Abstract
We describe the core Protein Production Platform of the Northeast Structural Genomics Consortium (NESG) and outline the strategies used for producing high-quality protein samples. The platform is centered on the cloning, expression and purification of 6X-His-tagged proteins using T7-based Escherichia coli systems. The 6X-His tag allows for similar purification procedures for most targets and implementation of high-throughput (HTP) parallel methods. In most cases, the 6X-His-tagged proteins are sufficiently purified (> 97% homogeneity) using a HTP two-step purification protocol for most structural studies. Using this platform, the open reading frames of over 16,000 different targeted proteins (or domains) have been cloned as > 26,000 constructs. Over the past nine years, more than 16,000 of these expressed protein, and more than 4,400 proteins (or domains) have been purified to homogeneity in tens of milligram quantities (see Summary Statistics, http://nesg.org/statistics.html). Using these samples, the NESG has deposited more than 900 new protein structures to the Protein Data Bank (PDB). The methods described here are effective in producing eukaryotic and prokaryotic protein samples in E. coli. This paper summarizes some of the updates made to the protein production pipeline in the last five years, corresponding to phase 2 of the NIGMS Protein Structure Initiative (PSI-2) project. The NESG Protein Production Platform is suitable for implementation in a large individual laboratory or by a small group of collaborating investigators. These advanced automated and/or parallel cloning, expression, purification, and biophysical screening technologies are of broad value to the structural biology, functional proteomics, and structural genomics communities.
Keywords: Structural Genomics, High throughput protein production, Construct optimization, Disorder prediction, Ligation independent cloning, Multiple Displacement Amplification, Laboratory Information Management System, Protein Structure Initiative, NMR, X-ray crystallography, T7 Escherichia Coli expression system, Wheat Germ Cell Free, NMR microprobe screening, Parallel protein purification, 6X-His tag, HDX-MS
Introduction
The NESG project (http://www.nesg.org), is one of the four National Institutes of Health (NIH)-funded structural genomics Large Scale Centers (LSC) of the National Institute of General Medical Sciences (NIGMS) Protein Structure Initiative (PSI). The primary goal of these structure production centers is to determine the three-dimensional (3D) atomic-level structures of hundreds of novel proteins and protein domains. The novel structural information generated can then be utilized in modeling thousands of additional proteins (or protein domains). In addition, these centers have a major focus on development and refinement of new technologies for high-throughput (HTP) protein production, X-ray crystallography, NMR spectroscopy, structural bioinformatics, and related supporting infrastructure. Overall, these centers aim to enrich the biological community by disseminating 3D structural information on important protein domain families, providing access to protein expression systems and protocols for protein sample preparation, and further enabling research by providing improved technology for the preparation of protein samples.
Nucleic acid-based genomic efforts have the advantage that the biophysical properties of the macromolecules studied are rather homogenous, allowing sample preparation that is highly standardized and amenable to high-throughput methods. By contrast, proteins often have diverse biophysical properties, making the preparation of suitable samples more difficult, especially when considering parallel HTP methods. Not surprisingly, one of the most critical issues facing structural genomics is the requirement to provide tens of milligram quantities of soluble, high purity, correctly folded, monodisperse protein samples. Adding additional complexity to this issue is the fact that the NESG Consortium utilizes both nuclear magnetic resonance (NMR) and X-ray crystallographic methods for protein structure determination (Montelione and Anderson, 1999), producing a similar number of structures by each method. Protein samples suitable for rapid three-dimensional (3D) structure determination by NMR generally require 13C,15N, and/or 2H isotope enrichment, while for X-ray crystallography we generally require selenomethionine labeling. Therefore the NESG protein production platform must be flexible enough to handle preparations of protein samples for both crystallization/crystallography and for heteronuclear NMR studies. Considering these challenges, one of the major contributions of the NESG is the development of new technologies in the areas of protein expression and purification to deliver protein samples suitable for both NMR, X-ray crystallography.
Here we describe our HTP cloning, protein expression and protein purification pipeline. This article emphasizes the recent technological advances that have been made during PSI-2, and builds on previous work describing our pipeline (Acton et al., 2005). This system is primarily based on E. coli T7 expression systems, which has to date proven to be the most productive, most efficient, and least expensive method to produce the quantities of protein required for structural studies. The description of this platform includes target selection, construct optimization, ligation-independent cloning (LIC), analytical scale expression and solubility screening, midi-scale expression, purification and biophysical characterization and large scale protein sample production (Fig. 1). Protein targets of the NESG project are either full length proteins or domain constructs. Currently, each week over one hundred protein targets are cloned and screened for expression, 50–75 expression constructs are fermented on a preparative (1 – 2 L) scale, and roughly 30 – 40 targets are purified in tens of milligram quantities for biophysical characterization, including NMR and/or crystallization screening. This platform is both scaleable and portable, and can be readily implemented by traditional structural biology laboratories, biotechnology industry, and various proteomics and functional genomics projects.
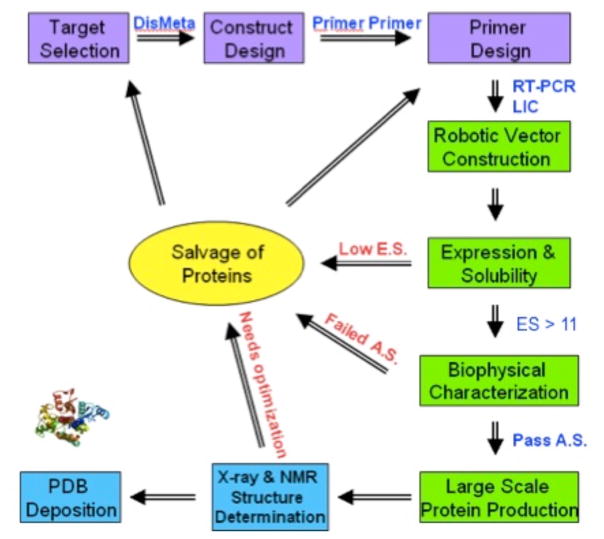
Fig. 1.
Protein Sample Production Platform currently used at the NESG. This diagram presents a schematic representation of the bioinformatics (purple); cloning, expression, purification, characterization, and sample preparation (green); structure determination (blue); and salvage strategies (yellow) used by the NESG Protein Sample Production Platform.
1. Bioinformatics Infrastructure and Target Curation
Protein targets, either full-length proteins or domain constructs, for structure determination are derived from three sources. The bulk of targets for the PSI LSCs are selected by a centralized PSI bioinformatics committee, including bioinformatics scientists nominated by each of the LCSs, and distributed amoung the four centers (Dessailly et al., 2009). These generally constitute large protein domain families with numerous members that have not been structurally characterized (BIG families), very large protein families with limited structural coverage (MEGA families), and domain families selected from metagenomic projects (META-families) such as the human gut microbiome project (Gill et al., 2006). The overall goal of targeting large protein domain familes is to provide the greatest novel leverage of structure space per target (Nair et al., 2009). Consequently this allows for pan-genomic targeting, taking advantage of the sequence differences and their concomitant biophysical characteristics within a domain family to isolate family members amenable for structure determination (Liu et al., 2004; Acton et al., 2005; Punta et al., 2009). Each LSC also is responsible for targets from a biomedical theme. The NESG pursues proteins from the Human Cancer Pathway Protein Interaction Network (HCPIN) (Huang et al., 2008) that we develop and curate (http://nesg.org:9090/HCPIN/). This is a collection of proteins involved in cancer associated signaling pathways and biological processes, together with their associated protein-protein interaction partners. Finally, the biomedical community nominates targets to the central committee, which distributes these Community Nominated Targets to the various PSI LSCs. Although protein target families are derived from these many sources, the focus of the NESG is on domain families represented in eukaryotic proteomes, including families that have exclusively eukaryotic members (e.g. the Ubiquitin Domain Mega family) and families that have both eukaryotic and prokaryotic members (e.g. the Start Domain Mega family).
One of the major goals of structural genomics is to increase the efficiency of structure production. More specifically, in the area of protein production, both experimental and bioinformatics studies have been published describing efforts to identify parameters and procedures that correlate with success, such as high levels of protein solubility or clone to PDB deposition rates (Dyson et al., 2004; Goh et al., 2004; Graslund et al., 2008a; Slabinski et al., 2007). We have developed numerous bioinformatics tools for the purpose of identifying the members of a protein domain family that are most amenable to protein production and structure determination. It is clear that the variation in protein sequence within a family can have great effects on its behavior with respect to protein production and biophysical properties. Using our extensive data set of proteins prepared in a similar fashion, we have identified primary sequence traits that correlate with (i) high levels of protein expression (E) and solubility (S) in our bacterial expression systems (PES) (unpublished results), (ii) greater probability of crystal structure determination based on protein sequence (PXS) (Price et al., 2009) and (iii) greater probability of amenability to NMR structure determination (PNMR) (unpublished results). These tools, together with our pan-genomic targeting strategy using our extensive list of over 175 Reagent Genomes (fully sequenced archeal, bacterial, and eukaryotic genomes and the corresponding genetic material for cloning) allows us to select several (4 – 6) proteins from each family for protein production by identifying those that are most likely to succeed.
Although we have made great efforts to enrich our protein production pipeline with amenable targets, one of the greatest enhancements to our pipeline in PSI-2 is our NESG Construct Optimization Software. A highly homogeneous protein sample with minimal numbers of disordered nonnative residues is generally required for successful protein crystallization and structure determination by X-ray crystallography (Sharma et al., 2009). While NMR can often be used successfully to study even fully disordered proteins, disordered segments of proteins can cause them to aggregate, and can be deleterious to NMR spectral quality. In addition, many targets are within multidomain proteins, which often misfold in prokaryotic systems (Netzer and Hartl, 1997), many multidomain proteins are also beyond the size limitations for high-throughput NMR structural determination techniques. To circumvent these problems domain parsing is often required. Obtaining soluble well-behaved domains with minimal disordered regions is challenging, and often cannot be accurately predicted. The NESG and others have taken an approach of producing several alternative constructs varying the termini of a targeted domain to identify the most amenable sequence (Graslund et al., 2008b; Chikayama et al., 2010). Briefly, the construct optimization software uses reports from the NESG DisMeta server, a metaserver providing a consensus analysis of sequence-based disorder predictors to predict disordered regions (www-nmr.cabm.rutgers.edu/bioinformatics/disorder), identify predicted secretion signal peptides, trans-membrane segments, possible metal binding sites, secondary structure, and interdomain disordered linkers. These structural bioinformatics data, together with multiple sequence alignments of homologous proteins and hidden Markov models (HMMs) characteristic of the targeted protein domain families (Dessailly et al., 2009) are used to identify possible structural domain boundaries. Based on this information, the software generates nested sets of alternative constructs, for full-length proteins, multidomain constructs, and single domain constructs. Thus for a single targeted region, we generally design multiple open reading frames varying the N and/or C-terminal sequences. Compared to only pursuing full-length proteins, these alternative constructs often possess significantly better expression, solubility and biophysical behavior, increasing the likelihood of success in crystallization and the efficiency of structure production.
Each of the proposed constructs are reviewed by a bioinformatics expert, and targets that pass this review are entered into our Protein Laboratory Information Management System (PLIMS). This JAVA-based Oracle database provides a detailed protein production data model, integrating closely with activities in the lab. A web-based application, PLIMS consists of four main modules: (i) Target Registration & Management, (ii) Molecular Biology & Protein Expression, (iii) Large-scale Fermentation, and (iv) Protein Purification. It is designed to capture all the information needed to completely reproduce the protein sample production process, interfacing where possible with robotics, and utilizing bar codes, PDAs, and wireless technology. Data from PLIMS is then uploaded to the internet-accessible NESG SPINE Structure Production Database (Bertone et al., 2001; Goh et al., 2003) to be shared across the consortium and with public databases.
Alternative construct DNA sequences are generated in the PLIMS database in a 96-well format. These sequences are then entered into the NESG Primer Prim’er software for automated primer design (Everett et al., 2004). This freely available web-based software (http://www.nesg.org/primer_primer) generates vector specific PCR primer sets designed to amplify and insert DNA targets into a vector of choice. Usually this vector is part of our “Multiplex Vector Kit”, a series of vectors with a common multiple cloning site designed to minimize the number of nonnative residues while adding a 6X-His tag (Acton et al., 2005). Affinity tags are generally required for high-throughput purification protocols (Sheibani, 1999; Crowe et al., 1994) however, large disordered tags found with many commercial vector systems can interfere with structural determination efforts. Although, both restriction endonuclese and viral recombination cloning strategies are supported in Primer Prim’er, we design ORF-specific primers with vector overlap regions for use with InFusion (Clonetech) Ligation Independent Cloning (LIC). Predominantly we clone into NESG-modified pET15 or pET21 T7 expression vector derivatives with N- (MGHHHHHHSH-) or C- (-LEHHHHHH) 6X-His affinity purification tags, respectively. The primer information in 96-well format is then entered into PLIMS, which produces the order forms for our oligonucleotide vendor.
2. High-Throughput Cloning for E. coli Expression
2.1 Methods for the Production of PCR Template
The first step in the cloning of our structural genomics targets involves PCR amplification of gene regions targeted in the construct design process described above. Oligonucleotide primers designed with Primer Prim’er are easily procured from a variety of vendors at inexpensive rates. However, PCR templates are not easily procured and are often expensive, and in the case of genomic DNA preparations from prokaryotic targets, of limited quantity. Further, our focus on eukaryotic protein families has the added complication that we must use cDNA in most cases in order to clone eukaryotic targets. Here we outline two alternative methods for generating template DNA for HTP 96-well PCR reactions.
The number of fully sequenced prokaryotes has increased at a rapid rate resulting in the elucidation of the genomic sequences of over 1,000 organisms, with even more in progress. As methods to predict success in expression, solubility, crystallization, and NMR spectral quality, based on primary sequence, are refined (Price et al., 2009; Price et al., 2010), it becomes possible to increase efficiency by selecting target proteins and domains from large domain families that are most likely to be successful. The protein sequences that arise from these prokaryotic sequencing projects are a rich source of targets that may be amenable to structural determination. However, genomic DNA preparations are commercially available for only a small fraction (~10%) of the sequenced prokaryotic strains. It is possible to produce genomic DNA by direct extraction from cultures, however many strains require specialized media and growth conditions which make such a strategy difficult and expensive. To circumvent these problems we have implemented Whole Genome Amplification (WGA) by Multiple Displacement Amplification (MDA) utilizing phi29 DNA polymerase (Lasken, 2007; Lasken, 2009; Kvist et al., 2007), to produce microgram quantities of genomic DNA suitable for use as cloning template (Dean et al., 2002). WGA by MDA is routinely used in metagenomic/environmental genome sequencing projects to prepare DNA templates from minute quantities of cells (Kvist et al., 2007; Lasken, 2009). As the vast majority of sequenced bacterial strains are available as inexpensive lyophilized cultures from ATCC (American Type Culture Collection), we routinely perform MDA on a small aliquot of freeze-dried cells to provide genomic DNA suitable for use as PCR template in cloning NESG target genes. This high-fidelity technique has proven to be extremely robust, and has successfully generated genomic DNA for more than 30 new bacterial and archaeal Reagent Genomes, including a number of human gut metagenome species, greatly expanding the range of proteomes that we can target (Acton et al., 2010).
One major advantage of bacterial targets for HTP cloning is the fact that they do not contain introns in their coding sequences. Therefore, genomic DNA can be used as template for PCR amplifying the coding region of a target for subsequent cloning into a bacterial expression vector. In a high-throughput setting, this also has the added advantage that a PCR “master mix”, containing the genomic DNA as template, can be added to multiple reactions, saving time and robotic liquid handling tips.
Conversely, eukaryotic organisms often have introns in the coding regions of their genes. E. coli does not have the ability to splice mRNA transcripts prohibiting the use of genomic DNA as PCR template for eukaryotic targets, a cDNA source is then necessary for amplification. There are numerous commercially available sources for cDNA. However, many have significant problems with the majority lacking full-length sequence verification, as such they can often contain polymorphisms. While there are full-length, fully sequenced cDNA sources such as the ORFeome collaboration clones (Open Biosystems) (Rual et al., 2004; Lamesch et al., 2007), these reagents are quite costly and these collections are not complete. Further, these individual clone libraries have logistical issues, they must be archived and rearrayed, before use as PCR template. In order to circumvent these problems, we have taken the approach of producing cDNA pools from various cell types using commercially available mRNA preparations (Clontech). In this strategy we use polyadenylated mRNA from various tissues, cell types, and developmental stages, including a considerable number of tumor cells and human cell lines, together with oligo dT or random primers, to carry out MMLV-mediated reverse transcriptase reactions (Acton et al., 2005). These cDNA pools are then mixed together and used as a common template that is added to each PCR reaction with target specific primers much like using bacterial genomic DNA. This greatly increases throughput and allows us to target genes that may not be in the available cDNA libraries. This strategy is quite effective. Analysis of our PLIMS database indicates that XX% of the RT-PCR amplification products are of the correct size. Although this approach may also generate clones with polymorphisms, we find it to be cost effective in comparison to cloning from commercially available cDNA sources. Recently we have expanded this strategy from mainly human targets to include Bos taurus, Mus musculus, Rattus norvegicus and Aribidopsis thaliana among others.
2.2. Ligation-Independent Cloning (LIC) and Automated Vector Construction with the Qiagen BioRobot 8000
The first step in the HTP production of proteins is the construction of vectors for expression of the target proteins. The NESG initially developed HTP approaches to cloning utilizing classical restriction endonuclease/ligase dependent methods in combination with our Multiplex Cloning Vector Set implemented in 96-well format using a QiaRobot 8000 (Acton et al., 2005). The vector system we created was designed to minimized the number of nonnative residues in the open reading frame while adding a 6X-His tag. Using this robust strategy we cloned nearly 7,000 target protein (or domains). Ligation Independent Cloning systems (LIC) are generally far more efficient, less time consuming and require less technical skill than ligase-dependent cloning (Aslanidis and de Jong, 1990; Haun and Moss, 1992). However, our view at the start of the project was that although the LIC technologies were promising, the technology was not developed enough in 2000 to meet the needs of a structural genomics project. For example, most of the early technologies resulted in the addition of a large number of nonnative residues to a protein coding sequence, which is not desirable for crystallization or NMR studies.
Although we have had great success with our classical system of cloning, LIC systems always held promise for our HTP applications. Recent advances, such as the InFusion cloning system (Clonetech) have negated the previous drawbacks. During PSI-2 we adapted the InFusion strategy to our HTP cloning pipeline. InFusion cloning only requires the addition of a 15 base pair tail to each of the gene specific PCR primers for a given target ORF; these base pairs are complimentary to the 5′ and 3′ regions of the vector multicloning site respectively (Zhu et al., 2007). After PCR amplification, the ORF DNA fragment now containing the region of vector overlap is incubated with the vector and the InFusion enzyme for thirty minutes and directly transformed into bacterial competent cells, resulting in a protein expression clone. LIC competent vector is produced by restriction endonuclease digestion in a nearly identical manner as described for ligase-dependent cloning (Acton et al., 2005). Briefly digestion with NdeI is followed by XhoI digestion, agarose gel purification, gel extraction and finally the concentration is normalized to 8 ng/μl. Further vector treatment against self-ligation is not necessary since the Infusion enzyme does not have ligase activity, and ligation by host enzymes appears inefficient with the minute overhangs produced by restriction digest. The greatest advantage of InFusion method is the substantial decrease in the number of cloning steps and the overall high efficiency of cloning. The restriction digest steps, long overnight ligation reactions, and several purification steps are no longer necessary. This results in a dramatic time savings, allowing the same number of technicians to nearly double their cloning output. In addition, this strategy is completely compatible with our Multiplex Cloning Vector Set (Acton et al. 2005), using the same exact vectors and the same strategy for minimizing nonnative residues. The removal of the restriction digestions steps also allows those ORFs with the most favored restriction sites internal to their coding sequence to be cloned while minimizing nonnative residues. Our modifications to the InFusion cloning system also allows this strategy to be cost efficient and actually below the cost of our ligase-dependent system. Using this new method we have cloned over 20,000 constructs of some 9,000 unique protein targets (multiple alternative constructs per target) into pET expression vectors, a dramatic increase in our previous rate of cloning.
We have automated each step of our vector construction strategy using a BioRobot 8000 to allow high throughput cloning in a 96-Well manner. Figure 2 outlines each of these steps of vector construction. Steps shown in blue typeface are automated while those in red are semiautomated, requiring some manual manipulations. A detailed protocol of the entire process can be downloaded (www-nmr.cabm.rutgers.edu/labdocuments/proteinprod/index.htm) and has been previously described (Acton et al., 2005). Each automated step is controlled by a custom Qiasoft 4.1 program developed in house. Initially, 50 uM concentrations for forward and reverse primers for each specific ORF (identical wells on two separate 96-well blocks) are placed on the Biorobot. From a separate position, the eight-channel pipette head transfers an appropriate PCR reaction mix to each well in a 96-well PCR plate [dNTPs, Advantage HF2 high-fidelity polymerase and buffer (Clontech), template DNA, and nuclease free H2O]. The BioRobot then transfers 100 pmol of the appropriate forward and reverse primers from the primer blocks into the corresponding well for each target in the PCR plate. A variety of Applied Biosystems thermocyclers are used for amplification with 35 total cycles. Each cycle contains a 10 s 94 °C melting step, a 20 s annealing step (50–55 °C), and a 3 min 68 °C elongation step, An annealing temperature step increase after 10 rounds of amplification is included taking advantage of the increased stability derived from the added recombination sites base pairs (Acton et al., 2005).
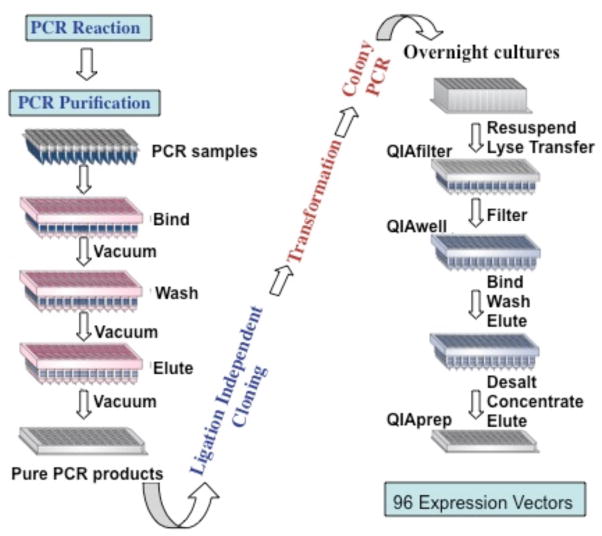
Fig. 2.
Schematic of the cloning process using the QIAGEN BioRobot 8000. Each step in the cloning strategy is indicated. Blue type denotes steps that are completely automated, and red type indicates steps that require some manual input. Procedures of QIAGEN-based protocols were modified, including Qiaquick Purification and DNA Mini-Prep protocols. However, most have been designed in the NESG Protein Production laboratory. A more detailed description of the robotic cloning procedure, as well as the automated protocols are provided elsewhere (Acton et al., 2005).
Our expansion of Reagent Genomes during PSI-2 has also increased the number of GC-rich genomes. As the GC content increases, PCR amplification becomes more problematic. In order to circumvent this problem, alternative thermostable polymerases and buffer conditions (such as the addtion of DMSO) must be utilized. Care must be taken to adjust buffer and annealing temperature conditions to maximize fidelity while increasing the likelihood of obtaining amplification product. GC-rich templates are often a problem with eukaryotic genes and although we have great success with the Advantage GC 2 Polymerase (Clontech), higher error rates will occur.
PCR products are visualized and separated on a 2% agarose gel, followed by Alpha Imager (Cell Biosciences) documentation, and entry into the PLIMS data management system. DNA fragments of the correct size are excised from the gel with a SafeXtractor and relocated into the appropriate well of a 96-well S-Block (Qiagen). Using reagents from the Qiagen Gel Extraction Kit and a QIAquick 96-well column PCR Cleanup plate, an automated 96-well gel extraction is performed on the BioRobot8000. The resulting purified PCR products are then subjected to LIC cloning into pET expression vectors, as described above. Following the InFusion enzyme activity, the resected and paired DNA fragments (vector and insert) are transformed into E. coli cells, using a 24-well format robotic transformation procedure. Briefly, a single microliter LIC product is transferred to the corresponding well of a fresh 96-well PCR plate prechilled at 0 °C on the robot deck. Each well of this plate contains 10 μl of XL-1 ultracompetent cells (Stratagene). A transformation procedure is then carried out on the robot deck keeping the PCR plate at 0 °C until a manual heat shock. SOC (100 μl) is added to each well, and the plate is incubated at 37 °C for 1h. The entire content of each well is transferred to a corresponding well in one of four 24-well blocks. The robot’s platform shaker spreads the mix via the 5–10 (3-mm-diameter) glass beads over the 2 ml of Luria broth (LB) medium/Agar with ampicillin in each well. Following overnight incubation at 37 °C, two colonies per ORF are harvested for colony PCR, using primers flanking the multiple cloning site (MCS). The results are visualized by agarose gel electrophoresis, documented into PLIMS, and the correct clones are subcultured overnight. Plasmid DNA is isolated using a completely automated Qiagen 96-well DNA mini-prep procedure and both the cultures and DNA constructs are archived in an NESG Reagent Repository.
3. Protein Expression, Solubility, and Biophysical Characterization
3.1. Analytical Scale Expression
The goal of analytical scale expression is to measure the expression and solubility level of each construct. Depending on the source of the protein target, roughly 30–50% of the clones will express soluble protein at the level needed for large-scale (1 – 3 L) fermentation in shake flasks, and purification. With this attrition rate, preparative-scale fermentation of each clone is not feasible. We have therefore developed a plate-based strategy to evaluate expression (E) and solubility (S) in a HTP fashion, while maintaining the highly aerated growth conditions found in later fermentation efforts. Figure 3 outlines this process starting with transformation into the codon-enhanced BL21(DE3)pMgK strain using a robotic transformation protocol and 24-well plates. Following overnight growth, individual colonies are inoculated into the corresponding well of a 96-well block containing 0.5 ml of LB per well. The pre-culture is incubated for 6 h at 37 °C, and preserving well assignment, subculterd robotically into a fresh 96-well block containing 0.5 ml of MJ9 minimal media (Jansson et al., 1996) for overnight growth. Growth in this same minimal media will be utilized in preparative-scale fermentations for isotope or selenomethionine enrichment. We have found that growth under minimal media conditions differs significantly from rich media, often affecting expression and solubility behavior. The BioRobot performs a 1:20 dilution of the saturated growth into one of four 24-square-well blocks (10 ml maximum volume/well) containing 2 ml of MJ9 media, preserving well assignment. Each block is sealed, covered with Airpore tape (Qiagen), and grown to mid-log phase (2–3 h growth, 0.5–1.0 OD600 units) with vigorous shaking at 37 °C. Expression is then induced with 1 mM IPTG, the temperature is shifted to 17 °C, and the cultures are grown overnight with vigorous shaking. The low temperature incubation often aids in producing soluble proteins (Shirano and Shibata, 1990), while the vigorous shaking with gas permeable tape allows for greater aeration rates like those that we obtain in our Midi-scale fermentor (described below) or large-scale fermentation in baffled flasks. Following overnight induction, cells are harvested by centrifugation, the pellets are resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM 2-mercaptoethanol) and robotically transferred to a 96-well PCR plate. A 96-probe sonicator (Misonix) is used for cell disruption. Total and soluble portions of the cell lysate are visualized by SDS-PAGE. Expression (E) and solubility (S) are scored, each on a scale of 0 (none) to 5 (max); i.e. the E x S (or ES value) ranges from 0 to 25. All data is documented in the PLIMS system.
Fig. 3.

High-throughput analytical scale protein expression screening using robotic methods. This schematic shows the step-by-step procedure used for small-scale expression screening. Completely automated steps are shown in blue, and partially automated steps are shown in red. Briefly, initial cultures are grown in 2.2 ml 96-well S-Blocks (Qiagen), followed by subculturing in 24 Well Block (Qiagen). Following overnight incubation the cultures are transferred into two separate S-Blocks (1 ml per respective well) and harvested by centrifugation (3000 × g, 10 minutes). The media is discarded and the cell pellet is resuspended in 100 μl lysis buffer and transferred to a 96-Well Round Bottom plate (Greiner). Following sonication a 30 μl aliquot of the total cellular lysate (Tot) is transferred to a new plate. The remainder is centrifuged for 10 minutes at 3000 × g, and a 30 μl aliquot of the supernatant (Sol) is transferred to a new plate. Equal amounts of Sol and Tot are added to adjacent wells for SDS-PAGE analysis.
3.2. Midi-Scale Protein Production and Characterization
Although all expression constructs with high expression and solubility levels (e.g., ES > 11) could be scaled-up on a preparative scale, a large fraction of the resulting samples turn out to be aggregated or even unfolded following preparative purification. As shown in Table 1, retrospective analysis of our earlier extensive data set (> 1,500 purified proteins) demonstrates that crystallization success rates are dramatically increased more than 10-fold for monodisperse protein samples in comparison with those polydisperse or aggregated (Price et al., 2009). Based on these results, and in order to maximize efficiency, we have developed a HTP Midi-scale Protein Production Pipeline, allowing production of hundreds-of-microgram quantities of protein, sufficient to characterize the biophysical properties of protein constructs before investing in large-scale expression and purification (Figure 4). This system utilizes (i) a 96-tube GNF fermentor (Genomics Institute of the Novartis Research Foundation) with O2 aeration allowing for high cell density protein expression at 60 ml scale; (ii) a His MultiTrap HP 96-well plate (GE Healthcare) for Ni-affinity protein purification; and (iii) Zeba™ 96-well desalting spin plate (Thermo Scientific) for buffer exchange. Typical yields of 0.2 – 1.0 mg of protein per 60 ml fermentation are achieved, with 96 fermentations carried out in parallel. These quantities of purified protein are sufficient for a series of analytical protein chemistry steps including: aggregation screening by analytical gel filtration with static light scattering (Acton et al., 2005), homogeneity analysis using Caliper microfluidics, target validation by MALDI-TOF mass spectrometry, concentration determination by a NanoDrop ND-8000 spectrophotometer, and NMR screening using a 1.7-mm micro cryo NMR probe (35 uL sample volume). Identification of aggregated/polydisperse proteins prior to scale-up allows us to screen multiple constructs of a target in order to find those most likely to succeed in crystallization and/or NMR experiments. NMR screening (Rossi et al., 2009), requiring 100 – 300 ug samples of protein, allow spectral evaluation by 1D 1H-NMR prior to isotopic enrichment. Further, the Midi-Scale process avoids scale-up of intractable protein targets, greatly increasing project efficiency
Table 1.
Analysis of Crystal Hits from NESG Protein Samples with Various Monodispersity (2001–2007)
| Year | Monodispese | Predominantly monodisperse | Mostly polydisperse | Polydisperse | Indeterminate | Aggregated |
|---|---|---|---|---|---|---|
| 2001 | 1/2 (50.0%) | |||||
| 2002 | 24/82 (29.3%) | 0/4 (0.0%) | ||||
| 2003 | 14/52 (26.9%) | 1/3 (33.3%) | 1/1 (100.0%) | 0/1 (0.0%) | ||
| 2004 | 42/112 (37.5%) | 6/19 (31.6%) | 0/11 (0.0%) | 0/110 (0.0%) | 0/35 (0.0%) | |
| 2005 | 37/148 (25.0%) | 2/31 (6.5%) | 1/9 (11.1%) | 0/24 (0.0%) | 0/20 (0.0%) | 0/27 (0.0%) |
| 2006 | 37/223 (16.6%) | 14/41 (34.1%) | 2/32 (6.3%) | 2/30 (6.7%) | 1/46 (2.2%) | 1/25 (4.0%) |
| 2007 | 47/277 (17.0%) | 7/57 (12.3%) | 0/26 (0.0%) | 0/19 (0.0%) | 1/69 (1.4%) | 1/70 (1.4%) |
| Total | 202/896 (22.5%) | 30/155 (19.4%) | 4/79 (5.1%) | 2/183 (1.1%) | 2/135 (1.5%) | 2/158 (1.3%) |
Proteins with crystal hits/Proteins provided for crystallization screening (% crystal hits)
Monodisperse: >90% monodispersity
Predominantly monodisperse: >80% but <90% monodispersity
Mostly polydisperse: >50% but <80% monodispersity and <3 peaks
Polydisperse: <50% monodispersity with > 3 peaks
Indeterminate: protein not in void volume (Vo), but obscured by ring-down from Vo
Aggregated: protein in Vo.
Fig. 4.
Midi-scale 96 sample protein expression, purification, and characterization. This system utilizes (i) a 96-tube GNF fermentor (Genomics Institute of the Novartis Research Foundation) with O2 aeration at 60 ml scale; (ii) a His MultiTrap HP 96-well plate (GE Healthcare) for Ni-affinity protein purification; and (iii) Zeba™ 96-well desalting spin plate (Thermo Scientific) for buffer exchange. Analytical protein chemistry steps include aggregation screening by analytical gel filtration with static light scattering, homogeneity analysis using Caliper LabChip® 90 system, target validation by MALDI-TOF mass spectrometry, concentration determination by a NanoDrop ND-8000 Spectrophotometer, and NMR screening using a 1.7-mm micro cryo NMR probe.
3.3. Midi-scale fermentation with GNF Fermentor system
To produce sufficient quantities of protein for biophysical characterization we have recently adapted a GNF 96-well Fermentor (Genomics Institute of the Novartis Research Foundation) to our Midi-scale pipeline. Using rich TB media (Peti et al., 2005), we routinely reach cell densities in the range of 15–20 OD600 units. This correlates to a quarter of the final cell mass obtained from 1 L of our large-scale protein expression in minimal media, which roughly produces to 3–5 OD600 units. Briefly this procedure starts with placing 500 ul of TB media with ampicillin and kanamycin into each well of a 96 well block. Expression clones scored with high (> 11) ES values are robotically transferred from their plate-based glycerol stocks into a PLIMS-directed unique well. Following an overnight incubation, the entire contents of each well are then transferred to a 100 ml test tube in the corresponding position of the GNF fermentor. Each tube contains 57 ml of TB and anti-foam, the air intake manifold is inserted into the rack of 96 tubes and placed in a water bath preheated to 37 °C. Using the manifold and its canulae, 100% oxygen is distributed to each well at a flow rate of ~3.5 cfm. This provides oxygen for growth as well as agitation for mixing the culture. We have found that the dual functioning canulae necessitate a high percentage of oxygen addition for the greatest yield, in turn requiring adequate system ventilation for safety. When OD600 reaches 5–6 units, IPTG is added for a final concentration of 1 mM. Concurrently, the water bath temperature is decreased to 17 °C using a refrigerated water circulator (VWR Scientific). Following 16 hours of incubation at this temperature and aeration with 100% oxygen, an aliquot is taken from each well to assay final cell density and for SDS-PAGE analysis of expression and solubility levels, and each is transferred to a labeled 50 mL conical tube, and centrifuged. The resulting data is documented in the PLIMS database.
3.4. Ni-affinity Protein Purification using His MultiTrap HP 96-well Plate
Cell pellets from each culture are resuspended in lysis buffer containing 1 X Cell Lytic B, 500 mg/ml lysozyme (freshly prepared), 100 units/ml RNAse, 100 units/ml DNAse, and 40 mM imidazole. Following a shaking incubation with 37 °C for 30 min, cell debris is cleared by centrifugation at 3,000 rpm for 20 min. 2 ml of each resulting supernatant is transferred to an empty 2.2-ml deep-well plate (Qiagen S-block). A Liquidator96 (Rainin) is used to transfer 400 μl from each well to the corresponding well of a His MultiTrap HP 96-well plate (GE Healthcare) for Ni-affinity protein purification. The plate is centrifuged for 4 min at 100 × g, the flow through is discarded, and the process repeated four more times to load the entire contents of each well. Each well in the Ni-NTA plate is washed three times with 500 μL of lysis buffer containing 40 mM imidazole (pH 7.5). Proteins are next eluted by adding 75 μl of lysis buffer containing 300 mM imidazole (pH 7.5) to each well, the plate is then incubated at room temperature for 5 min and centrifuged at 100 × g for 4 min. The Ni-affinity purified proteins are then immediately transferred to Zeba™ 96-well desalting spin plate (Thermo Scientific) for buffer exchange into appropriate buffers for biophysical characterization.
3.5. Biophysical characterization
(i) Homogeneity analysis using Caliper LabChip® 90 system
To assay the purity of proteins from the Midi-scale purification we have incorporated the use of a LabChip® 90 system (Caliper). This microfluidic device uses the same electrophoresis separation principle as SDS-PAGE (Bousse et al., 2001). However, the LabChip® 90 system has higher sensitivity, lower volume (1–2 μl) requirements, 96-well format compatibility with the BioRobot8000, and is less time consuming (~90 min per plate). These make it ideal for our Midi-scale purification and characterization platform. Briefly, samples are prepared in a 96-well plate by mixing 2 μl of protein sample with 7 μl of denaturing buffer. Following heat denaturation (5 min. @ 95 °C), 35 μl of water is added to each well. The LabChip 90 automation system then loads the Protein Express chip, following separation and detection the software reports the size, relative concentration and purity of the proteins. Although this system provides high quality results, lower molecular weight proteins (<12 kDa) cannot be accurately analyzed with this system. All data is archived in our PLIMS database
(ii) Target validation by MALDI-TOF mass spectrometry
Samples are prepared by mixing 1 μl of the protein sample from each well with 10 microliters of sinapinic acid (SA) matrix solution (10 mg/ml SA in 50% acetonitrile/50% 0.1% TFA). Spectra are collected for each protein spot, corresponding to a well, on a MALDI-TOF/TOF (ABI-MDS SCIEX 4800) in single TOF mode. The spectrum of each well is compared to the expected size of the purified protein; species differing from their expected mass by greater than 500 Daltons likely represent invalid targets, and are discontinued and further investigated in order to validate the protein sequence.
(iii) Aggregation screening by analytical gel filtration with static light scattering
It is now well established that proteins that are monodisperse in solution are more likely to produce crystals during screening trials than polydisperse or aggregated samples (add Price et al ref) (Klock et al., 2008; Ferre-D’Amare and Burley, 1994; Ferre-D’Amare and Burley, 1997). Analytical gel filtration followed by multi-angle static light scattering (SEC-LS) is an extremely sensitive method for detecting the distribution of oligomers and/or aggregates in a protein sample. Briefly, an Agilent 1200 series HPLC system with an automated 96 well sample changer is used with a Shodex KW-802.5 HPLC size-exclusion column to separate the protein species in solution. A miniDAWN TREOS detector (Wyatt technologies) simultaneously measures light scattering at three different angles (45°, 90°, and 135°). Refractive index is also measure using an Optilab rEX Refractometer (Wyatt Technology). Together, the analysis of this data provides the shape-independent weight-average molecular mass of each species in the gel filtration effluent, and their relative distributions. As shown in top panel of Figure 5, the light scattering trace for NESG target HR3580C indicates peaks corresponding to monomer, dimer, higher oligomers, and aggregates of the protein. The bottom panel traces the refractive index and indicates that the majority of mass is contained as a monomer. Further data analysis indicates that roughly 75% of the mass is monomeric in nature, with significant mass in other species. This suggests that further construct optimization or other “salvage” efforts are required before promotion to large-scale fermentation and purification.
Fig. 5.

Aggregation screening using analytical gel filtration with static light scattering, target HR3580C. Data was collected on a miniDAWN Light Scattering instrument (Wyatt Technology) at λ= 690 nm and at 30°C on a sample of target HR3580C. The elution profile as detected by static light scattering at 90° (LS) (red-trace) and refractive index (blue-trace) is illustrated.
(iv) Concentration determination by a NanoDrop ND-8000 Spectrophotometer
Traditional spectrophotometry requires placing samples into cuvettes or capillaries. This is impractical due to the limited sample volumes generated by the Midi-scale system. The NanoDrop™ 8000 spectrophotometer enables the quantification of samples in volumes as low as 0.5–2 microliters without dilution. Using an eight-channel pipette to transfer the purified proteins from the 96-well plate to a linear array (96-well spacing) of pedestals allows the measurement of 96 samples in less than six minutes. The protein concentration in each well is calculated automatically using its respective extinction coefficient. The accurate protein concentrations derived from this assay are used for the light-scattering data analysis, sample preparation for NMR screening, and for calculating process yield. All of the information generated in this step is recorded in the Spine database.
(iv) Microprobe NMR Screening
Recent advancements in NMR microprobe technology have greatly decreased the amount of protein necessary for study. Typically, only 10–200 micrograms of protein in a volume of 35 ul is sufficient for screening with a Bruker 600 MHz and TXI 1.7 mm MicroCryoprobe. Our microscale protein NMR sample screening pipeline has been discussed elsewhere (Zhang et al., 2008; Rossi et al., 2010). In the context of the Midi-scale expression and purification procedure, there are a few changes. The 96 purified proteins are buffer exchanged into NMR buffer (typically, 20 mM MES, 200 mM NaCl, 10 mM DTT, 5 mM CaCl2 at pH 6.5) using a Zeba™ 96-well desalting spin plate (Thermo Scientific) and aliquots are transferred to 1.7 mm SampleJet Tubes (Bruker) using a Gilson 96 liquid handler. The rich TB broth does not allow for isotope enrichment, therefore only 1D 1H NMR spectra can be preformed. However, this screen can detect the dispersion of amide protons and upfield-shifted methyl protons that are indicative of aromatic and methyl stacking (folded protein core). Proteins exhibiting these traits, well folded and disperse amide protons, are more than likely amenable for structure determination by NMR and can be scaled up for fermentation and purification with isotope enrichment.
4. Preparative-Scale Protein Sample Production
Proteins with high expression and solubility levels, high monodispersity, and/or good 1D 1H NMR spectra are promoted for large-scale expression, purification, and sample preparation.
4.1. Preparative Protein Fermentation
Although recent technical advances have in some cases allowed structural determination with as little as 75 micograms of protein (Aramini et al., 2007), the amount of protein required for crystallization screening and/or structure determination by NMR is typically 5 – 50 mg, with greater than 95% purity. Our process for preparative-scale (large-scale) protein expression has been designed to optimize conditions with respect to yield, cost, throughput, and the different structural determination approaches. A strategy based on fermentation in 2-liter baffled Furnbach flasks was chosen because of its simplicity, the low cost of the associated equipment such as shakers, and ease of parallelization (Acton et al., 2005). In addition, NMR structural determination requires enrichment with 15N, 13C, and/or 2H isotopes. Conversely, high-throughput X-ray crystallography of proteins is most efficient using single (SAD) and multiple anomalous diffraction (MAD) methods (Hendrickson, 1991) requiring selenomethionine substituted protein samples. In order to achieve this we have developed a fermentation system based on growth with MJ9 minimal media (Jansson et al., 1996). This allows both isotopic (i.e. 15N, 13C, and/or 2H) enrichment or selenomethionine labeling while achieving adequate cell density and protein expression levels for structural biology studies.
Briefly, protein expression constructs that pass analytical scale characterization are identified through the PLIMS database, and their appropriate glycerol stock plate and well position reported (each plate has a unique bar code identification). An aliquot is transferred to 500 μl of LB with ampicillin and kanamycin and incubated for six hours at 37 °C. This preculture (40 μl) is then used to inoculate a 250 ml flask containing 40 ml of MJ9 minimal media, and incubated overnight at 37 °C. For producing isotope-enriched proteins for NMR (e.g. U-13C,U-15N-enriched proteins), the entire volume of overnight culture is then used to inoculate a 2-liter baffled flask containing 1.0 liter of MJ9 supplemented with uniformly (U)-13C glucose and (U)-15NH4 salts as the sole source of carbon and nitrogen. For X-ray crystallography, non-isotope enriched carbon and ammonia sources are used. In both cases, the cultures are incubated at 37 °C until the OD600 reaches of 0.8–1.0 units, equilibrated to 17 °C, and induced with IPTG (1 mM final concentration). As a slight modification, in selenomethionine labeling, induction is done 15 min after addition of several amino acids to the medium to down regulate methionine synthesis (lysine, phenylalanine, and threonine at 100 mg/liter, isoleucine, leucine, and valine at 50 mg/liter, L-selenomethionine at 60 mg/liter) (Doublie et al., 1996). The use of a methionine auxotroph is often used for selenomethionine incorporation (Walden, 2010). However our strategy, repressing methionine synthesis, routinely results in 75–80 % selenomethionine substitution and allows for the same expression host to be utilized for both NMR and X-ray crystallography sample production. Incubation with vigorous shaking in a 17 °C room continues overnight followed by harvesting through centrifugation. An aliquot of cells at harvest is used for determining final cell density and for SDS-PAGE analysis of expression and solubility. An aliquot is also taken and sequence analyzed for quality control. During PSI-2, we acquired an Avanti centrifuge (Beckman) with the Harvestliner bag system, these centrifuge bags allow for storage in minimal space, as well as ease of cell resuspension in subsequent steps. All data is uploaded in the PLIMS database; select information useful for sharing across the NESG consortium and/or with the public databases is transferred to our project-wide Spine database (Bertone et al., 2001). Add Goh et al ref.
4.2. Large-Scale Parallel Protein Purification Using ÄKTAxpress Systems
For both X-ray crystallography and NMR structural studies, it is imperative that the protein samples are highly homogeneous. The need to produce protein samples of sufficient purity while retaining high throughput is challenging. For preparing samples for either NMR or X-ray crystallography, the centrifuge bags are thawed on ice and 30 ml of lysis buffer containing protease inhibitors (Complete, Mini, EDTA-free, Roche) are used to resuspend the cells. The bag contents are then transferred to a metal sonication cup and sonicated in an ice water bath for 5 × 60s cycles (10s on/10s off). The supernatant is cleared by centrifugation at 27,000 × g for 30 min, followed by filtering through a 0.2 μm filter. The supernatant is then loaded onto an ÄKTAxpress system (GE Healthcare) and a two-step automated purification protocol is performed, comprised of a Ni-affinity column (HisTrap HP, 5 ml), and a gel filtration column (Superdex 75 26/60, GE Healthcare) in a linear series using the preinstalled default settings (AF-GF). Briefly, the 6X His-tagged proteins are eluted from the HisTrap column using 5 column volumes of Elution Buffer (50 mM Tris-HCl, 500 mM NaCl, 500 mM imidazole, 1 mM TCEP, pH 7.5) at 4 ml/min. The proteins are automatically detected by monitoring absorbance at 280 nm, and fractions above the designated threshold (major peaks) are collected into internal storage loops. The major peaks are then automatically injected onto the Superdex 75 gel filtration column equilibrated with low salt buffer (20 mM Tris-HCL, 100 mM NaCl, 5 mM DTT, pH 7.5) or Standard NMR Buffers (Table 2). Resulting protein fractions above the designated absorbance threshold are collected into 2 ml 96-well blocks and the purification trace for each protein is archived into the Spine database. The ÄKTAxpress system is modular in design with four HisTrap HP columns and one size-exclusion column per module allowing four separate two-step purifications in less than twelve hours. Overall, we have found this system to be extremely robust.
Table 2.
Buffers used for NMR buffer optimization screening.
| Buffer ID | PH | Recipe |
|---|---|---|
| MJ001 | 6.5 | 20 mM MES†, 100 mM NaCl, 5 mM CaCl2, 10 mM DTT‡, 0.02% NaN3, protease inhibitor# 1x, 10% D2O |
| MJ002 | 5.5 | 20 mM NH4OAc, 100 mM NaCl, 5 mM CaCl2, 10mM DTT, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ003 | 4.5 | 20mM NH4OAc, 100mM NaCl, 5mM CaCl2, 10mM DTT, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ004 | 5.0 | 50 mM NH4OAc, 10 mM DTT, 50 mM Arginine, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ005 | 5.0 | 50 mM NH4OAc, 10 mM DTT, 5% CH3CN, 0.02% NaN3, protease inhibitor 1x, 10%D2O |
| MJ006 | 6.0 | 50 mM MES, 10 mM DTT, 50 mM Arginine, 0.02% NaN3, protease inhibitor 1x, 10%D2O |
| MJ007 | 6.0 | 50 mM MES, 10 mM DTT, 5% CH3CN, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ008 | 6.5 | 25 mM Na2PO4, 450 mM NaCl, 10 mM DTT, 20 mM ZnSO4, protease inhibitor 1x, 0.01% NaN3 (pH 6.5), 10% D2O |
| MJ009 | 6.5 | 20 mM MES, 100 mM NaCl, 5% CH3CN, 10mM DTT, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ010 | 6.5 | 20 mM MES, 100 mM NaCl, 50 mM Arginine, 10mM DTT, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
| MJ011 | 6.5 | 20 mM MES, 100 mM NaCl, 1% Zwitter§, 10 mM DTT, 0.02% NaN3, 10% D2O |
| MJ012 | 6.5 | 20 mM MES, 100 mM NaCl, 50 mM ZnSO4, 10 mM DTT, 0.02% NaN3, protease inhibitor 1x, 10% D2O |
DTT: Dithiothreitol,
MES: 2-(N-morpholino)ethanesulfonic acid,
Zwitter: ZWITTERAGENT® 3–12 Detergent cat. 963015 (CALBIOCHEM),
TCEP: tris(2-carboxyethyl)phosphine,
Tris: tris(hydroxymethyl)aminomethane,
Protease inhibitor: Protease inhibitor cocktail tablets cat. 11836170001 (ROCHE).
4.3. Sample Preparation
The fractions produced on the ÄKTAxpress are analyzed by SDS-PAGE and pooled followed by concentration using Amicon ultrafiltration concentrators (Millipore). The preparation is then subjected to a series of quality control and analytical protein chemistry steps including aggregation screening by analytical gel filtration with static light scattering, homogeneity analysis using SDS-PAGE, molecular weight validation by MALDI-TOF mass spectrometry, and concentration determination by a NanoDrop ND-8000 Spectrophotometer. This data is then archived into Spine for use by researchers throughout the NESG.
For NMR sample preparation, the fractions produced on the ÄKTAxpress are analyzed by SDS-PAGE and pooled, followed by concentration using Amicon ultrafiltration concentrators (Millipore). All samples are spiked with 50 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) as an internal reference, 1X Complete Protease cocktail (Roche) and 10% 2H2O. For NMR microprobe screening, aliquots (8 or 35 μl) are then transferred to 1.0-mm or 1.7-mm SampleJet Tubes (Bruker), respectively, using a Gilson 96-well liquid-handler. Samples destined for structure determination are transferred into a 5 mm Shigemi tube (BP50) with a volume of 300 μl, and stored at 4 °C.
For preparation of crystallization screening samples, selenomethionine labeled proteins are concentrated to ~10 mg/ml in low salt buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM d,l-dithiothreitol). Protein samples are aliquoted in 100 ul portions, flash-frozen in liquid nitrogen, and stored at −80 °C. These samples are then used for crystallization screening (Luft et al., 2003).
In addition to archiving the data generated during the purification procedure, SPINE also serves to direct shipments of protein samples to NESG researchers outside of the protein production lab. The majority of crystal screening and structure determination is performed outside of the protein production lab and NMR samples are also shipped for structural determination by various NESG researchers. The Spine database coordinates this effort with bar code based registration of shipment tubes and automatically tracks shipments through the FedEx database.
5. Protein Salvage Strategies
For proteins providing marginal quality (e.g. “Promising”) HSQC spectra or crystal screening hits that cannot be optimized to provide diffraction quality crystals, several “salvage” processes have been developed to provide more tractable protein samples. Some of the most effective strategies include sample buffer optimization using microprobe NMR screening (Rossi et al., 2010) and further construct optimization using amide hydrogen deuterium exchange with mass spectrometry detection (HDX-MS) (Sharma et al., 2009).
5.1. Buffer Optimization
Proteins are identified for buffer optimization during the initial screening process carried out with a standard NMR buffer at pH 6.5 (or pH 4.5) (Table 2). If a protein sample is deemed adequate for structure determination but unstable such that prohibitive precipitation would occur during the data acquisition time period, or provided marginal quality HSQC spectra, then the sample is directed to buffer optimization. Briefly, a purified protein for a given target is exchanged into twelve buffer conditions (Table 2) using a Zeba™ 96-well desalting spin plate (Thermo Scientific) and loaded into separate corresponding NMR microprobe tubes. The tubes are scored for precipitation following a set time interval equal to the average data acquisition time for an NMR study. This is followed by NMR screening. This identifies the most stable buffer conditions and future protein samples for this protein are prepared in the identified buffer. A more detailed description of sample preparation for buffer optimization has been previously described (Rossi et al., 2010).
5.2. Construct Optimization using Amide Hydrogen Deuterium Exchange with Mass Spectrometry Detection (HDX-MS)
The disorder prediction methods described above using the DisMeta server have improved the efficiency of our production pipeline. In some instances, however these predictions do not reliably identify the disordered regions of the protein. In these cases, NMR screening can identify that some regions of the protein are disordered, even without determining the resonance assignments and location of the disordered regions. In order to refine such constructs, we have implemented and automated the HDX-MS procedure (Sharma et al., 2009; Englander, 2006; Woods and Hamuro, 2001) for the experimental identification of the boundaries of disordered protein segments. Once identified, alternate constructs are designed to delete these regions, and reintroduced into the protein production pipeline for recloning, protein purification and attempts for structural determination. As an example, in a recent pilot study on a small set of targets from the NESG (Sharma et al., 2009), we demonstrated the feasibility of using HDX-MS to design truncated constructs yielding NMR spectra that are more amenable for NMR structure determination and crystallization.
5.3. Alternative expression systems
5.3.1 Wheat Germ Cell Free Protein Expression
A recent analysis of expression and solubility data derived from the NESG pipeline attests to the difficulty of producing eukaryotic proteins in E. coli. Whereas 49% of bacterial target domains were solubly expressed, only 32% of eukaryotic target domains were solubly expressed in bacteria. Previous studies have shown that eukaryotic cell-free systems may permit successful production of proteins that undergo proteolysis or accumulate in inclusion bodies during bacterial expression (Vinarov et al., 2006). To further explore and develop this technology, we have modified the Promega TnT wheat germ cell-free (WGCF) vector to allow ligase independent cloning of the PCR products used in our normal cloning pipeline. A pilot study (Zhao et al., 2010) using this system with 66 non-secreted human targets that were problematic in the prokaryotic expression system produced very promising results. Following wheat germ cell-free expression we have found that 9 out of 13 bacterially expressed but insoluble proteins were solubly expressed in WGCF. In total, 34 of the 66 non-secreted human targets (52%) were solubly expressed in WGCF. The major drawback to this system is the fact that only small quantities of protein can be produced. However, recent advances in NMR microprobe technology have made it possible to determine structures with the roughly 100 microgram levels of protein routinely produced in this system (Aramini et al., 2007; Rossi et al., 2010).
5.3.2 Chaperone enhanced E. coli expression strains
The NESG has also developed E. coli strains in which one or more E. coli chaperones (including GroEL/ES, trigger factor, DnaK/J, GrpE, and ClpB) can be induced during recombinant protein expression. These systems exploit the pACYCDuet vector system (Novagen), in which multiple T7lac promoters control expression of the chaperones. These vectors are compatible with our modified pET15 and pET21 vectors, the workhorse plasmids of the NESG, and can co-exist within BL21(DE3) in a stable fashion. We have found that these chaperones can aid in producing soluble protein expression. As a case study, protein target ER58 (COAE_ECOLI) has limited solubility when expressed in our normal T7 system. However, co-expression with trigger factor in a pDuet system greatly enhances solubility. Using this approach resulted in the structure of protein target ER58 (pdb_id 1spv).
6. Future Perspectives
The attrition rate in protein production of human and other eukaryotic targets is considerably higher and continues through each step in the pipeline. However, we view eukaryotic protein targets as very high value in spite of the added difficulty. This will also be a driving force for technology development going forward. Although challenges exist, the current NESG Protein Production Pipeline has proven very robust for both producing some eukaryotic proteins in a form amenable to 3D structure determination. To date, the NESG has deposited into the Protein Data Bank (PDB) some 80 eukaryotic protein structures, including some 50 human protein structures. This represents approximately ten percent of the total NESG structure count of over 900 PDB submissions.
Although eukaryotic targets have higher attrition rates than prokaryotic targets we have made considerable progress as shown in Figure 6. During PSI-2 we have cloned over 1200 human protein domains as 2850 constructs (2–10 constructs per domain based on the number of predicted disordered regions in the target). Roughly 4% of cloned human domains have progressed to structural determination, and many more of these samples will yield structures in the coming months. This success rate greatly eclipses the rate we found with over 1000 full-length proteins human proteins (<1%) in PSI-1. A large factor in this increased efficiency is no doubt a result of the disorder based construct optimization software. As shown in the bar graph in Figure 6, it appears that NMR has a significantly higher success rate than X-ray crystallography in producing structures from these optimized human constructs. Conversely, many other protein families such as the Meta domains and Tol-B mega-family among others are more successful as X-ray crystallography targets. This is consistent with previous reports indicating the complementary nature of the structural determination approaches (Snyder et al., 2005; Yee et al., 2005). Although we have greatly increased efficiency in producing protein samples of human domains amenable for structural determination, prokaryotic targets continue to yield higher success rates (~6% “In PDB”/cloned).
Fig. 6.

The percentage of PDB depositions relative to cloned targets from various origins and target classes. The graph is further broken down into targets solved by NMR and X-ray crystallography. Target classes are from left to right Prokaryotic (Prok), Eukaryotic (Euk), Human, Metagenomic, Ubiqutitin, OB-Fold (2.40.50.140), Start Domains (3.30.530.40), MCSG Salvage (small soluble proteins that failed to crystallize), BIG-Family, P-loop (3.40.50.300), NADP- binding (3.40.50.720), Tol-B (2.120.10.30), Aldolase (3.20.20.70), Polysaccaride synthesis, (3.90.550.10), Oxidoreductase (3.20.20.70) and VP39 (3.40.50.150). CATH superfamily indentification numbers (Greene et al., 2007) are indicated in parenthesis where appropriate.
Realizing the need for producing eukaryotic proteins in eukaryotic hosts, the NESG is investing in many promising technologies for eukaryotic protein sample production, such as WGCF coupled with NMR microprobe technology, as described above. Unfortunately, the yield of proteins from WGCF is often not sufficient for X-ray crystallography studies in a relatively cost effective manner. Accordingly, we are also exploring other technologies that produce the larger amounts of eukaryotic proteins necessary for crystallization studies. NESG researchers have explored a Pichia pastoris expression system that has been successful in producing soluble secreted human proteins. Fermentation yields from this organism can often be in the tens of milligram range. Another promising eukaryotic expression system we are exploring is a human HEK293T cell-based expression systems in conjunction with new bioreactor technology. This technology involves large-scale production of recombinant secreted proteins using the “BelloCell” oscillating bioreactor, which provides the surface area of over 20 one-liter roller bottles (Wang et al., 2006; Ho et al., 2004). Using this technology, NESG researchers have produce 3–4 mg samples of secreted glycosylated human proteins, amounts suitable for crystallization studies, while greatly reducing media cost.
Going forward we will continue to develop the NESG Protein Production Pipeline with the goal of increased efficiency and broader applicability to eukaryotic proteins and protein complexes. There are several areas that can be improved as technology is developed. One area is increased use of total gene synthesis. Although prohibitively expensive to use as a primary source of PCR template today, it continues to decline in cost. Gene optimization for expression in E. coli has in our hands resulted in increased expression levels. In many cases it is the difference between no expression with the natural sequence to high levels of expression with the codon optimized synthetic genes. Although we typically employ rare codon enhanced strains of BL21, not all genes can be rescued by this strategy. Total gene synthesis will no doubt play a large role in biology research going forward. Additionally, we will develop eukaryotic host systems, more robust vectors, and improved purification procedures for the eukaryotic targets. Often, the lower expression rates and more limited solubility of eukaryotic proteins leads to protein purifications that are less homogenous following our current high-throughput procedures, and various approaches are being explored to improve the purification process for these targets.
Conclusions
This paper illustrates the overall sample production and screening technologies of the NESG. It is both scalable and efficient in regard to cost (initial setup of equipment and supplies) and time. We have outlined our strategies and platform for HTP production of high-quality protein samples using E. coli expression systems. The platform contains powerful bioinformatics tools (software, database, and servers), HTP ligation-independent cloning using a BioRobot 8000, HTP Midi-scale fermentation for biophysical characterization, and multi-module large-scale protein purification system using AKTAxpress systems, among other notable technology. Detailed protocols can be found at (www-nmr.cabm.rutgers.edu/labdocuments/proteinprod/index.htm) and are freely available.
Unlike many other reported protein production pipelines, this system is easily scalable, adding more equipment or personnel will easily increase output. Perhaps even more important, this system was engineered to work with commercially available equipment and reagents, such that it can be duplicated in any structural biology lab or core facility. Much of the technology designed for this project will likely prove useful for other structural biologists, such as the disorder-based construct design, robotic cloning and analytical expression.
With the low cost of primers and relatively inexpensive Qiagen lab automation the barrier to this strategy is low. One of the main goals of this paper to inform the community about our technology and to make it accessible to other researchers. Overall, this pipeline has allowed us to clone and express nearly 17,000 different protein targets, purify over 4,400 proteins in tens of milligram quantities, and deposit over 900 new protein structures to the PDB over the past ten years (http://nesg.org/statistics.html). Our current production rates are about 30–40 purified protein targets in tens of milligram quantities per week. This enables us to achieve success in producing not only enough protein samples but also 3D structures. We hope that these improved automated and/or parallel cloning, expression, protein production, and biophysical screening technologies will of value to genomic biologists, biochemists, and structural biologists.
Acknowledgments
We thank Profs. C. Arrowsmith, J. Hunt, M. Gerstein, M. Inouye, J. Marcotrigiano, and L. Tong, along with all the members of the NESG Consortium, for valuable advice in the development of the NESG Protein Sample Production Platform. This work was supported by a grant from the National Institute of General Medical Sciences Protein Structure Initiative U54-GM074958 (to GTM).
Abbreviations
- 6X-His
hexa-histidine polypeptide sequence tag
- 3D
three-dimensional
- HCPIN
Human Cancer Pathway Protein Interaction Network
- HDX-MS
amide hydrogen deuterium exchange with mass spectrometry detection
- HMM
hidden Markov model
- HTP
high throughput
- LIC
ligation independent cloning
- LSC
Large Scale Centers
- MALDI-TOF
matrix-assisted laser-desorption-induced time-of-flight
- MCS
multiple cloning site
- MDA
Multiple Displacement Amplification
- MMLV
Moloney Mouse Leukemia Virus
- NESG
Northeast Structural Genomics Consortium
- NIGMS
National Institute of General Medical Sciences
- NMR
nuclear magnetic resonance spectroscopy
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- PLIMS
Protein Laboratory Information Management System
- PSI-2
Protein Structure Initiative-2
- SDS-PAGE
sodium dodecylsulfate polyacrylamide electrophoresis
- WGA
Whole Genome Amplification
References
- Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, Baran MC, Chiang YW, Climent T, Cooper B, Denissova NG, Douglas SM, Everett JK, Ho CK, Macapagal D, Rajan PK, Shastry R, Shih LY, Swapna GV, Wilson M, Wu M, Gerstein M, Inouye M, Hunt JF, Montelione GT. Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- Acton TB, Lee DY, Wang H, Montelione GT. An Alternative Method for Generating Genomic DNA PCR Template and Expansion of ‘Reagent Genomes’. 2010. in preparation. [Google Scholar]
- Aramini JM, Rossi P, Anklin C, Xiao R, Montelione GT. Microgram-scale protein structure determination by NMR. Nat Methods. 2007;4(6):491–493. doi: 10.1038/nmeth1051. [DOI] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Kluger Y, Lan N, Zheng D, Christendat D, Yee A, Edwards AM, Arrowsmith CH, Montelione GT, Gerstein M. SPINE: an integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res. 2001;29(13):2884–2898. doi: 10.1093/nar/29.13.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousse L, Mouradian S, Minalla A, Yee H, Williams K, Dubrow R. Protein sizing on a microchip. Anal Chem. 2001;73(6):1207–1212. doi: 10.1021/ac0012492. [DOI] [PubMed] [Google Scholar]
- Chikayama E, Kurotani A, Tanaka T, Yabuki T, Miyazaki S, Yokoyama S, Kuroda Y. Mathematical model for empirically optimizing large scale production of soluble protein domains. BMC Bioinformatics. 2010;11:113. doi: 10.1186/1471-2105-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe J, Dobeli H, Gentz R, Hochuli E, Stuber D, Henco K. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol Biol. 1994;31:371–387. doi: 10.1385/0-89603-258-2:371. [DOI] [PubMed] [Google Scholar]
- Dean FB, Hosono S, Fang L, Wu X, Faruqi AF, Bray-Ward P, Sun Z, Zong Q, Du Y, Du J, Driscoll M, Song W, Kingsmore SF, Egholm M, Lasken RS. Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci U S A. 2002;99(8):5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessailly BH, Nair R, Jaroszewski L, Fajardo JE, Kouranov A, Lee D, Fiser A, Godzik A, Rost B, Orengo C. PSI-2: structural genomics to cover protein domain family space. Structure. 2009;17(6):869–881. doi: 10.1016/j.str.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S, Kapp U, Aberg A, Brown K, Strub K, Cusack S. Crystallization and preliminary X-ray analysis of the 9 kDa protein of the mouse signal recognition particle and the selenomethionyl-SRP9. FEBS Lett. 1996;384(3):219–221. doi: 10.1016/0014-5793(96)00316-x. [DOI] [PubMed] [Google Scholar]
- Dyson MR, Shadbolt SP, Vincent KJ, Perera RL, McCafferty J. Production of soluble mammalian proteins in Escherichia coli: identification of protein features that correlate with successful expression. BMC Biotechnol. 2004;4:32. doi: 10.1186/1472-6750-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17(11):1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett JK, Acton TB, Montelione GT. Primer Prim’r: A web based server for automated primer design. Journal of Functional and Structural Genomics. 2004;5(1–2):13–21. doi: 10.1023/B:JSFG.0000029238.86387.90. [DOI] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Burley SK. Use of dynamic light scattering to assess crystallizability of macromolecules and macromolecular assemblies. Structure. 1994;2(5):357–359. doi: 10.1016/s0969-2126(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Ferre-D’Amare AR, Burley SK. Methods in Enzymology. Vol. 276. New York: Academic Press; 1997. Dynamic Light Scattering in Evaluating Crystallizability of Macromolecule; pp. 157–166. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh CS, Lan N, Douglas SM, Wu B, Echols N, Smith A, Milburn D, Montelione GT, Zhao H, Gerstein M. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. J Mol Biol. 2004;336(1):115–130. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- Goh CS, Lan N, Echols N, Douglas SM, Milburn D, Bertone P, Xiao R, Ma LC, Zheng D, Wunderlich Z, Acton T, Montelione GT, Gerstein M. SPINE 2: a system for collaborative structural proteomics within a federated database framework. Nucleic Acids Res. 2003;31(11):2833–2838. doi: 10.1093/nar/gkg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008a;5(2):135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graslund S, Sagemark J, Berglund H, Dahlgren LG, Flores A, Hammarstrom M, Johansson I, Kotenyova T, Nilsson M, Nordlund P, Weigelt J. The use of systematic N- and C-terminal deletions to promote production and structural studies of recombinant proteins. Protein Expr Purif. 2008b;58(2):210–221. doi: 10.1016/j.pep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, Sillitoe I, Yeats C, Thornton JM, Orengo CA. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35(Database issue):D291–297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun RS, Moss J. Ligation-independent cloning of glutathione S-transferase fusion genes for expression in Escherichia coli. Gene. 1992;112(1):37–43. doi: 10.1016/0378-1119(92)90300-e. [DOI] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254(5028):51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Ho L, Greene CL, Schmidt AW, Huang LH. Cultivation of HEK 293 cell line and production of a member of the superfamily of G-protein coupled receptors for drug discovery applications using a highly efficient novel bioreactor. Cytotechnology. 2004;45(3):117–123. doi: 10.1007/s10616-004-6402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Hang D, Lu LJ, Tong L, Gerstein MB, Montelione GT. Targeting the human cancer pathway protein interaction network by structural genomics. Mol Cell Proteomics. 2008;7(10):2048–2060. doi: 10.1074/mcp.M700550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Li YC, Jendenberg L, Anderson S, Montelione GT. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. Journal of Biomolecular NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71(2):982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- Kvist T, Ahring BK, Lasken RS, Westermann P. Specific single-cell isolation and genomic amplification of uncultured microorganisms. Appl Microbiol Biotechnol. 2007;74(4):926–935. doi: 10.1007/s00253-006-0725-7. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, Rogers J, Lawlor S, McLaren S, Dricot A, Borick H, Cusick ME, Vandenhaute J, Dunham I, Hill DE, Vidal M. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89(3):307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol. 2007;10(5):510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Lasken RS. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem Soc Trans. 2009;37(Pt 2):450–453. doi: 10.1042/BST0370450. [DOI] [PubMed] [Google Scholar]
- Liu J, Hegyi H, Acton TB, Montelione GT, Rost B. Automatic target selection for structural genomics on eukaryotes. Proteins. 2004;56(2):188–200. doi: 10.1002/prot.20012. [DOI] [PubMed] [Google Scholar]
- Luft JR, Collins RJ, Fehrman NA, Lauricella AM, Veatch CK, DeTitta GT. A deliberate approach to screening for initial crystallization conditions of biological macromolecules. J Struct Biol. 2003;142(1):170–179. doi: 10.1016/s1047-8477(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Montelione GT, Anderson S. Structural genomics: keystone for a Human Proteome Project. Nat Struct Biol. 1999;6(1):11–12. doi: 10.1038/4878. [DOI] [PubMed] [Google Scholar]
- Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, Montelione GT, Rost B. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10(2):181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388(6640):343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- Peti W, Page R, Moy K, O’Neil-Johnson M, Wilson IA, Stevens RC, Wuthrich K. Towards miniaturization of a structural genomics pipeline using micro-expression and microcoil NMR. J Struct Funct Genomics. 2005;6(4):259–267. doi: 10.1007/s10969-005-9000-x. [DOI] [PubMed] [Google Scholar]
- Price WN, 2nd, Chen Y, Handelman SK, Neely H, Manor P, Karlin R, Nair R, Liu J, Baran M, Everett J, Tong SN, Forouhar F, Swaminathan SS, Acton T, Xiao R, Luft JR, Lauricella A, DeTitta GT, Rost B, Montelione GT, Hunt JF. Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data. Nat Biotechnol. 2009;27(1):51–57. doi: 10.1038/nbt.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price WN, Handelman SK, Everett J, Tong SN, Bracic A, Acton T, Xiao R, Rost B, Montelione GT, Hunt JF. Large scale studies show unexpected amino acid effects on protein expression and solubility. 2010. in preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Love J, Handelman S, Hunt JF, Shapiro L, Hendrickson WA, Rost B. Structural genomics target selection for the New York consortium on membrane protein structure. J Struct Funct Genomics. 2009;10(4):255–268. doi: 10.1007/s10969-009-9071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi P, Swapna GV, Huang YJ, Aramini JM, Anklin C, Conover K, Hamilton K, Xiao R, Acton TB, Ertekin A, Everett JK, Montelione GT. A microscale protein NMR sample screening pipeline. J Biomol NMR. 2010;46(1):11–22. doi: 10.1007/s10858-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, Clingingsmith TR, Hartley JL, Esposito D, Cheo D, Moore T, Simmons B, Sequerra R, Bosak S, Doucette-Stamm L, Le Peuch C, Vandenhaute J, Cusick ME, Albala JS, Hill DE, Vidal M. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14(10B):2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Zheng H, Huang YJ, Ertekin A, Hamuro Y, Rossi P, Tejero R, Acton TB, Xiao R, Jiang M, Zhao L, Ma LC, Swapna GV, Aramini JM, Montelione GT. Construct optimization for protein NMR structure analysis using amide hydrogen/deuterium exchange mass spectrometry. Proteins. 2009;76(4):882–894. doi: 10.1002/prot.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheibani N. Prokaryotic gene fusion expression systems and their use in structural and functional studies of proteins. Prep Biochem Biotechnol. 1999;29(1):77–90. doi: 10.1080/10826069908544695. [DOI] [PubMed] [Google Scholar]
- Shirano Y, Shibata D. Low temperature cultivation of Escherichia coli carrying a rice lipoxygenase L-2 cDNA produces a soluble and active enzyme at a high level. FEBS Lett. 1990;271(1–2):128–130. doi: 10.1016/0014-5793(90)80388-y. [DOI] [PubMed] [Google Scholar]
- Slabinski L, Jaroszewski L, Rychlewski L, Wilson IA, Lesley SA, Godzik A. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics. 2007;23(24):3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- Snyder DA, Chen Y, Denissova NG, Acton T, Aramini JM, Ciano M, Karlin R, Liu J, Manor P, Rajan PA, Rossi P, Swapna GV, Xiao R, Rost B, Hunt J, Montelione GT. Comparisons of NMR spectral quality and success in crystallization demonstrate that NMR and X-ray crystallography are complementary methods for small protein structure determination. J Am Chem Soc. 2005;127(47):16505–16511. doi: 10.1021/ja053564h. [DOI] [PubMed] [Google Scholar]
- Vinarov DA, Loushin Newman CL, Markley JL. Wheat germ cell-free platform for eukaryotic protein production. FEBS J. 2006;273(18):4160–4169. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- Walden H. Selenium incorporation using recombinant techniques. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):352–357. doi: 10.1107/S0907444909038207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IK, Hsieh SY, Chang KM, Wang YC, Chu A, Shaw SY, Ou JJ, Ho L. A novel control scheme for inducing angiostatin-human IgG fusion protein production using recombinant CHO cells in a oscillating bioreactor. J Biotechnol. 2006;121(3):418–428. doi: 10.1016/j.jbiotec.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Woods VL, Jr, Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J Cell Biochem Suppl. 2001;(Suppl 37):89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- Yee AA, Savchenko A, Ignachenko A, Lukin J, Xu X, Skarina T, Evdokimova E, Liu CS, Semesi A, Guido V, Edwards AM, Arrowsmith CH. NMR and X-ray crystallography, complementary tools in structural proteomics of small proteins. J Am Chem Soc. 2005;127(47):16512–16517. doi: 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Horst R, Geralt M, Ma X, Hong WX, Finn MG, Stevens RC, Wuthrich K. Microscale NMR screening of new detergents for membrane protein structural biology. J Am Chem Soc. 2008;130(23):7357–7363. doi: 10.1021/ja077863d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Zhao K, Hurst R, Slater M, Acton TB, Swapna GVT, Shastri R, Kornhaber GJ, Montelione GT. Engineering of a wheat germ expression system to provide compatibility with a high throughput pET-based cloning platform. J Struct Funct Genomics. 2010 doi: 10.1007/s10969-010-9093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Cai G, Hall EO, Freeman GJ. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques. 2007;43(3):354–359. doi: 10.2144/000112536. [DOI] [PubMed] [Google Scholar]