Abstract
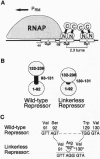
The bacteriophage lambda repressor binds cooperatively to pairs of adjacent sites in the lambda chromosome, one repressor dimer binding to each site. The repressor's amino domain (that which mediates DNA binding) is connected to its carboxyl domain (that which mediates dimerization and the interaction between dimers) by a protease-sensitive linker region. We have generated a variant lambda repressor that lacks this linker region. We show that dimers of the variant protein are deficient in cooperative binding to sites at certain, but not all, distances. The linker region thus extends the range over which carboxyl domains of DNA-bound dimers can interact. In particular, the linker is required for cooperative binding to a pair of sites as found in the lambda chromosome, and thus is essential for the repressor's physiological function.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu Rev Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978 Jan;13(1):65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- Beckett D., Burz D. S., Ackers G. K., Sauer R. T. Isolation of lambda repressor mutants with defects in cooperative operator binding. Biochemistry. 1993 Sep 7;32(35):9073–9079. doi: 10.1021/bi00086a012. [DOI] [PubMed] [Google Scholar]
- Benson N., Adams C., Youderian P. Genetic selection for mutations that impair the co-operative binding of lambda repressor. Mol Microbiol. 1994 Feb;11(3):567–579. doi: 10.1111/j.1365-2958.1994.tb00337.x. [DOI] [PubMed] [Google Scholar]
- Burz D. S., Beckett D., Benson N., Ackers G. K. Self-assembly of bacteriophage lambda cI repressor: effects of single-site mutations on the monomer-dimer equilibrium. Biochemistry. 1994 Jul 19;33(28):8399–8405. doi: 10.1021/bi00194a003. [DOI] [PubMed] [Google Scholar]
- Bushman F. D., Shang C., Ptashne M. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell. 1989 Sep 22;58(6):1163–1171. doi: 10.1016/0092-8674(89)90514-x. [DOI] [PubMed] [Google Scholar]
- Carra J. H., Schleif R. F. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993 Jan;12(1):35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustance R. J., Bustos S. A., Schleif R. F. Reaching out. Locating and lengthening the interdomain linker in AraC protein. J Mol Biol. 1994 Sep 30;242(4):330–338. doi: 10.1006/jmbi.1994.1584. [DOI] [PubMed] [Google Scholar]
- Guarente L., Nye J. S., Hochschild A., Ptashne M. Mutant lambda phage repressor with a specific defect in its positive control function. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2236–2239. doi: 10.1073/pnas.79.7.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler M., Bickle T. A. Increased protein flexibility leads to promiscuous protein--DNA interactions in type IC restriction-modification systems. EMBO J. 1991 Apr;10(4):951–957. doi: 10.1002/j.1460-2075.1991.tb08029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Dunn T., Schleif R. Upstream repression and CRP stimulation of the Escherichia coli L-arabinose operon. J Mol Biol. 1984 Nov 25;180(1):61–72. doi: 10.1016/0022-2836(84)90430-3. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Interaction at a distance between lambda repressors disrupts gene activation. Nature. 1988 Nov 24;336(6197):353–357. doi: 10.1038/336353a0. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Kornfeld K., Saint R. B., Beachy P. A., Harte P. J., Peattie D. A., Hogness D. S. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev. 1989 Feb;3(2):243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- Kuldell N., Hochschild A. Amino acid substitutions in the -35 recognition motif of sigma 70 that result in defects in phage lambda repressor-stimulated transcription. J Bacteriol. 1994 May;176(10):2991–2998. doi: 10.1128/jb.176.10.2991-2998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Moyle H., Susskind M. M. Target of the transcriptional activation function of phage lambda cI protein. Science. 1994 Jan 7;263(5143):75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Maurer R., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J Mol Biol. 1980 May 15;139(2):163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Binari R., Perkins L. A., Bender W. Alternative RNA products from the Ultrabithorax domain of the bithorax complex. EMBO J. 1988 Feb;7(2):435–445. doi: 10.1002/j.1460-2075.1988.tb02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T., Sturtevant J. M., Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer R. T., Pabo C. O., Meyer B. J., Ptashne M., Backman K. C. Regulatory functions of the lambda repressor reside in the amino-terminal domain. Nature. 1979 May 31;279(5712):396–400. doi: 10.1038/279396a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Smith D. L., Johnson A. D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 1988 Jul;2(7):807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- Smith D. L., Johnson A. D. A molecular mechanism for combinatorial control in yeast: MCM1 protein sets the spacing and orientation of the homeodomains of an alpha 2 dimer. Cell. 1992 Jan 10;68(1):133–142. doi: 10.1016/0092-8674(92)90212-u. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Karplus M., Patel D. J., Sauer R. T. Solution NMR studies of intact lambda repressor. J Biomol Struct Dyn. 1983 Oct;1(1):151–157. doi: 10.1080/07391102.1983.10507431. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Whipple F. W., Kuldell N. H., Cheatham L. A., Hochschild A. Specificity determinants for the interaction of lambda repressor and P22 repressor dimers. Genes Dev. 1994 May 15;8(10):1212–1223. doi: 10.1101/gad.8.10.1212. [DOI] [PubMed] [Google Scholar]
- Yu X. M., Reznikoff W. S. Deletion analysis of the CAP-cAMP binding site of the Escherichia coli lactose promoter. Nucleic Acids Res. 1984 Jul 11;12(13):5449–5464. doi: 10.1093/nar/12.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]