukuyama-type congenital muscular dystrophy and its origin in Japanese and Northeast Asian populations. Am. J. Med. Genet. A, 138, 344–348 [DOI] [PubMed] [Google Scholar]
- 67. Conley M.E., et al. (2005). Two independent retrotransposon insertions at the same site within the coding region of BTK. Hum. Mutat., 25, 324–325 [DOI] [PubMed] [Google Scholar]
- 68. Chen J.M., et al. (2010). Genomic rearrangements in inherited disease and cancer. Semin. Cancer Biol., 20, 222–233 [DOI] [PubMed] [Google Scholar]
- 69. Hastings P.J., et al. (2009). Mechanisms of change in gene copy number. Nat. Rev. Genet., 10, 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kvikstad E.M., et al. (2010). The (r)evolution of SINE versus LINE distributions in primate genomes: sex chromosomes are important. Genome Res., 20, 600–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Abrusán G., et al. (2006). The distribution of L1 and Alu retroelements in relation to GC content on human sex chromosomes is consistent with the ectopic recombination model. J. Mol. Evol., 63, 484–492 [DOI] [PubMed] [Google Scholar]
- 72. Smith T.M., et al. (1996). Complete genomic sequence and analysis of 117kb of human DNA containing the gene BRCA1. Genome Res., 6, 1029–1049 [DOI] [PubMed] [Google Scholar]
- 73. Puget N., et al. (2002). Distinct BRCA1 rearrangements involving the BRCA1 pseudogene suggest the existence of a recombination hot spot. Am. J. Hum. Genet., 70, 858–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Puget N., et al. (1997). A 1-kb Alu-mediated germ-line deletion removing BRCA1 exon 17. Cancer Res., 57, 828–831 [PubMed] [Google Scholar]
- 75. Peixoto A., et al. (2013). Genomic characterization of two large Alu-mediated rearrangements of the BRCA1 gene. J. Hum. Genet., 58, 78–83 [DOI] [PubMed] [Google Scholar]
- 76. Bannert N., et al. (2004). Retroelements and the human genome: new perspectives on an old relation. Proc. Natl Acad. Sci. USA, 101, 14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shin W., et al. (2013). Human-specific HERV-K insertion causes genomic variations in the human genome. PLoS One, 8, e60605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hughes J.F., et al. (2001). Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet., 29, 487–489 [DOI] [PubMed] [Google Scholar]
- 79. Barahona A. (1997). Barbara McClintock and the transposition concept. Arch. Int. Hist. Sci. (Paris)., 46, 309–329 [PubMed] [Google Scholar]
- 80. Wang T., et al. (2007). Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc. Natl Acad. Sci. USA, 104, 18613–18618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bourque G., et al. (2008). Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res., 18, 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ponicsan S.L., et al. (2010). Genomic gems: SINE RNAs regulate mRNA production. Curr. Opin. Genet. Dev., 20, 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Han J.S., et al. (2004). Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature, 429, 268–274 [DOI] [PubMed] [Google Scholar]
- 84. Yelin R., et al. (2003). Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol., 21, 379–386 [DOI] [PubMed] [Google Scholar]
- 85. Carninci P., et al. (2005) The transcriptional landscape of the mammalian genome. Science, 309, 1559–1563 [DOI] [PubMed] [Google Scholar]
- 86. Faulkner G.J., et al. (2009). The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet., 41, 563–571 [DOI] [PubMed] [Google Scholar]
- 87. Irvin-Wilson C.V., et al. (2005). Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res., 7, R563–R569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Singer G.A., et al. (2008). Genome-wide analysis of alternative promoters of human genes using a custom promoter tiling array. BMC Genomics, 9, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Thorsen K., et al. (2011). Tumor-specific usage of alternative transcription start sites in colorectal cancer identified by genome-wide exon array analysis. BMC Genomics, 12, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wolff E.M., et al. (2010). Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet., 6, e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pagani F., et al. (2004). Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet., 5, 389–396 [DOI] [PubMed] [Google Scholar]
- 92. Castle J.C., et al. (2008). Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet., 40, 1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang E.T., et al. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature, 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shen S., et al. (2011). Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl Acad. Sci. USA, 108, 2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Athanasiadis A., et al. (2004). Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol., 2, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Van de Lagemaat L.N., et al. (2006). Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 7, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Eswaran J., et al. (2013). RNA sequencing of cancer reveals novel splicing alterations. Sci. Rep., 3, 1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang J., et al. (2013). Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov., 3, 1228–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Watson I.R., et al. (2013). Emerging patterns of somatic mutations in cancer. Nat. Rev. Genet., 14, 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. David C.J., et al. (2010). Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Linker S., et al. (2013). Linear decay of retrotransposon antisense bias across genes is contingent upon tissue specificity. PLoS One, 8, e79402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ling J., et al. (2002). The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J. Virol., 76, 2410–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Pi W., et al. (2010). Long-range function of an intergenic retrotransposon. Proc. Natl Acad. Sci. USA, 107, 12992–12997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cruickshanks H.A., et al. (2013). Expression of a large LINE-1-driven antisense RNA is linked to epigenetic silencing of the metastasis suppressor gene TFPI-2 in cancer. Nucleic Acids Res., 41, 6857–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Smalheiser N.R., et al. (2005). Mammalian microRNAs derived from genomic repeats. Trends Genet., 21, 322–326 [DOI] [PubMed] [Google Scholar]
- 106. Gogvadze E., et al. (2009). Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J. Virol., 83, 6098–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Greaves M., et al. (2012). Clonal evolution in cancer, Nature, 481, 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Perkel J.M. (2013). Visiting “noncodarnia”. Biotechniques, 54, 301, 303–304 [DOI] [PubMed] [Google Scholar]
- 109. Carninci P., et al. (2005). The transcriptional landscape of the mammalian genome. Science, 309, 1559–1563 [DOI] [PubMed] [Google Scholar]
- 110. Wang K.C., et al. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell, 43, 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Ji P., et al. (2003). MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene, 22, 8031–8041 [DOI] [PubMed] [Google Scholar]
- 112. Gibb E.A., et al. (2011). The functional role of long non-coding RNA in human carcinomas. Mol. Cancer, 10, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Huarte M., et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell, 142, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yildirim E., et al. (2013). Xist RNA is a potent suppressor of hematologic cancer in mice. Cell, 152, 727–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ganesan S., et al. (2002). BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell, 111, 393–405 [DOI] [PubMed] [Google Scholar]
- 116. Yu W., et al. (2008). Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature, 451, 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mighell A.J., et al. (2000). Vertebrate pseudogenes. FEBS Lett., 468, 109–114 [DOI] [PubMed] [Google Scholar]
- 118. Balakirev E.S., et al. (2003). Pseudogenes: are they “junk” or functional DNA? Annu. Rev. Genet., 37, 123–151 [DOI] [PubMed] [Google Scholar]
- 119. Nie L., et al. (2012). Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res., 4, 127–150 [PMC free article] [PubMed] [Google Scholar]
- 120. Pei B., et al. (2012). The GENCODE pseudogene resource. Genome Biol., 13, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Patnala R., et al. (2014). Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res. Treat., 143, 239–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Salmena L., et al. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Sumazin P., et al. (2011). An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell, 147, 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Poliseno L., et al. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465, 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hawkins P.G., et al. (2010). Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription, 1, 165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Duret L., et al. (2006). The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science, 312, 1653–1655 [DOI] [PubMed] [Google Scholar]