Abstract
The c-myc proto-oncogene product, Myc, is a transcription factor that binds thousands of genomic loci1. Recent work suggested that rather than up- and down-regulating selected groups of genes1-3, Myc targets all active promoters and enhancers in the genome (a phenomenon termed “invasion”) and acts as a general amplifier of transcription4,5. However, the available data did not readily discriminate between direct and indirect effects of Myc on RNA biogenesis. We addressed this issue with genome-wide chromatin immunoprecipitation and RNA expression profiles during B-cell lymphomagenesis in mice, in cultured B-cells and fibroblasts. Consistent with long-standing observations6, we detected general increases in total RNA or mRNA copies per cell (hereby termed “amplification”)4,5 when comparing actively proliferating cells with control quiescent cells: this was true whether cells were stimulated by mitogens (requiring endogenous Myc for a proliferative response)7,8 or by deregulated, oncogenic Myc activity. RNA amplification and promoter/enhancer invasion by Myc were separable phenomena that could occur without one another. Moreover, whether or not associated with RNA amplification, Myc drove the differential expression of distinct subsets of target genes. Hence, while having the potential to interact with all active/poised regulatory elements in the genome4,5,9-11, Myc does not directly act as a global transcriptional amplifier4,5. Instead, our results imply that Myc activates and represses transcription of discrete gene sets, leading to changes in cellular state that can in turn feed back on global RNA production and turnover.
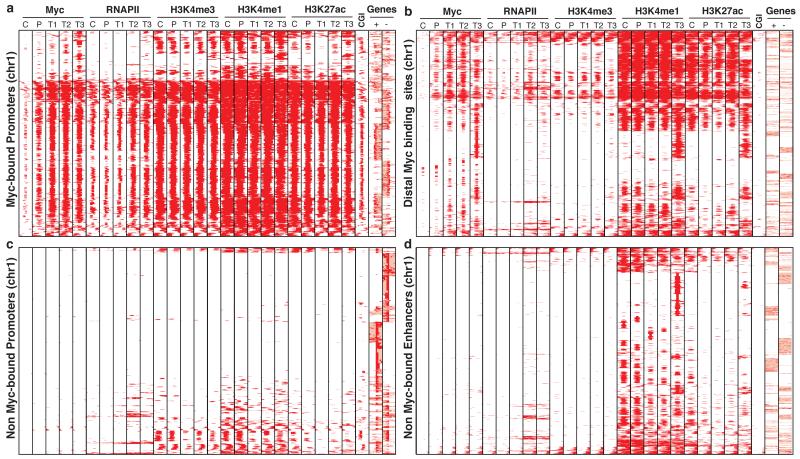
We first analyzed the genomic distribution of Myc during B-cell lymphomagenesis in vivo. To this aim, we generated ChIP-seq profiles in B-cells from young non-transgenic (Control, “C”) and Eμ-myc transgenic littermates (Pre-tumoral, “P”), and in lymphomas arising in adult Eμ-myc animals (Tumor, “T”) (Extended Data Fig. 1a-j). Consistent with progressive increases in Myc mRNA and proteins levels, both binding intensity and the total number of binding sites progressively increased (ca. 7,000 in C, 17,000 in P, 30,000 in T). Two thirds of the Myc peaks in C were proximal to an annotated Transcription Start Site (-2 to +1 kb from the TSS, henceforth “promoter”). While the numbers of proximal and distal peaks both increased in P and T, most of the new binding sites were distal, with equal proportions of intra- and extra-genic locations, and increasing distances from the nearest TSS. To chart active promoters and enhancers12,13, we profiled RNA Polymerase II (RNAPII) and the histone marks H3K4me3, H3K4me1 and H3K27ac: as expected, these features marked virtually all Myc-bound promoters (Fig. 1a, Extended Data Fig. 2a)9,10. Taking TSS annotations and H3K4me3 as references, Myc bound ca. 34% of active promoters in C, 66% in P, and 87-94% in T. Instead, most unbound promoters showed no active histone marks or RNAPII (Fig. 1c). Distal Myc-binding sites bore H3K4me1, the activation mark H3K27ac and to a lesser extent RNAPII, and showed the high H3K4me1/H3K4me3 ratios characteristic of enhancers (Fig. 1b, Extended Data Fig. 2b, c, 3a). Out of 20 H3K4me1-positive regions characterized as active enhancers in mouse B-cells14, 2 were bound by Myc in C, 9 in P and 18 in T (Extended Data Fig. 3b). Instead, distal enhancers with no Myc showed H3K4me1 but little or no H3K27ac or RNAPII (Fig. 1d), indicative of an inactive state. At both proximal and distal sites, the RNAPII and chromatin patterns in naive B-cells (sample C) were similar to those in P and T, preceding Myc at the same sites (Fig. 1a, b), and their intensities correlated with those of Myc binding (Extended Data Fig. 2d). In summary, Myc associated with regulatory elements that pre-existed in a poised/active state in naive B-cells, the characteristic chromatin profiles of these sites anticipating Myc binding9. A majority of these active elements was ultimately targeted in tumors, consistent with the concept of invasion4.
Fig. 1. Increased Myc levels during lymphoma progression lead to invasion of accessible regulatory elements in the genome.
a. The heatmap shows the distribution of Myc at annotated promoters. Each row represents a different genomic interval (6 kb width centered on Myc peaks). The panel includes every annotated promoter in chromosome 1 that was called as Myc-associated by ChIP-seq in at least one of the experimental samples (C, P, T1, T2, T3). For the same intervals, the distributions of RNAPII, H3K4me3, H3K4me1, H3K27ac, CpG Islands (CGIs) and annotated genes (exons in red, introns in pink; + sense, - antisense strand) are also shown. b-d. As in a., for b. distal (non promoter) Myc-binding sites, d. Non Myc-bound promoters (6 kb width centered on the TSS) and c. Non Myc-bound enhancers, identified as distal H3K4me1-positive elements (6 kb width centered on H3K4me1 peaks).
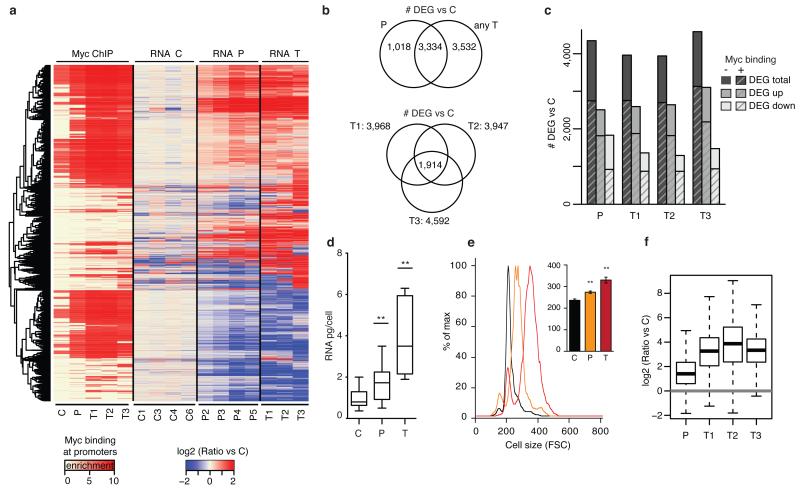
We used RNA-seq to profile mRNA levels during tumor progression (Fig. 2a, Extended Data Fig. 4a). Normalizing to mean expression values yielded ca. 4,300 differentially expressed genes (DEGs) in P and 3,900-4,600 in each tumor relative to C (Fig. 2b, c, Extended Data Fig. 4b; Supplementary Table 1). Most of the DEGs in P were also DEGs in a least one tumor, with an equivalent amount of T-specific DEGs. Of all DEGs in lymphomas, 1,914 (27.8%) were common to the three samples (Fig. 2b). In all instances more than half of DEGs showed increased expression, and ca. 2/3 of either class (up or down-regulated) had Myc bound to the promoter (Fig. 2c). Interpreting changes in mRNA levels is confounded by the fact that Myc can enhance total cellular RNA content4,15, an effect that was confirmed in our P and T samples (note the parallel increases in cell size16, Fig. 2d, e). To account for this feature, we selected 754 mRNAs from our RNA-seq data and quantified them digitally with NanoString technology. This readily validated RNA-seq results, either as absolute or as differential expression relative to C (Extended Data Fig. 5a, b). Normalizing the NanoString counts per cell equivalents revealed upward shifts in the P and T samples (Extended Data Fig. 5 c, d): as a consequence, very few down-regulated mRNAs were left, most mRNAs showing modest to strong up-regulation of mRNA copies per cell (Fig. 2f). Most importantly, this was observable whether or not Myc was bound to the promoter. Two implications follow from these data: first, rather than directly activating every promoter4,5, Myc indirectly induced RNA amplification; second, direct regulatory cues (here the up- and down-regulation of selected Myc-target genes) need to be discerned from global changes in RNA levels: this is achieved by normalizing RNA-seq profiles to mean expression, and not to cell equivalents15.
Fig. 2. Transcriptional amplification co-exists with selective up- and down-regulation of specific Myc targets genes.
a. Heatmap of differentially expressed genes (DEGs) together with the indication about enrichment of Myc binding at the corresponding promoter. All DEGs in P or T are relative to C. b. Venn diagrams showing the number of common DEGs between P and T samples (top) and between the three tumors (bottom). c. Bar plot showing the number of DEGs (total, up and down) divided as Myc-bound and unbound, as indicated. d. Quantification of the total RNA per cell (n=21C, 20P, 9T mice, Student’s t-test). e. FACS analysis of cell size distribution for representative C, P and T samples; inset: mean ± s.d. of n=9C, 10P and 11T mice, Student’s t-test). f. Box plot showing gene expression changes in P and T relative to C samples, as measured by NanoString with normalization to cell equivalents (n=3 mice each). **P< 0.001
The invasion of active regulatory elements by Myc and RNA amplification were described upon activation of a tet-myc transgene in the human B-cell line P493-6 (ref. 4): both of these effects were confirmed in our experiments (Extended Data Fig. 6a-c: 0h, 1h, 24h, High). However, tet-repressed P493-6 cells (0h) were quiescent, and Myc activation induced cell growth and proliferation17 (Extended Data Fig. 6d, e). P493-6 cells can also proliferate, albeit more slowly, upon activation of a viral EBNA2-ER fusion protein, eliciting expression of endogenous Myc (“Low”) at lower levels than with tet-Myc (“High”)18 and intermediate levels of RNA amplification (Extended Data Fig. 6a, f-h). Thus, RNA amplification correlated not only with Myc levels, but also with cellular activation and growth rates, precluding clear conclusions on cause-to-effect relationships. The same limit may apply to tumor cell lines proliferating with different Myc levels4.
RNA amplification was also reported upon stimulation of B-cells with LPS, and was attributed to a direct effect of endogenous Myc5. We repeated these experiments in B-cells homozygous for a conditional knockout allele (c-mycf/f), allowing deletion of the gene prior to LPS treatment. Within the first 12h, we confirmed a moderate increase in total RNA that was slightly reduced in the absence of Myc, as reported5 (Extended Data Fig. 6). This was accompanied by a similar effect on cell size, followed by a Myc-dependent proliferative response7 (Extended Data Fig. 6j, k). It ought to be noted however that at 24h and 48h both RNA contents and cell size underwent dramatic increases, concomitant with plasma cell differentiation6,19, which still occurred in the absence of Myc (Extended Data Fig. 6l)20. Thus, once again, neither of the above experiments allow us to discriminate between direct and indirect RNA amplification by Myc, the latter following from the up- and down-regulation regulation of specific gene sets in those cells (e.g. ref. 21).
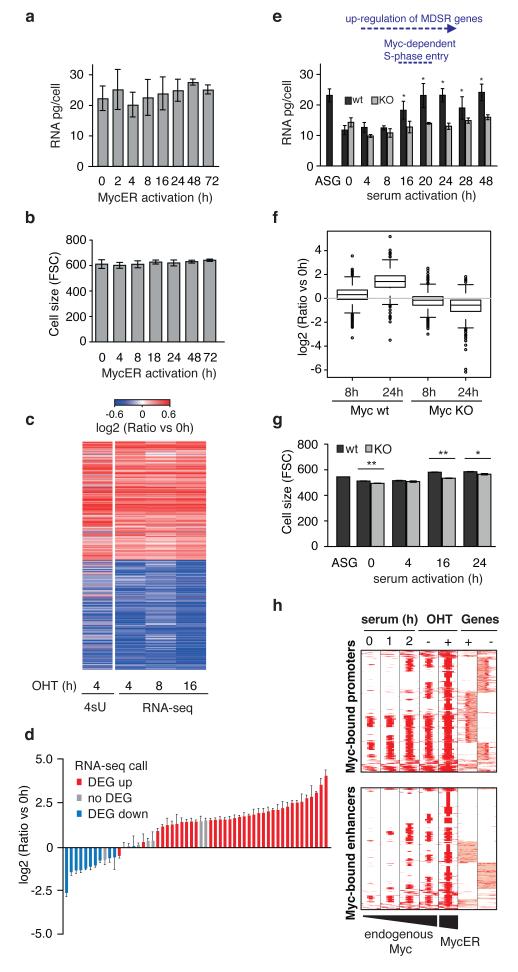
To further address this issue, we used mouse 3T9 fibroblasts expressing a conditional Myc-estrogen receptor chimaera (3T9MycER). Endogenous Myc levels in exponentially growing 3T9MycER cells were sufficient to bind most of the active promoters (76%), as well as a sizeable number of active enhancers (33%): these were invaded further upon MycER activation (95% and 59%, respectively), while inactive elements remained unbound (Extended Data Fig. 7a-c). Unlike seen in B-cells, MycER activation for up to 72 hours caused no increase in cellular RNA content or cell size in fibroblasts (Fig. 3a, b). RNA-seq at 4, 8 and 16 hours post-activation revealed differential regulation of 1,400 to 2,300 genes, of which up to 46% were down-regulated (Fig. 3c, Supplementary Table 2). A time-course with RT-qPCR on several genes showed that expression changes were rapid and at plateau before 8h (Extended Data Fig. 7d). NanoString analysis validated the suppression and induction of selected mRNAs on a per-cell basis, without the general up-regulation seen in B-cells (Fig. 3d). Most importantly, these changes in gene expression were transcriptional in nature, as selective sequencing of newly synthesized RNA (4sU-seq) yielded a pattern highly consistent with RNA-seq (Fig. 3c). Hence, in either cell type, Myc contributed to direct transcriptional activation and repression of distinct groups of genes1,2.
Figure 3. RNA amplification and chromatin invasion are separable events in murine fibroblasts.
a. Quantification of total RNA per cell (mean ± s.d; n=3) and b. cell size (FSC as mean ± s.d; n=3) after treatment of 3T9MycER cells with OHT (h). c. Relative levels of nascent RNA (4sU-seq, single sample) and total mRNA (RNA-seq, n=3) for up- and down-regulated genes following MycER activation. d. NanoString validation of differentially expressed genes (DEG) 8h after OHT treatment (mean ± s.d.; n=3; for the gene list, see Methods). e. Quantification of total RNA per cell of asynchronously growing (ASG) or serum starved and released fibroblasts (mean ± s.d; n=3, Student’s t-test, p-values of induced total cellular RNA levels were calculated relative to 0h). “KO” indicates Myc deletion prior to serum stimulation. As indicated on top, up-regulation of Myc-dependent serum-response (MDSR) genes is observed from 4h onwards, and Myc-dependent S-phase entry at ca. 16h8. f. Box plot showing gene expression changes 8h and 24h after serum stimulation relative to starved cells (0h), as measured by NanoString with normalization to cell equivalents (single sample). g. Cell size after serum stimulation (h). Values are FSC mean ± s.d.; n=3; Student’s t-test). h. Distribution of Myc binding at Myc-bound promoters and distal enhancers (as in Fig. 1). *P<0.05, **P<0.001.
We then deleted the endogenous c-mycf/f gene in quiescent 3T9 fibroblasts, followed by serum stimulation: these cells showed a series of Myc-dependent effects, starting with the selective activation of ca. 300 Myc-dependent serum response (MDSR) genes in early G1, followed by S-phase entry8 and, as shown here, concomitant increases in either total RNA (back to the level observed in asynchronously growing cells: compare Fig. 3e and 3a) or mRNA molecules per cell as assayed by NanoString (Fig. 3f). Serum also induced a moderate increase in size that was attenuated in the absence of Myc (Fig. 3g). ChIP-seq analysis showed that endogenous Myc progressively binds a small subset of the sites bound by active MycER (Fig. 3h). Hence, the RNA “amplification” effect seen in stimulated B-cells5 also occurs in fibroblasts, but without complete invasion of active regulatory elements.
Several aspects of our data address the nature of chromatin “invasion” by over-expressed Myc. First, the binding hierarchies among different promoters were conserved at different Myc levels in either B-cells or fibroblasts (Extended Data Fig. 8a). Second, while all targeted promoters - even the weakest - were enriched for CpG islands and the H3K4me3 mark8-10,22, only the strongest targets were enriched for consensus DNA binding motifs8,9 (Extended Data Fig. 8b). The same was true at active enhancers, considering the H3K27ac mark. Third, MycER-bound sites in fibroblasts were hypersensitive to DNAse I digestion, indicating higher accessibility (Extended Data Fig. 7c)23. Hence, invasion reflects increased interaction of over-expressed Myc with accessible chromatin domains, possibly favored by recruitment (or tethering) via low-affinity protein-protein interactions4,8,9,11, followed by non-specific engagement on DNA, and only at the strongest sites by sequence-specific DNA binding. The same order of events is likely to determine genome recognition by Myc or other transcription factors at physiological levels11,23. These observations imply that (i.) Myc cannot act as a pioneer factor23 but instead requires prior opening of the targeted elements, as also observed in reprogramming experiments10, and (ii.) invasion shall not a priori be equated with a productive engagement of Myc on all cross-linked elements11.
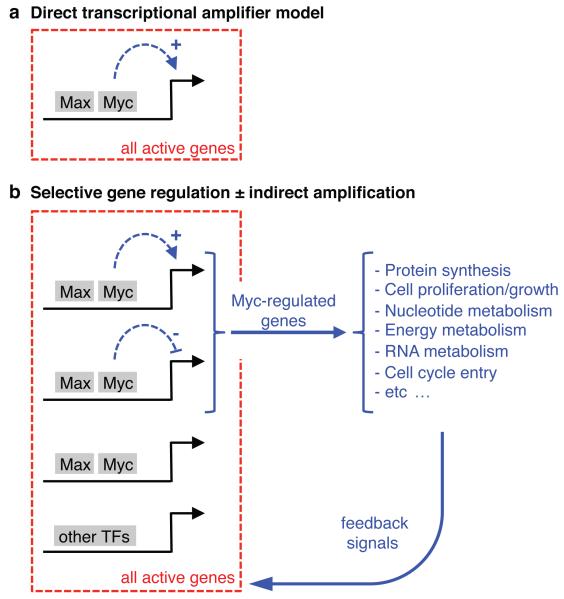
Our results altogether show that promoter/enhancer invasion and RNA amplification are separable and functionally unrelated consequences of Myc over-expression. Thus, rather than the direct up-regulation of every active gene (Fig. 4a)4,5, RNA amplification - when it occurs - is an indirect effect of Myc, which acts primarily through the differential regulation of specific groups of genes (Fig. 4b). Among the functional categories enriched by Myc-regulated genes, we find a variety of DNA- and RNA-associated processes (Extended Data Fig. 9) that may be instrumental for RNA amplification either co- or post-transcriptionally, as exemplified by the Myc-repressed RNA-stabilizing factor ZFP36/TTP24. Other cellular processes controlled by Myc and its target genes, such as nucleotide and energy metabolism, mitochondrial biomass, ribosome biogenesis, or cell growth/size3,16,17,25-27 may feed back on general transcriptional activity28,29 (Fig. 4b): these feedback mechanisms are highly conserved, pre-date Myc in evolution and rely in part on the global regulation of transcriptional elongation (or stalling), a phenomenon that was also directly attributed to Myc4,30. Analysis of RNAPII distribution along Myc-induced genes in lymphomas and fibroblasts revealed composite increases in promoters and gene bodies, albeit without consistent effects on stalling indexes (Extended Data Fig. 10), warranting further discrimination between direct and indirect effects of this pervasive transcription factor. An essential corollary of our work is that the biology of Myc in physiology and disease must still be understood through exhaustive mapping of its target genes, as achieved here during B-cell lymphomagenesis. Our data constitute a unique resource for the functional characterization of these genes in tumor progression and maintenance.
Figure 4. An alternative mechanism for Myc-induced RNA amplification.
a. Direct transcriptional amplification4,5. b. Indirect RNA amplification: here, selective gene regulation by Myc leads to a variety of changes in cellular state, which in the appropriate physiological settings feed back on general RNA biogenesis and turnover (see text). This scenario predicts that RNA amplification may happen in the absence of enhancer/promoter invasion by Myc and, conversely, invasion shall not systematically be associated with RNA amplification, as verified here.
Methods
Primary mouse B cells and cell lines
C57/Bl6 Eμ-myc transgenic mice31 were monitored twice a week for lymphoma development by peripheral lymph node palpation32. In all experiments, we used gender- and age-matched mice (both males and females) without randomization or blinding. Tumors and lymphoid organs were dissected and processed for molecular analysis as described33. Briefly, for Control (C) and Pre-tumoral (P) samples, spleens of 6–8 weeks old mice with no infiltration of peripheral lymph nodes were used. We obtained single-cell suspensions by pressing the spleen through nylon cell strainers and subsequent hypotonic lysis of red blood cells. To isolate B cells, we incubated single-cell suspensions with B220 MicroBeads (Miltenyi Biotech) and enriched them by magnetic cell sorting (MACS), according to the manufacturer instructions (Miltenyi Biotech). Lymphoma samples (or Tumors, T) composed primarily of tumor cells, were dissected from infiltrated lymph nodes and not purified further. At this stage, all samples were fixed directly for chromatin immunoprecipitation (see below) without any in vitro treatment or culture. Experiments involving animals were performed in accordance with the Italian Laws (D.L.vo 116/92 and following additions), which enforce EU 86/609 Directive (Council Directive 86/609/EEC of 24 November 1986 on the approximation of laws, regulations and administrative provisions of the Member States regarding the protection of animals used for experimental and other scientific purposes).
P493-6 cells34 were cultured in RPMI medium with 10% Tetracycline-free serum, 2 mM L-Gln 1% penicillin/streptomycin and non essential amino acids (NEAA) with either (i.) 0.2 μg/ml Tetracycline (Sigma, T7660), (ii.) no additive, or (iii.) 200 nM OHT and 0.2 μg/ml Tetracycline, corresponding to the repressed (or 0h), induced (or High Myc) and Low Myc conditions, respectively18.
Primary B-cells from wild type and homozygous c-mycf/f conditional knockout animals35 were purified by negative selection with the B Cell Isolation Kit (Miltenyi Biotech). After exposure to a recombinant tat-Cre protein (50 μg/ml for 1h in optimem + 1% serum)36 in order to induce deletion of the c-mycf/f allele, splenocytes were stimulated with LPS (50 μg/ml; Sigma, L6237) in DMEM and IMDM (ratio 1:1), 10% serum, 1% glutamine, 1% NEAA, 1% penicillin/streptomycin and 25 μM β-mercaptoethanol37. Deletion efficiency was checked by qPCR on genomic DNA.
3T9 c-mycf/f and 3T9MycER fibroblasts were grown in DMEM medium supplemented with 10% serum, penicillin/streptomycin and 2 mM L-Gln. The role of endogenous Myc in 3T9 c-mycf/f cells was studied by deleting the c-mycf/f allele through activation of a conditional CreER chimera in quiescent cells prior to serum stimulation, as previously described8. Deletion efficiency was checked by qPCR on genomic DNA. To generate the 3T9MycER line, the 3T9 c-mycf/f cells were infected with a pBabe-Bleo retrovirus38 encoding the MycER chimaera39. Cells were selected with 400 μg/ml zeocin (Invitrogen, 46-0509) for one week and kept in zeocin-free medium for subsequent experiments. Single-cell clones were derived and tested for ‘leakiness’, i.e. activation of MycER in the absence of OHT: a non-leaky clone (#14) was used for all experiments described here. MycER was activated by the addition of 400 nM 4-hydroxytamoxifen (OHT) or ethanol (vehicle control). All cell lines were tested for Mycoplasma in our tissue culture facility before usage.
Chromatin immunoprecipitation
Fixation of cultured cells and their processing for chromatin immunoprecipitation (ChIP) were performed as described40, except for blocking proteinA-Sepharose beads with tRNA (Sigma) instead of salmon sperm and purifying immunoprecipitated DNA through Qiaquick columns (Qiagen) instead of phenol-chloroform extraction. The following adjustments were made for in vivo analysis: MACS-sorted splenic B cells or dissected lymphomas were resuspended in PBS with 0.5% BSA and 2 mM EDTA at room temperature and fixed by addition of 1% formaldehyde for 10 min. Fixation was stopped by addition of 0.125 M glycine. Cells were washed three times in PBS, resuspended in SDS buffer (50 mM Tris at pH 8.1, 0.5% SDS, 100 mM NaCl, 5 mM EDTA, and protease inhibitors) and stored at −80 °C before further processing for ChIP40. To minimize inter-individual variation we decided to process ChIP samples of control and pre-tumoral mice in pools of at least ten animals, while due to the clonal nature of lymphomas we kept tumor samples separated. For ChIP-Seq analysis of Myc and RNAPII, lysates from 30×106 B-cells or 50×106 3T9 fibroblasts were immunoprecipitated with 10 μg of the corresponding antibody (see below). For histone marks, lysates from 5×106 B-cells or 3T9MycER fibroblasts were immunoprecipitated with 3 to 5 μg of the corresponding antibody pre-bound to G-protein coupled paramagnetic beads (Dynabeads) in PBS/BSA 0.5%. After overnight incubation beads were washed 6 times in a modified RIPA buffer (50 mM Hepes pH 7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-deoxycholate) and once in TE containing 50 mM NaCl. DNA was eluted in TE/2% SDS and crosslink reversed by incubation overnight at 65 °C. DNA was then purified by Qiaquick columns (Qiagen) and quantified using PicoGreen (Invitrogen). 2-10 ng ChIP DNA was prepared for Solexa Genome Analyzer or HiSeq2000 sequencing with TruSeq ChIP Sample Prep Kit (Illumina) following manufacturer instructions, except for the H3K4me3 ChIP-seq in P493-6 cells for which libraries were produced as published41.
Antibodies
The following antibodies were used for ChIP: H3K4me1 (Abcam, ab8895), H3K27ac (Abcam, ab4729), H3K4me3 (Active Motif, #39159), Myc N262 (Santa Cruz, sc-764) and RNAPII N20 (Santa Cruz, sc-899). Normal rabbit IgG (Santa Cruz, sc-2027) was used as background control. All antibodies were ChIP-grade, as specified by the manufacturer. For western blot: Myc Y69 (Abcam, ab32072), Vinculin (Sigma, V9264).
RNA extraction and analysis
Total RNA was purified onto RNeasy columns (Qiagen) and treated on-column with DNase (Qiagen). Complementary DNA (cDNA) was produced using the reverse-transcriptase ImPromII (Promega). 10 ng of cDNA were used for Real-time PCR reactions with FAST SYBR Green Master Mix (Applied Biosystems). 100 ng of total RNA were processed for NanoString analysis as described by the manufacturer. For RNA-seq, total RNA from 107 cells was purified using Trizol (Invitrogen), treated with Turbo DNase (Ambion) and purified with RNA Clean XP (Agencourt). 5 μg of purified RNA were then treated with Ribozero rRNA removal kit (Epicentre) and EtOH precipitated. RNA quality and removal of rRNA were checked with the Agilent 2100 Bioanalyser (Agilent Technologies). Libraries for RNA-Seq were then prepared with the TruSeq RNA Sample Prep Kits v2 (Illumina) following manufacturer instruction (except for skipping the first step of mRNA purification with poly-T oligo-attached magnetic beads).
4sU-labeling
Treatment of 3T9MycER fibroblasts with 4-thio-Uridine (4sU, Sigma T4509), isolation and sequencing of 4sU-labeled RNA were performed as previously described42 with minor modifications. Briefly, cells were labeled with 300 μM 4sU for 10 min. Medium was removed and cells were put on ice, washed 3 times, scraped and pelleted in ice-cold PBS. RNA was extracted with the Qiagen miRNeasy kit according to the manufacturer including the recommended DNase I digest. 50 μg of total RNA was used for the biotinylation reaction: RNA was diluted in 100 μl of RNAse free water. 100 μl of biotinylation buffer (2.5× stock: 25 mM Tris pH 7.4, 2.5 mM EDTA) and 50 μl of EZ-link Biotin-HPDP (1 mg/ml in DMF; Pierce/Thermo Scientific 21341) were added and incubated for 2h at room temperature. RNA was precipitated and unbound Biotin-HPDP was removed by a combination of chloroform/isoamylalcohol (24:1) precipitation with purification using MaXtract high density tubes from Qiagen. Biotinylated RNA was purified using Dynabeads MyOne Streptavidin T1 (Invitrogen). Before addition of RNA, 50 μl of beads were washed 2 times in washing buffer A (100 mM NaOH, 50 mM NaCl) and once in washing buffer B (100 mM NaCl). Beads were resuspended in 100 μl of buffer C (2 M NaCl, 10 mM Tris pH 7.5, 1 mM EDTA, 0.1% Tween-20) to a final concentration of 5 μg/μl. RNA was added in an equal volume and rotated at room temperature for 15 min. Beads were washed 3 times with washing buffer D (1 M NaCl, 5 mM Tris pH 7.5, 0.5 mM EDTA, 0.05% Tween-20). RNA was eluted from the beads in 100 μl of 10 mM EDTA in 95% formamide (65 °C, 10 min). RNA was extracted with the RNeasy MinElute Spin columns from Qiagen according to the manufacturer and eluted in 14 μl of RNase free water. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). The 4sU-sequencing library was prepared with the TruSeq RNA Sample Prep Kits v2 (Illumina) following the manufacturer instructions starting from the RNA fragmentation step.
DNase I hypersensitivity
Genome-wide sequencing of DNase I hypersensitive sites (DNase I seq) was performed as described43,44. Briefly, 3T9MycER fibroblasts were treated with OHT for 4h. Cells were washed with PBS, trypsinized, pelleted (1300 rpm, 5 min, 4 °C) and washed once more with PBS. Pipetting in the following steps was performed with cut tips to avoid DNA breaks due to pipetting force. Cells were resuspended in Buffer A (15 mM Tris-HCl pH 8, 15 mM NaCl, 60 mM KCl, 1 mM EDTA pH 8, 0.5 mM EGTA pH 8, freshly supplemented with 0.5 mM spermidine and 0.15 mM spermine). An equal volume of lysis buffer (Buffer A with 0.1% NP-40) was added and the cells were incubated on ice for 10 min. Nuclei were pelleted, washed once with buffer A and then resuspended at a concentration of 50×106 nuclei/ml. 107 nuclei were diluted with an equal volume of 2× DNase I reaction buffer (Roche). DNase I (Roche, 04716728001) was added at increasing concentrations (0, 100, 200, 300, 400, 500 U/ml) and DNA was digested for 10 min at 37 °C. An equal volume of Stop buffer (50 mM Tris-HCl pH 8, 100 mM NaCl, 0.1% SDS, 100 mM EDTA pH 8, freshly supplemented with 0.5 mM spermidine, 0.15 mM spermine and 10 μg/ml of RNAse A) was added. Samples were incubated at 55 °C for 30 min (220 rpm agitation). 0.2 μg/μl of Proteinase K were added and samples were incubated at 55 °C overnight (220 rpm). DNA was extracted using a standard phenol/chloroform extraction protocol, dissolved in 100 μl of TE (55°C, 2h). 300 ng of DNA of each digested sample was checked on an agarose gel for the appearance of a smear of slightly digested DNA. Small molecular weight DNA was purified using AMPure beads (Agencourt AMPure XP Reagent, A63881). The digested DNA samples (100 μl) were supplemented with 50 μl of AMPure beads, 150 μl of 20% PEG buffer (20% PEG8000, 2.5 M NaCl) and incubated for 15 min at RT. Beads were separated on a magnet, washed twice with 80% EtOH and small molecular weight DNA was eluted in 100 μl of 5.5% PEG buffer. The eluted DNA was purified once more (20 μl of beads; 120 μl of 20% PEG buffer) and after washing eluted in 20 μl of H2O. DNase I performance was checked by qPCR and samples for sequencing were selected based on the highest signal to noise ratio based on selected genomic regions (200-300 U/ml of DNase I). Chosen samples were size-selected on an agarose gel, small molecular weight DNA (<500 bp) was eluted from the gel with a Qiagen Gel purification kit according to the manufacturer. Up to 10 ng DNA was prepared for HiSeq2000 sequencing with TruSeq ChIP Sample Prep Kit (Illumina) following manufacturer’s instructions.
Primer Design and List of Primers
Primers for ChIP and mRNA analysis were designed by using computer assisted primer design software (Primer3). The complete list of primers used in this study is shown in Supplementary Table 3.
Immunoblot analysis
5-10×106 B-cells were lysed with RIPA Buffer (20 mM HEPES at pH 7.5, 300 mM NaCl, 5 mM EDTA, 10% Glycerol, 1% Triton X-100, supplemented with protease inhibitors (Mini, Roche) and phosphatase inhibitors 0.4 mM Ortovanadate, 10 mM NaF) and sonicated. Cleared lysates were electrophoresed and immunoblotted with the indicated primary antibodies. Chemiluminescent detection, after incubation of the membranes with appropriate secondary antibodies, was done through a CCD camera using the ChemiDoc System (Bio-Rad). Quantification of protein levels was done using the Image Lab Software (Bio-Rad, version 4.0).
Proliferation and Cell Size analysis
To measure cell size, 500,000 live cells were resuspended in 500 μl of PBS and 40,000 total events were collected using a FACsCalibur machine (Becton Dickinson). Propidium iodide (PI) staining solution was added to exclude dead cells from the analysis. Data were then analyzed by using FlowJo software (TreeStar) and the mean of PI negative population scored. BrdU incorporation was analyzed as described.45 Cell proliferation was monitored using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
NanoString Analysis
For quantitative mRNA measurements on the NanoString platform46, we used four nCounter Reporter CodeSets. (i.) A custom CodeSet was used for monitoring gene expression in the Eμ-myc model (Supplementary Table 4, Fig. 2f), for which we selected 754 genes (among which 458 were bound at their promoter by Myc in T) covering the whole expression range and regulatory patterns seen by RNA-seq, including 25 genes classified as non expressed and 5 housekeeping genes (Crocc, Sdha, Tbp, Tubb1, Tubb4). (ii.) A custom CodeSet was used for monitoring gene expression in 3T9 c-mycf/f cells (Supplementary Table 5, Fig. 3f), with 446 genes covering the whole expression range mapped by RNA-seq in 3T9MycER fibroblasts, and including 30 Myc-dependent serum response (MDSR) genes and 20 Myc-independent serum response (MISR) genes8. (iii.) A custom CodeSet was used for monitoring MycER-responsive genes in 3T9MycER cells (Fig. 3d). This CodeSet includes the following 55 genes: Arntl, Ddx58, Olfml2b, Ypel5, Lasp1, Vwa5a, Hsd17b11, Clec2d, Ctso, Prmt2, Myc, Capg, Crocc, Ubb, Dusp6, Rplp0, Car12, Tbp, Mycn, Cdca7l, Hpdl, Ifrd2, Hapln4, Efna3, Polr1b, Slc16a1, Slc19a1, Elovl4, Tfrc, Nnolc1, Wdr73, Zc3h8, Polr3g, Adi1, Fam136a, Bzw2, Wdr55, Taf4b, Mars2, Rrp9, Rragb, Slc25a33, Pdxp, Ica1, Smpdl3b, Dyrk3, Dgat2, GluI, Ifi30, Nr1d1, Reep6, Slc38a3, St6galnac4, Ankrd6, Smtnl2. Data for these genes are provided in exactly the same order (sorted from left to right) in Fig. 3d. (iv.) A pre-designed NanoString CodeSet, the Human Cancer Reference Kit (GXA-CR1), was used for the experiments with P493-6 cells (Extended Data Fig. 6c).
Dedicated nCounter software was used for data analysis, and raw counts were normalized on the geometric mean of the internal positive control probes included in each CodeSet. Data were plotted either without further normalization, or normalized to cell equivalents (based on the total RNA recovered per cell in each single sample).
Statistical analysis
All the experiments were performed on biological replicates unless otherwise specified. Sample size was not predetermined and is reported in the respective figure legends. Two-tailed Student’s t-test was used to calculate p-values; significant values are specified in the figure legends.
Computational analysis
NGS data filtering and quality assessment
ChIP-seq and RNA-seq NGS reads sequenced with the Illumina HiSeq2000 were filtered using the fastq_quality_trimmer (setting the options to −Q33 −t 20 −l 10) and fastq_masker (setting the options to −q 20 −r N) tools of the FASTX-Toolkit suite (http://hannonlab.cshl.edu/fastx_toolkit/). Their quality was evaluated and confirmed using the FastQC application (www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Analysis of ChIP-seq data
ChIP-seq NGS reads were aligned to the mm9 (Eμ-myc and 3T9 fibroblasts data) and hg19 (P493-6) genomes through the BWA aligner using default settings47. Peaks were called using the widely–used MACS software (v1.4)48. Only peaks with p-value <1e-8 were retained (Positive peaks). MACS was also used to perform saturation analysis (as a control of False Negatives) and to determine an estimated False Discovery Rate (as a control of False Positives) for each experiment. In the saturation analysis, the fraction of peaks confirmed with 80% of the reads was determined: for most of the samples this fraction was higher than 60%. False Discovery Rate was determined as the proportion of Negative vs. Positive peaks, where Negative peaks were identified by calling MACS on the input samples, using the ChIP as reference. False Discovery Rates were typically lower than 5-10%.
Normalized reads count within a genomic region was determined as the number of reads per million of library reads (total number of reads in the sequencing library). Peak enrichment was determined as log2 (ChIPw - inputw), where ChIPw and inputw is the normalized count of reads in the peak region in the ChIP and in the corresponding input sample. Myc P binding sites are defined as the union of peaks over 3 ChIP-seq experiments on independent P samples. Enhancers are defined as distal H3K4me1 peaks, i.e. peaks not overlapping with promoters (-2 kb to +1 kb from TSS) and not associated with CGIs.
The RNA Polymerase II Stalling Index (SI, also called Elongation Rate)30,49 was calculated as SI = Prom/GB, where Prom refers to the read counts on the promoter (TSS ± 300 bp interval) and GB to the read counts in the gene body (the interval between TSS +301 and 3,000 bp after the transcription termination site): these values were normalized both to library size (total number of reads) and to the length of the interval, and only genes with GB>600 and with a RNAPII ChIP-seq peak in the region [TSS – 2,000; TSS + 1,000 bp] were considered.
RNA-seq data analysis
RNA-seq NGS reads were aligned to the mm9 mouse reference genome using the TopHat aligner (version 2.0.6) with default parameters50.
For Eμ-myc data read counts were associated to each gene (based on UCSC derived mm9 GTF gene annotations) using the HTSeq software (http://www.huber.embl.de/users/anders/HTSeq/doc/overview.html) setting the options -q --mode=intersection-nonempty --stranded=no. Absolute gene expression was defined determining RPKM as previously described51. Differentially expressed genes (DEGs) were identified using the Bioconductor52 package DESeq53 based on read counts, considering genes whose q-value relative to the control is lower than 0.05 and whose maximum expression over all samples is higher than RPKM of 3.
For 3T9MycER data read counts were defined for each gene isoform as the sum of the read counts over all exons. Exon read counts were determined based on the TxDb.Mmusculus.UCSC.mm9.knownGene Bioconductor52 annotation library using the countOverlaps Bioconductor52 method. We calculated the read counts of a gene with more than one isoform as the rounded mean of the counts of its isoforms. The absolute expression of a gene in both RNA-Seq and 4sU-seq was estimated through RPKM as previously described51, defining total library size as the number of reads mapping to exons only. This is due to the fact that 4sU-seq library shows a marked enrichment in the reads mapping over intronic features compared to RNA-Seq library42. Therefore, in order to compare the expression of genes in both types of libraries, we measured library sizes using the reads mapping over exonic features only. Differentially expressed genes (DEGs) were identified using the Bioconductor52 package DESeq2, considering genes whose q-value relative to the control is lower than 0.01 and whose minimum expression in all samples is higher than RPKM of 1.
For both Eμ-myc and 3T9MycER RNA-seq data, samples were normalized using DESeq and DESeq2 based on scaling factors, assuming that most genes are not differentially expressed. For each gene the scaling factor is computed as the median of the ratio of its read counts over its geometric mean across all samples.
Other bioinformatic analysis
Bioinformatics and statistical analysis, including heatmaps of ChIP-seq data, GeneOntology enrichment, and hierarchical clustering of RNA-seq data were performed using R and Bioconductor packages. R scripts, data, and the R code used to reproduce bioinformatics and statistical analyses are available at http://genomics.iit.it/supplementalData/SaboNature2014.
Supplementary Material
Extended Data Fig. 1| Intensity and frequency of Myc binding events increase during tumor progression, along with their distance from promoters. a. Quantification of c-myc mRNA levels by RT-qPCR (mean ± s.d. of n=6C, 8P, 6T mice; Student’s t-test). b. Myc protein levels were measured by quantitative immunoblotting (mean ± s.d. of n=4C, 4P, 3T mice; Student’s t-test). c. qPCR analysis of Myc ChIP in C, P and T samples at the indicated promoters. AchR is a negative control, while Ncl, Ubfd1 and Dusp6 are known Myc target genes. ChIP data are quantified as % of input DNA40. d. ChIP-seq tracks at the same target genes. The maxima of the tracks are 100 for Ncl, 50 for Ubfd1 and 30 for Dusp6. e-g. ChIP-seq quality controls in the Eμ-myc dataset: e. ChIP-seq saturation analysis for Myc peaks of different enrichment levels for a representative tumor (T1). In this analysis a random subset of the reads is removed (down-sampling) and the number of identified peaks in the down-sampled dataset is compared to the full dataset. If most of the peaks are confirmed upon down-sampling, the ChIP-seq is considered to be well saturated. f. Distribution of read density (library size normalized) for Myc-bound or unbound promoters, as indicated (Myc+, Myc-). 20,000 random 3 kb regions not overlapping with promoters were used to assess background. The Myc signal in unbound promoters (red) is close to the background (gray). g. Validation of ChIP-seq peaks by qPCR. We designed PCR amplicons at 42 sites that correspond to the following categories in Eμ-myc: (i.) genes that are either not or barely expressed (RPKM<4) and show no Myc binding in their promoter according to our ChIP-seq analysis (cyan), (ii.) genes that are expressed (RPKM>4) but not bound by Myc (red), (iii.) genes that are both expressed and bound (black). An IgG antibody was used as negative control: the gray dotted lines indicate the 20th and 80th percentiles of the IgG signal. Left panel: enrichment determined by ChIP-qPCR relative to that in ChIP-seq. ChIP DNA from C, P and T samples was used to amplify each test site. Right panel: distribution of signals obtained in ChIP-qPCR for the different categories of genes along with the signal obtained for all amplicons with the IgG antibody (gray). h. Pie charts showing the number and percentage of promoter, intergenic, and intragenic Myc peaks in C, P and T samples (T1, T2, T3), based on the genomic position of each peak’s midpoint. The number above each chart gives the total amount of peaks. i. Overlap between Myc ChIP-seq peaks. For each column the percentage of peaks overlapping (at least 1 bp) with the reference samples is reported. j. Box plot showing the net distance of Myc from the nearest TSS (upstream and downstream sites together). *P<0.05, **P<0.001.
Extended Data Fig. 2| Myc binding to open chromatin. a. Percentages of Myc peaks associated with the indicated combinations of chromatin features at promoters. b. Same as a. but for distal Myc binding sites. c. H3K4me1/K4me3 ratios of library size normalized read counts over promoter-proximal (black) and distal (red) Myc-binding sites in a representative tumor (T1). d. Top: Lowess regression between the intensity of Myc binding (X-axis) and level of RNAPII, H3K4me1, H3K4me3 and H3K27ac (Y-axis) at Myc bound promoters for C, P and T samples (T1, T2, T3); on both axes read density is determined as the number of reads within 6 kb regions centered at the TSS, divided by the maximum value observed for each mark. Bottom: as above, for Myc-bound distal sites (6 kb centered at Myc peaks).
Extended Data Fig. 3| Myc binds active enhancers. a. Representative snapshots of Myc-bound regions in the genome. ChIP-seq tracks are shown for Myc, RNAPII, H3K4me3, H3K4me1 and H3K27ac in the indicated samples (C, P and T1 as a representative Tumor). CGI: CpG Island. Promoter and Enhancer regions, based on annotation and chromatin features, are indicated. b. List of the domains (Amplicon) previously tested and validated (last column) as B-cell enhancers14. Bound C, P, T: Myc-binding status in our dataset (y: yes; n: no). Note that the two domains listed in bold are also displayed next to the table (Bst1 and Bcl7a loci).
Extended Data Fig. 4| Gene expression data. a. Hierarchical clustering of C, P and T samples based on the RNA-seq data shown in Fig. 2a. C and P samples showed relatively homogeneous profiles, while T samples were more variable, consistent with the clonal nature of lymphomas. b. Scatter plots with differential gene expression; our calls for up- and down-regulated DEGs are reported in red and blue, respectively, and their corresponding numbers reported as inset.
Extended Data Fig. 5| Comparison of NanoString and RNA-seq gene expression data. All scatter plots confront NanoString and RNA-seq data on the Y- and X-axes, respectively. a. Absolute expression levels (NanoString counts versus RNA-seq RPKM units). Red and blue dashed lines indicate ±2 and ±4 differences, respectively. b. The data in a., expressed as fold-change in the corresponding P and T samples relative to C (log2 FC). c. Same as a. normalized to cell equivalents. d. Same as b. normalized to cell equivalents.
Extended Data Fig. 6| RNA amplification correlates with cell cycle entry and cell growth in P493-6 and primary murine B-cells. a-h. P493-6 cells. Cells were cultured in tet-containing medium (0h: tet-Myc repressed) or Tet-free medium (tet-Myc on) for 1h, 24h or multiple passages (High Myc), as indicated. Low Myc: cells were passaged in Tet- and OHT-containing medium (see text). a. Chromatin profiles of Myc-bound promoters (left panel) and distal sites (right panel) on chromosome 1 (as in Fig. 1). b. Quantification of the total RNA per cell (mean ± s.d, n=6 per group; Student’s t-test). c. Box plot showing gene expression changes relative to 0h, as measured by NanoString normalized to cell equivalents (n=3). d. FACS analysis of cell size distribution (mean ± s.d., n=3 per group; Student’s t-test). e. Percentage of BrdU positive cells (mean of two experiments). f. Measurement of Myc protein levels by quantitative western blot (mean ± s.d., n=3 per group; Student’s t-test). g. Growth curve (CellTiter-Glo assay; technical triplicate). h. Quantification of the total RNA per cell (mean ± s.d., n=6, 6, 11; Student’s t-test). i-l. Primary murine B-cells. Wild-type (wt) or c-mycf/f primary murine B-cells were treated with tat-Cre (see Methods) and then with LPS for the indicated periods of time. i. Quantification of total RNA per cell (mean ± s.d, n=3 pools of mice per genotype; Student’s t-test). j. FACS analysis of cell size distribution (mean ± s.d., n=3 mice per genotype; Student’s t-test); the bar plot reports the percentage of cells with FSC higher than 600, as illustrated in the representative graph on the right. k. Growth curve, based on direct cell counts (mean ± s.d, n=3 mice per genotype). l. RT-qPCR analysis of genes regulated during differentiation in plasma cells (mean ± s.d., n=3 pools of mice per genotype; Student’s t-test). Blimp1 and Xbp1 are induced, while Bcl6 is repressed during plasma cell differentiation20,54. The data show that these regulatory patterns are intact in c-myc-deleted cells. P-values are shown either relative to 0h, or between wt and c-mycf/f cells, as indicated. *P<0.05, **P<0.001.
Extended Data Fig. 7| MycER binds active promoters and enhancers. a. ChIP-seq tracks at two representative loci for the indicated proteins and histone marks as well as DNase I hypersensitive sites (HS) in 3T9MycER fibroblasts before (0h OHT, endogenous Myc) and after MycER activation (4h OHT). Maxima in the Myc tracks are 80 for Ncl, and 20 for Twist2. CGI: CpG Island. b. Annotation of Myc binding events, as in Extended Data 1h. c. Upper panels: chromatin profiles of Myc-bound promoters and distal sites on chromosome 1; lower panels: same at non-bound promoters and distal enhancers (as in Fig. 1). d. RT-qPCR validation on a per-cell-basis of up-, down- and non-regulated genes after OHT treatment in 3T9MycER fibroblast relative to untreated cells (mean log2 FC, n=3). RNA was extracted from 50,000 cells at each time-point.
Extended Data Fig. 8| Myc binding hierarchy. a. Heatmaps showing library size-normalized ChIP-seq read counts for Myc bound promoters in Eμ-myc and 3T9 samples, as indicated. Promoters are ranked by read density. b. For all the Eμ-myc samples (C, P, T1, T2, T3) Myc binding events were divided in 5 equally sized groups based on their enrichment: within each group the graphs report the percentage of Myc peaks overlapping (at least 1 bp) with the indicated histone marks, CGI, canonical and variant E-boxes or only the canonical E-box within a 100 bp window around Myc peak summits. The upper and lower graphs refer to Myc peaks on promoters and enhancers, respectively.
Extended Data Fig. 9| Gene Ontology enrichment analysis. a. GeneOntology (GO) terms enriched in up- and down-regulated genes in Eμ–μψχ lymphomas (T) vs. C, and in OHT-treated (4h) vs untreated 3T9MycER cells. Genes differentially expressed in at least one tumor were considered for Eμ–μψχ. GO terms significantly enriched (Fisher test p-value < 1e-10) in at least one gene set were selected (rows in the heatmap) and the p-values for each GO term in each gene set (columns) were color-coded as indicated. Only GO terms in the Biological Process ontology that are assigned to less than 2,000 and more than 10 genes in the mouse genome were considered. b. Venn diagram showing the overlaps among genes up- and down-regulated in Eμ-myc lymphomas and 3T9MycER fibroblasts.
Extended Data Fig. 10| Stalling Index analysis. The RNAPII Stalling Index, also called Traveling Ratio, is defined as the ratio of RNAPII read counts on promoter and gene body for all genes having an RNAPII peak on their promoters30,49. Each row shows the Stalling Index (left panels), the promoter (central panels) and the gene body (right panels) RNAPII counts for the indicated population of genes in the Eμ-myc and 3T9MycER models: a, b. All genes; c, b. Myc-bound up-regulated genes (DEG up); e, f. Myc-bound down-regulated genes (DEG down). For 3T9MycER cells the 4sU-seq data were used, in order to consider only transcriptional changes, but similar results were obtained when using the RNA-seq dataset (not shown).
These observations can be summarized as follows: first, considering all genes, the Stalling Index followed different trends after Myc activation in different setting: a decrease along tumor progression in Eμ-myc mice (a.) and an increase upon MycER activation in fibroblasts (b.). However, the density of RNAPII on the promoter and gene body revealed that both of these measures increased in both systems upon Myc activation, yet in different proportions, resulting in the opposite variations in stalling index. Thus, the stalling index, while capturing the ratio between RNAPII counts on promoter and gene body, conveys no information on the changes of either quantity per se, and may prove mechanistically misleading55. Second, on Myc-induced genes, the increases in RNAPII on promoter and gene body were both larger (c, d.), suggesting that Myc can up-regulate transcription by increasing both RNAPII recruitment (or re-loading) and elongation30,56. As expected, these effects were lost on Myc-repressed genes (e, f.).
Acknowledgments
We thank Andrea Piontini, Paola Nicoli, Alberto Gobbi and Manuela Capillo for their help with the management of mouse colonies, Salvatore Bianchi, Luca Rotta and Thelma Capra for assistance with the Illumina HiSeq and NanoString platforms, Sara Barozzi, Amanda Oldani and Dario Parazzoli for assistance with imaging technologies, Silvia Bonifacio and Giuseppe Diaferia for discussions about DNase I sequencing, and Gioacchino Natoli for comments on the manuscript. We are grateful to Chia-Lin Wei and collaborators (Genome institute of Singapore) for the sequencing of ChIP-seq samples in a preliminary phase of this study. T.R.K. was supported by the Structured International Post Doc program of the European School of Molecular medicine. Work in the Amati lab was supported by grants from the European Research Council, the EU-FP7 Program (EuroSystem and MODHEP consortia), the Association for International Cancer Research (AICR), the Italian health ministry, Fondazione Cariplo and the Italian Association for Cancer Research (AIRC).
Footnotes
The Authors declare no competing financial interests.
Note added in proof: Cunningham et al. (Cell 157, 1088-1103, 2014) recently reported a mechanism that could underlie indirect transcriptional amplification via a metabolic increase in total RNA production in the same Myc-transgenic model used here: in pre-tumoral Eμ-myc B-cells, induction of the translation factor eIF4E led to augmented production of the nucleotide biosynthetic enzyme PRPS2, which in turn was required for the observed increase in RNA synthesis.
References
- 1.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herkert B, Eilers M. Transcriptional repression: the dark side of myc. Genes & cancer. 2010;1:580–6. doi: 10.1177/1947601910379012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darzynkiewicz Z, Traganos F, Melamed MR. New cell cycle compartments identified by multiparameter flow cytometry. Cytometry. 1980;1:98–108. doi: 10.1002/cyto.990010203. [DOI] [PubMed] [Google Scholar]
- 7.de Alboran IM, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 8.Perna D, et al. Genome-wide mapping of Myc binding and gene regulation in serum-stimulated fibroblasts. Oncogene. 2012;31:1695–709. doi: 10.1038/onc.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 10.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabo A, Amati B. Genome Recognition by MYC. Cold Spring Harbor perspectives in medicine. 2014;4 doi: 10.1101/cshperspect.a014191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews. Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 13.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Molecular cell. 2013;49:825–37. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nature immunology. 2010;11:635–43. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–82. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci U S A. 1999;96:13180–5. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuhmacher M, et al. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–1258. doi: 10.1016/s0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 18.Yustein JT, et al. Induction of ectopic Myc target gene JAG2 augments hypoxic growth and tumorigenesis in a human B-cell model. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3534–9. doi: 10.1073/pnas.0901230107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzine F, et al. Global regulation of promoter melting in naive lymphocytes. Cell. 2013;153:988–99. doi: 10.1016/j.cell.2013.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murn J, et al. A Myc-regulated transcriptional network controls B-cell fate in response to BCR triggering. BMC genomics. 2009;10:323. doi: 10.1186/1471-2164-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J, et al. Time-dependent c-Myc transactomes mapped by Array-based nuclear run-on reveal transcriptional modules in human B cells. PloS one. 2010;5:e9691. doi: 10.1371/journal.pone.0009691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez P, et al. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guertin MJ, Lis JT. Mechanisms by which transcription factors gain access to target sequence elements in chromatin. Current opinion in genetics & development. 2013;23:116–23. doi: 10.1016/j.gde.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rounbehler RJ, et al. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell. 2012;150:563–74. doi: 10.1016/j.cell.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graves JA, et al. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PloS one. 2012;7:e37699. doi: 10.1371/journal.pone.0037699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu YC, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marguerat S, Bahler J. Coordinating genome expression with cell size. Trends in genetics: TIG. 2012;28:560–5. doi: 10.1016/j.tig.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 29.das Neves RP, et al. Connecting variability in global transcription rate to mitochondrial variability. PLoS biology. 2010;8:e1000560. doi: 10.1371/journal.pbio.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–45. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–8. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 33.Gorrini C, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–7. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- 34.Pajic A, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–93. doi: 10.1002/1097-0215(20000915)87:6<787::aid-ijc4>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Trumpp A, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 36.Peitz M, Pfannkuche K, Rajewsky K, Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc Natl Acad Sci U S A. 2002;99:4489–94. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerondakis S, Grumont RJ, Banerjee A. Regulating B-cell activation and survival in response to TLR signals. Immunology and cell biology. 2007;85:471–5. doi: 10.1038/sj.icb.7100097. [DOI] [PubMed] [Google Scholar]
- 38.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–96. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nature protocols. 2013;8:539–54. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 42.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nature biotechnology. 2011;29:436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He HH, et al. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Research. 2012;22:1015–25. doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nature methods. 2006;3:511–8. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 45.Campaner S, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–9. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 46.Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26:317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 52.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaffer AL, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 55.Ehrensberger AH, Kelly GP, Svejstrup JQ. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–5. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M. Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. Embo J. 2004;23:2830–2840. doi: 10.1038/sj.emboj.7600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Fig. 1| Intensity and frequency of Myc binding events increase during tumor progression, along with their distance from promoters. a. Quantification of c-myc mRNA levels by RT-qPCR (mean ± s.d. of n=6C, 8P, 6T mice; Student’s t-test). b. Myc protein levels were measured by quantitative immunoblotting (mean ± s.d. of n=4C, 4P, 3T mice; Student’s t-test). c. qPCR analysis of Myc ChIP in C, P and T samples at the indicated promoters. AchR is a negative control, while Ncl, Ubfd1 and Dusp6 are known Myc target genes. ChIP data are quantified as % of input DNA40. d. ChIP-seq tracks at the same target genes. The maxima of the tracks are 100 for Ncl, 50 for Ubfd1 and 30 for Dusp6. e-g. ChIP-seq quality controls in the Eμ-myc dataset: e. ChIP-seq saturation analysis for Myc peaks of different enrichment levels for a representative tumor (T1). In this analysis a random subset of the reads is removed (down-sampling) and the number of identified peaks in the down-sampled dataset is compared to the full dataset. If most of the peaks are confirmed upon down-sampling, the ChIP-seq is considered to be well saturated. f. Distribution of read density (library size normalized) for Myc-bound or unbound promoters, as indicated (Myc+, Myc-). 20,000 random 3 kb regions not overlapping with promoters were used to assess background. The Myc signal in unbound promoters (red) is close to the background (gray). g. Validation of ChIP-seq peaks by qPCR. We designed PCR amplicons at 42 sites that correspond to the following categories in Eμ-myc: (i.) genes that are either not or barely expressed (RPKM<4) and show no Myc binding in their promoter according to our ChIP-seq analysis (cyan), (ii.) genes that are expressed (RPKM>4) but not bound by Myc (red), (iii.) genes that are both expressed and bound (black). An IgG antibody was used as negative control: the gray dotted lines indicate the 20th and 80th percentiles of the IgG signal. Left panel: enrichment determined by ChIP-qPCR relative to that in ChIP-seq. ChIP DNA from C, P and T samples was used to amplify each test site. Right panel: distribution of signals obtained in ChIP-qPCR for the different categories of genes along with the signal obtained for all amplicons with the IgG antibody (gray). h. Pie charts showing the number and percentage of promoter, intergenic, and intragenic Myc peaks in C, P and T samples (T1, T2, T3), based on the genomic position of each peak’s midpoint. The number above each chart gives the total amount of peaks. i. Overlap between Myc ChIP-seq peaks. For each column the percentage of peaks overlapping (at least 1 bp) with the reference samples is reported. j. Box plot showing the net distance of Myc from the nearest TSS (upstream and downstream sites together). *P<0.05, **P<0.001.
Extended Data Fig. 2| Myc binding to open chromatin. a. Percentages of Myc peaks associated with the indicated combinations of chromatin features at promoters. b. Same as a. but for distal Myc binding sites. c. H3K4me1/K4me3 ratios of library size normalized read counts over promoter-proximal (black) and distal (red) Myc-binding sites in a representative tumor (T1). d. Top: Lowess regression between the intensity of Myc binding (X-axis) and level of RNAPII, H3K4me1, H3K4me3 and H3K27ac (Y-axis) at Myc bound promoters for C, P and T samples (T1, T2, T3); on both axes read density is determined as the number of reads within 6 kb regions centered at the TSS, divided by the maximum value observed for each mark. Bottom: as above, for Myc-bound distal sites (6 kb centered at Myc peaks).
Extended Data Fig. 3| Myc binds active enhancers. a. Representative snapshots of Myc-bound regions in the genome. ChIP-seq tracks are shown for Myc, RNAPII, H3K4me3, H3K4me1 and H3K27ac in the indicated samples (C, P and T1 as a representative Tumor). CGI: CpG Island. Promoter and Enhancer regions, based on annotation and chromatin features, are indicated. b. List of the domains (Amplicon) previously tested and validated (last column) as B-cell enhancers14. Bound C, P, T: Myc-binding status in our dataset (y: yes; n: no). Note that the two domains listed in bold are also displayed next to the table (Bst1 and Bcl7a loci).
Extended Data Fig. 4| Gene expression data. a. Hierarchical clustering of C, P and T samples based on the RNA-seq data shown in Fig. 2a. C and P samples showed relatively homogeneous profiles, while T samples were more variable, consistent with the clonal nature of lymphomas. b. Scatter plots with differential gene expression; our calls for up- and down-regulated DEGs are reported in red and blue, respectively, and their corresponding numbers reported as inset.
Extended Data Fig. 5| Comparison of NanoString and RNA-seq gene expression data. All scatter plots confront NanoString and RNA-seq data on the Y- and X-axes, respectively. a. Absolute expression levels (NanoString counts versus RNA-seq RPKM units). Red and blue dashed lines indicate ±2 and ±4 differences, respectively. b. The data in a., expressed as fold-change in the corresponding P and T samples relative to C (log2 FC). c. Same as a. normalized to cell equivalents. d. Same as b. normalized to cell equivalents.
Extended Data Fig. 6| RNA amplification correlates with cell cycle entry and cell growth in P493-6 and primary murine B-cells. a-h. P493-6 cells. Cells were cultured in tet-containing medium (0h: tet-Myc repressed) or Tet-free medium (tet-Myc on) for 1h, 24h or multiple passages (High Myc), as indicated. Low Myc: cells were passaged in Tet- and OHT-containing medium (see text). a. Chromatin profiles of Myc-bound promoters (left panel) and distal sites (right panel) on chromosome 1 (as in Fig. 1). b. Quantification of the total RNA per cell (mean ± s.d, n=6 per group; Student’s t-test). c. Box plot showing gene expression changes relative to 0h, as measured by NanoString normalized to cell equivalents (n=3). d. FACS analysis of cell size distribution (mean ± s.d., n=3 per group; Student’s t-test). e. Percentage of BrdU positive cells (mean of two experiments). f. Measurement of Myc protein levels by quantitative western blot (mean ± s.d., n=3 per group; Student’s t-test). g. Growth curve (CellTiter-Glo assay; technical triplicate). h. Quantification of the total RNA per cell (mean ± s.d., n=6, 6, 11; Student’s t-test). i-l. Primary murine B-cells. Wild-type (wt) or c-mycf/f primary murine B-cells were treated with tat-Cre (see Methods) and then with LPS for the indicated periods of time. i. Quantification of total RNA per cell (mean ± s.d, n=3 pools of mice per genotype; Student’s t-test). j. FACS analysis of cell size distribution (mean ± s.d., n=3 mice per genotype; Student’s t-test); the bar plot reports the percentage of cells with FSC higher than 600, as illustrated in the representative graph on the right. k. Growth curve, based on direct cell counts (mean ± s.d, n=3 mice per genotype). l. RT-qPCR analysis of genes regulated during differentiation in plasma cells (mean ± s.d., n=3 pools of mice per genotype; Student’s t-test). Blimp1 and Xbp1 are induced, while Bcl6 is repressed during plasma cell differentiation20,54. The data show that these regulatory patterns are intact in c-myc-deleted cells. P-values are shown either relative to 0h, or between wt and c-mycf/f cells, as indicated. *P<0.05, **P<0.001.
Extended Data Fig. 7| MycER binds active promoters and enhancers. a. ChIP-seq tracks at two representative loci for the indicated proteins and histone marks as well as DNase I hypersensitive sites (HS) in 3T9MycER fibroblasts before (0h OHT, endogenous Myc) and after MycER activation (4h OHT). Maxima in the Myc tracks are 80 for Ncl, and 20 for Twist2. CGI: CpG Island. b. Annotation of Myc binding events, as in Extended Data 1h. c. Upper panels: chromatin profiles of Myc-bound promoters and distal sites on chromosome 1; lower panels: same at non-bound promoters and distal enhancers (as in Fig. 1). d. RT-qPCR validation on a per-cell-basis of up-, down- and non-regulated genes after OHT treatment in 3T9MycER fibroblast relative to untreated cells (mean log2 FC, n=3). RNA was extracted from 50,000 cells at each time-point.
Extended Data Fig. 8| Myc binding hierarchy. a. Heatmaps showing library size-normalized ChIP-seq read counts for Myc bound promoters in Eμ-myc and 3T9 samples, as indicated. Promoters are ranked by read density. b. For all the Eμ-myc samples (C, P, T1, T2, T3) Myc binding events were divided in 5 equally sized groups based on their enrichment: within each group the graphs report the percentage of Myc peaks overlapping (at least 1 bp) with the indicated histone marks, CGI, canonical and variant E-boxes or only the canonical E-box within a 100 bp window around Myc peak summits. The upper and lower graphs refer to Myc peaks on promoters and enhancers, respectively.
Extended Data Fig. 9| Gene Ontology enrichment analysis. a. GeneOntology (GO) terms enriched in up- and down-regulated genes in Eμ–μψχ lymphomas (T) vs. C, and in OHT-treated (4h) vs untreated 3T9MycER cells. Genes differentially expressed in at least one tumor were considered for Eμ–μψχ. GO terms significantly enriched (Fisher test p-value < 1e-10) in at least one gene set were selected (rows in the heatmap) and the p-values for each GO term in each gene set (columns) were color-coded as indicated. Only GO terms in the Biological Process ontology that are assigned to less than 2,000 and more than 10 genes in the mouse genome were considered. b. Venn diagram showing the overlaps among genes up- and down-regulated in Eμ-myc lymphomas and 3T9MycER fibroblasts.
Extended Data Fig. 10| Stalling Index analysis. The RNAPII Stalling Index, also called Traveling Ratio, is defined as the ratio of RNAPII read counts on promoter and gene body for all genes having an RNAPII peak on their promoters30,49. Each row shows the Stalling Index (left panels), the promoter (central panels) and the gene body (right panels) RNAPII counts for the indicated population of genes in the Eμ-myc and 3T9MycER models: a, b. All genes; c, b. Myc-bound up-regulated genes (DEG up); e, f. Myc-bound down-regulated genes (DEG down). For 3T9MycER cells the 4sU-seq data were used, in order to consider only transcriptional changes, but similar results were obtained when using the RNA-seq dataset (not shown).
These observations can be summarized as follows: first, considering all genes, the Stalling Index followed different trends after Myc activation in different setting: a decrease along tumor progression in Eμ-myc mice (a.) and an increase upon MycER activation in fibroblasts (b.). However, the density of RNAPII on the promoter and gene body revealed that both of these measures increased in both systems upon Myc activation, yet in different proportions, resulting in the opposite variations in stalling index. Thus, the stalling index, while capturing the ratio between RNAPII counts on promoter and gene body, conveys no information on the changes of either quantity per se, and may prove mechanistically misleading55. Second, on Myc-induced genes, the increases in RNAPII on promoter and gene body were both larger (c, d.), suggesting that Myc can up-regulate transcription by increasing both RNAPII recruitment (or re-loading) and elongation30,56. As expected, these effects were lost on Myc-repressed genes (e, f.).