Abstract
Wide-spread hypomethylation of CpG dinucleotides is characteristic of many cancers. Retrotransposons have been identified as potential targets of hypomethylation during cellular transformation. We report the results of an preliminary examination of the methylation status of CpG dinucleotides associated with the L1 and HERV-W retrotransposons in benign and malignant human ovarian tumors. We find a reduction in the methylation of CpG dinucleotides within the promoter regions of these retroelements in malignant relative to non-malignant ovarian tissues. Consistent with these results, we find that relative L1 and HERV-W expression levels are elevated in representative samples of malignant vs. non-malignant ovarian tissues.
Findings
In the genome of differentiated somatic human cells, it is estimated that ~80% of all CpG dinucleotides are methylated [1]. Methylation of CpG dinucleotides located in or near promoter regions is typically associated with transcriptional repression [e.g, [2]]. Most retrotransposons, satellite DNA sequences, imprinted genes, X-chromosome inactivated genes, and many genes that are expressed tissue-specifically and that are transcriptionally silenced in differentiated human cells are associated with highly methylated (hypermethylated) CpG dinucleotides [3,4]. In contrast, CpG dinucleotides associated with the promoter regions of genes normally expressed in differentiated human cells, including tumor suppressor genes and genes involved in DNA repair and apoptosis, are associated with under-methylated (hypomethylated) CpG dinucleotides [e.g, [5]]. A number of recent studies have demonstrated that dramatic changes in these characteristic patterns of methylation are typically associated with cellular transformation [3]. For example, wide-spread (global) hypomethylation of up to 25% of CpG dinucleotides normally methylated in differentiated cells has been shown to be characteristic of many cancer cells, including those isolated from prostate, liver, ovarian and breast carcinomas [6]. In many instances, these increases in levels of global hypomethylation have been observed to be correlated with an increase in more localized patterns of hypermethylation associated with a subset of genes that includes tumor suppressor genes and other genes involved in DNA repair and apoptosis. The consequent reduction in expression of these genes is believed to initiate a cascade of events leading to cellular transformation [7].
Relatively little is currently known concerning the specific targets of global increases in hypomethylation associated with cellular transformation, although they frequently include repetitive sequences located in proximity to centromeres [8]. One major class of middle repetitive sequences believed to be targets of hypomethylation during cellular transformation are retrotransposons [9,10].
At least 40% of the human genome is comprised of retrotransposons [11]. Moreover, retrotransposons are widely dispersed throughout the human genome and, as alluded above, these elements are generally hypermethylated and transcriptionally silenced in most postnatal somatic cells. Retrotransposons are classified into three subtypes: long interspersed elements (LINEs) comprising 16–21% of the genome, short interspersed elements (SINEs) comprising ~13% of the genome and LTR [long terminal repeat] retrotransposons comprising 5–8% of the genome [12].
A significant increase in the expression of a variety of retrotransposons has been previously observed in a number of carcinomas including lymphocytic leukemia [13,14], lung [15], renal cell [9], colorectal [16] and breast [17] carcinomas. Direct evidence for the hypomethylation of retrotransposons has been observed in hepatocellular [18], prostate [19], lymphocytic leukemia [20], renal cell [9], and urinary bladder [21] carcinomas. In this paper, we report the results of a preliminary examination of the methylation status of CpG dinucleotides associated with two representative families of retrotransposons in benign and malignant human ovarian tumors. L1 is the most abundant family of human LINE elements comprising about 17% of the genome [22]. Human Endogenous Retrovirus-W (HERV-W) is a family LTR retrotransposons consisting of ~140 full-length or truncated elements randomly dispersed throughout the human genome [23]. Our results demonstrate that large numbers of both families of retrotransposons are hypomethylated in ovarian carcinomas. We further demonstrate that relative levels of both L1 and HERV-W expression are elevated in representative samples of malignant vs. non-malignant ovarian tissues. Our findings are consistent with the hypothesis that retrotransposons are a major target of global hypomethylation associated with cellular transformation.
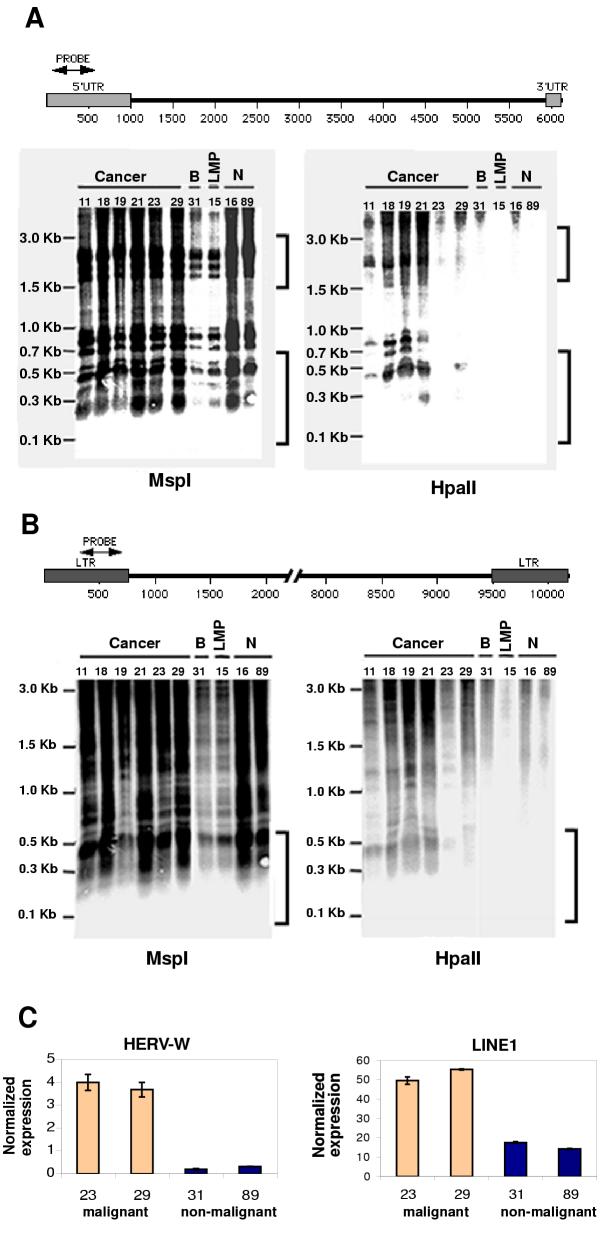
To test the hypothesis that L1 and HERV-W elements may experience reduced methylation in malignant ovarian carcinomas, we utilized a restriction-enzyme based assay to compare the methylation status of CpG dinucleotides located within the promoter regions of these elements in a series of malignant and non-malignant ovarian tissues. The restriction enzymes MspI and HpaII both recognize the sequence CCGG but HpaII only cuts when the recognition sequence is unmethylated at the inner cytosine (i.e., CCGG) while MspI is indifferent to the methylation status of the inner cytosine
Figure 1A &1B displays Southern blots of HpaI and MspI digested genomic DNA isolated from tissue samples and hybridized against probes homologous to regions encompassing the promoter regions of each family of elements. The HpaII/MspI restriction sites located within the promoter regions of both L1 and HERV-W elements are polymorphic among family members. By aligning the promoter regions of both families of elements present in the consensus human genome http://genome.ucsc.edu/ and identifying the HpaII/MspI sites present, we estimated that the expected size range of restriction fragments within the elements to be between ~100 – 700 bp and ~1500 – 3000 bp for L1 elements and between ~100 – 500 bp for HERV-W elements. Larger sized fragments representing partial digestions and/or polymorphic HpaII/MspI sites located within the elements or in regions flanking the elements are also visible.
Figure 1.
Hypomethylation and expresion of L1 and HERV-W elements in ovarian cancer. Genomic DNA was digested either with MspI (left) or HpaII (right), and hybridized with probes specific for the promoter regions of L1 (A) or HERV-W (B) elements. The restriction enzymes MspI and HpaII recognize the sequence CCGG but HpaII only cuts when the recognition sequence is unmethylated at the inner cytosine (i.e., CCGG) while MspI is indifferent to the methylation status of the inner cytosine. Brackets indicate bands from restriction cut sites internal to the elements (B = benign cystic mass; LMP = low-malignancy potential or borderline tumor; N = normal ovary. (C) Real time RT-PCR was performed to determine expression levels of LINE-1 and HERV-W elements in representative malignant and non-malignant samples. Normalized values (retroelement expression value divided by expression value of the RPS27A control gene. Shown is the average of 3 replicate assays per sample ± SE. RPS27A expression has been previously determined to be unchanged between the malignant and non-malignant samples examined in this study (see text for details).
The results presented in Figure 1A &1B show that MspI-generated bands within the expected size range of internal fragments were visible in digestions of DNA from all tissue samples. In contrast, HpaII-generated fragments within the expected size range were only visible in digestions of DNA from the malignant samples. These results are indicative of a consistent reduction in the methylation of CpG dinucleotides within the promoter regions of both L1 and HERV-W elements in the malignant tissue. The fact that the number and intensity of HpaII generated bands in the malignant samples is significantly less than generated by MspI digestion indicates that some L1 and HERV-W elements remain hypermethlyated in the malignant samples. The reasons for this variable response among members of the same retrotransposon family are currently under investigation. Regardless, so far as we know, this is the first report of the hypomethylation of L1 elements in ovarian carcinomas and of the hypomethylation of HERV-W in any human cancer.
As noted above, hypomethylation of retroelement promoter regions may be expected to result in a localized relaxation of chromatin structure and a corresponding increased element expression [e.g., [10]]. In order to test this prediction in our samples, we extracted total RNA from representative samples of two malignant and two non-malignant ovarian tissues and conducted quantitative Real Time RT-PCR. Two replicate assays were run for each tissue sample. The results shown in Figure 1C indicate a significant average increase in both L1 and HERV-W expression in the malignant vs. non-malignant ovarian tissues examined.
If further work supports the generality of these and other similar findings [10,19-22], the implications would be important. Hypomethylation is generally associated with the relaxation of chromatin structure, an increased accessibility of transcription factors and a consequent elevation in levels of expression [27]. Our findings are generally consistent with these prior results. Since transcription is a rate limiting step in retrotransposition [11], hypomethylation might be expected to result in an increase in retrotransposon insertion mutations. While there have been occasional reports of L1 and other retrotransposon insertion mutations implicated in cancer development in humans [e.g, [28]], this may not be as significant a factor as it apparently is in the mouse [29], perhaps because most L1 and other retrotransposon sequences in the human genome are believed to be truncated or otherwise transpositionally defective [30].
Another possible consequence of the hypomethylation of retroelements in humans is the opportunity it provides for ectopic pairing and recombination among homologous elements dispersed throughout the genome. The unequal-crossover events typically associated with ectopic recombination might well account for at least some of the various chromosomal aberrations and aneuploid events characteristic of human malignancies. Indeed, direct evidence of such an effect has recently been documented in mice [31,32]. In humans, L1 retrotransposition events have been shown to induce various forms of chromosomal instabilities [33] and L1 and other retrotransposon sequences have frequently been linked with a variety of chromosomal aberrations associated with human cancers [e.g, [34]].
A third possible consequence of the hypomethylation of retroelements in cancer cells is the potential regulatory impact of the release of methylation complexes known to be bound to these elements in post-embryonic somatic cells [e.g, [35]]. Although little is currently understood concerning the factors that determine the relative affinity of methylation complexes for DNA target sequences, retrotransposons are known to be high affinity targets [e.g, [10]]. Complexes released from retroelements may initiate a cascade of regulatory changes by binding to other lower affinity target sites and possibly resulting in the down regulation of genes essential for DNA repair and genome stability.
Methods
Tissue samples, DNA extraction, Southern hybridization
Bulk ovarian tissue samples were surgically removed and placed in RNA later (Ambion, Austin, TX) in the operating room within 1 minute of removal from the patients. The pathological and clinical information of each sample is as follows: Sample #11 (Age 43), Adenocarcinoma (papillary serous, poorly differentiated, Stage IIc); Sample #18 (Age 34), Adenocarcinoma (endometroid, well differentiated, Stage IIb); Sample #19 (Age 57), Adenocarcinoma (papillary serous, poorly differentiated, Stage IIc); Sample #21 (Age 80), Malignant mixed mullerian; Sample #23 (Age 52), Adenocarcinoma (papillary serous, poorly differentiated, Stage IIa); Sample #29 (Age 66), Adenocarcinoma (papillary serous, poorly differentiated, Stage III); Sample #15 (Age 54), Serous borderline /low-malignancy potencial; Sample #31 (Age 40), Benign cystic masses; Sample #16 (Age 53), Normal ovary; Sample #89 (Age 53), Normal ovary. This study was approved by the Institutional Review Board of the University of Georgia and of Northside Hospital (Atlanta), from which the samples were obtained.
Genomic DNA was extracted by proteinase K digestion of 20–25 mg of bulk ovarian tissue and phenol-chlorophorm extraction. DNA was ethanol precipitated and re-suspended in water. Ten micrograms of genomic DNA were digested overnight at 37°C with 10 to 16 excess amount of either HpaII [methylation sensitive restriction enzyme] or MspI [not sensitive for methylation at internal cytosine]. These enzymes recognize the sequence CCGG, which is found in diverse positions in the promoter regions of these retroelements. Digested DNA was resolved on an agarose gel and transferred to a nylon membrane (Hybond N; Amersham-Biosciences, Piscataway, NJ) with NaOH. Membranes were prehybridized for 1 hour with 10 mg/ml of herring sperm DNA in Church buffer [0.5 M NaH2PO4, 7% SDS and 10 M EDTA] and hybridized overnight at 65°C in the same buffer with 100–200 ng of probe DNA labeled with [α-32P]dCTP using a Nick Translation Kit (Roche, Indianapolis, IN). Filters were washed twice for 15 min in 2 × SSC and 0.1% SDS and then twice for 30 min in 1 × SSC and 0.1% SDS at 65°C and exposed to Phosphorimager screens (Molecular Dynamics, Sunnyvale, CA).
The HERV-W probe was designed in the LTR region, downstream of the putative TTAAAT box. PCR was performed on genomic DNA with forward primer HERVF 5'-CCACCACTGCTGTTTGCCAC-3 ' and reverse primer HERVR 5 '-GCCTCGTGTTCTCTGACCTGGGG-3', producing a 304 bp fragment. The LINE1 probe for the promoter region was designed according to Takai et al [18]. PCR was performed on genomic DNA with forward primer L1F 5'-CGGGTGATTTCTGCATTTCC-3' and reverse primer L1R 5'-GACATTTAAGTCTGCAGAGG-3', giving a product of 540 bp. PCR products were cloned into pCR2.1-TOPO and transformed into TOP10 E. coli cells (Invitrogen, Carlsbad, CA). Plasmids were extracted (Qiaprep Spin Miniprep Kit, Qiagen, Valencia, CA) and sequenced. Subsequent PCR reactions were performed on cloned plasmid DNA for both HERV-W and LINE1, and gel extracted PCR products were used as hybridization probes.
RNA extraction, quantitative real time RT-PCR
Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA) and 2–5 μg of total RNA were reverse transcribed into first-strand cDNA using the Thermoscript RT-PCR system (Invitrogen, Carlsbad, CA) in a final volume of 20 μl. The HERV-W primers used were: forward; 5'-TTGGCGGTATCACAACCTCT-3' reverse; 5'-GTGACGATTCCGGATTGA-3'; (product size:230 bp) based on the HERV-W sequence (GeneBank accession no. AC000064). The LINE-1 primers were: forward 5'-TCATAAAGCAAGTCCTCAGTGACC-3'; reverse 5 '-GGGGTGGAGAGTTCTGTAGATGTC-3' (product size:165 bp) based on the LINE-1 sequence (GeneBank accession no. M80343). Real-time monitoring of PCR reactions was performed using the DNA Engine Opticon 2 System (MJ Research, Waltham, MA) and the SYBR Green iQ dye (BioRad, Hercules, CA) [24]. For each reaction, the amount of a target and of an endogenous control (Ribosomal Protein S27A) were determined using a calibration curve and the amount of target molecule was divided by the amount of endogenous reference to obtain a normalized target value [25]. RPS27A has been previously identified as a valid control gene in expression studies conducted among human malignant and control tissues [26]. In addition, we independently verified by microarray analyses that RPS27A expression levels are constant among the samples examined in this study (data not shown). Separate calibration (standard) curves for RPS27A, HERV-W and LINE-1 were constructed using serial dilutions of total cDNA from normal human ovarian tissue (purchased from Ambion, Austin, TX). Standards for HERV-W, LINE-1 and RPS27A were defined to contain an arbitrary starting concentration, and serial dilutions were used to construct the standard curve. Standard curve calibrations were included in each assay.
List of abbreviations
HERV-W, human endogenous retrovirus-W; LINEs, L1, long interspersed elements; LTR, long terminal repeat; RPS27A, ribosomal protein S27A; PCR, polymerase chain reaction; RT-PCR, reverser transcription PCR
Authors' contributions
LM performed all assays. LM and JM collectively designed the study, analyzed the results and drafted the manuscript. BB provided tissue samples and suggestions for finalization of the manuscript. All authors read and approved the manuscript.
Acknowledgments
Acknowledgments
We thank Dr. Nina Schubert for valuable comments and critical reading of the manuscript. The research was supported by the Ovarian Cancer Institute [Atlanta].
Contributor Information
Laura Menendez, Email: lmenen@uga.edu.
Benedict B Benigno, Email: AMorgan@sjha.org.
John F McDonald, Email: mcgene@uga.edu.
References
- Bird AP, Taggart MH. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980;8:1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461–467. doi: 10.1093/carcin/21.3.461. [DOI] [PubMed] [Google Scholar]
- Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- Tycko B. DNA and alterations in cancer: genetic and epigenetic alterations. In: M E, editor. In. Natick: Eaton Publishing; 2000. pp. 333–349. [Google Scholar]
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg962. [DOI] [PubMed] [Google Scholar]
- Qu G, Dubeau L, Narayan A, Yu MC, Ehrlich M. Satellite DNA hypomethylation vs. overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat Res. 1999;423:91–101. doi: 10.1016/S0027-5107(98)00229-2. [DOI] [PubMed] [Google Scholar]
- Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Schubeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol Cell Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Patzke S, Lindeskog M, Munthe E, Aasheim HC. Characterization of a novel human endogenous retrovirus, HERV-H/F, expressed in human leukemia cell lines. Virology. 2002;303:164–173. doi: 10.1006/viro.2002.1615. [DOI] [PubMed] [Google Scholar]
- Depil S, Roche C, Dussart P, Prin L. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia. 2002;16:254–259. doi: 10.1038/sj.leu.2402355. [DOI] [PubMed] [Google Scholar]
- Andersson AC, Svensson AC, Rolny C, Andersson G, Larsson E. Expression of human endogenous retrovirus ERV3 (HERV-R) mRNA in normal and neoplastic tissues. Int J Oncol. 1998;12:309–313. doi: 10.3892/ijo.12.2.309. [DOI] [PubMed] [Google Scholar]
- Debniak T, Gorski B, Cybulski C, Jakubowska A, Kurzawski G, Kladny J, Lubinski J. Comparison of Alu-PCR, microsatelite instability, and immunohistochemical analyses in finding features characteristic for hereditary nonpolyposis colorectal cancer. J Cancer Res Clin Oncol. 2001;127:565–569. doi: 10.1007/s004320100261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T. Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Jpn J Clin Oncol. 2000;30:306–309. doi: 10.1093/jjco/hyd079. [DOI] [PubMed] [Google Scholar]
- Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–174. doi: 10.1002/(SICI)1097-0045(19990515)39:3<166::AID-PROS4>3.3.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Dante R, Dante-Paire J, Rigal D, Roizes G. Methylation patterns of long interspersed repeated DNA and alphoid repetitive DNA from human cell lines and tumors. Anticancer Res. 1992;12:559–563. [PubMed] [Google Scholar]
- Jurgens B, Schmitz-Drager BJ, Schulz WA. Hypomethylation of L1 LINE sequences prevailing in human urothelial carcinoma. Cancer Res. 1996;56:5698–5703. [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Kim HS, Lee WH. Human endogenous retrovirus HERV-W family: chromosomal localization, identification, and phylogeny. AIDS Res Hum Retroviruses. 2001;17:643–648. doi: 10.1089/088922201300119752. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Herrmann MG, Moss AA, Rasmussen RP. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 1997;22:130–138. doi: 10.2144/97221bi01. [DOI] [PubMed] [Google Scholar]
- Bieche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–1156. [PubMed] [Google Scholar]
- Lee PD, Sladek R, Greenwood CM, Hudson TJ. Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res. 2002;12:292–297. doi: 10.1101/gr.217802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler LA, Jones PA. Hypomethylation of DNA in the regulation of gene expression. Dev Biol (N Y 1985) 1988;5:335–349. doi: 10.1007/978-1-4615-6817-9_12. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Kuff EL. Intracisternal A particles in mouse neoplasia. Cancer Cells. 1990;2:398–400. [PubMed] [Google Scholar]
- Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH., Jr Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455–455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/S0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Schmiesing JA, Dong Y, Barak OG, Speicher DW, Yokomori K, Shiekhattar R. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]