SUMMARY
CD4+ T cell differentiation is regulated by specialized antigen-presenting cells. Dendritic cells (DCs) produce cytokines that promote naive CD4+ T cell differentiation into T helper 1 (Th1), Th17, and inducible T regulatory (iTreg) cells. However, the initiation of Th2 cell responses remains poorly understood, although it is likely that more than one mechanism might be involved. Here we have defined a specific DC subset that is involved in Th2 cell differentiation in vivo in response to a protease allergen, as well as infection with Nippostrongylus brasiliensis. We have demonstrated that this subset is controlled by the transcription factor interferon regulatory factor 4 (IRF4), which is required for their differentiation and Th2 cell-inducing function. IRF4 is known to control Th2 cell differentiation and Th2 cell-associated suppressing function in Treg cells. Our finding suggests that IRF4 also plays a role in DCs where it controls the initiation of Th2 cell responses.
INTRODUCTION
Dendritic cells (DCs) play a key role in linking innate immune recognition to the generation of adaptive immune responses (Reis e Sousa, 2001). Upon innate immune recognition of infection, DCs produce cytokines that promote naive CD4+ T cell differentiation into T helper 1 (Th1), Th17, and inducible T regulatory (iTreg) cells (O’Shea and Paul, 2010; Zhou et al., 2009). In contrast, it remains unclear which instructive signals are necessary for Th2 cell differentiation and which cell types are their relevant sources (Paul and Zhu, 2010; Pulendran and Artis, 2012).
DCs are a heterogeneous population of professional antigen-presenting cells (Hashimoto et al., 2011). Based on expression of surface markers, they can be divided into distinct subpopulations with different functions. In the skin-draining lymph nodes, these include epidermal Langerhans cells (langerin+ CD11blo CD103−) and dermal DCs, which can be divided into at least three subsets: CD103+ dermal DCs (langerin+ CD11blo CD103+), CD11bhi dermal DCs (langerin−CD11bhi CD103−), and CD11blo dermal DC (langerin−CD11blo CD103− DC) (Henri et al., 2010). Epidermal Langerhans cells are critical for inducing Th17cell responses, whereas CD103+ dermal DCs are important for Th1 cell responses (Haley et al., 2012; Igyarto et al., 2011; King et al., 2010). Although much is known about the DC subsets that control Th1 and Th17 cell-mediated immunity, the DCs involved in the initiation of Th2 cell responses remain poorly characterized.
IRF4, a member of the interferon-regulatory family of transcription factors (Tamura et al., 2008), is expressed by Th2 cells and is critical for their differentiation and function (Lohoff et al., 2002; Rengarajan et al., 2002). Expression of this transcription factor is also important for the ability of Foxp3+ regulatory T cells to specifically inhibit Th2 cell-mediated responses (Zheng et al., 2009). IRF4 was shown to regulate the development of CD11bhi CD8α− DCs in the spleen (Suzuki et al., 2004) and the migration of DCs in the skin (Bajana et al., 2012). IRF4 also controls plasma cell differentiation and class switch recombination (Klein et al., 2006). In addition, IRF4 has been found to control M2 polarization of macrophages (Satoh et al., 2010). Finally, IRF4 inhibits TLR-induced inflammatory gene expression in macrophages (Negishi et al., 2005).
Here we have identified a DC subset that appears to be specialized for regulation of Th2 cell responses. These DCs have unique phenotypic markers and their differentiation is dependent on IRF4. Finally, elimination of this DC subset in DC-specific Irf4−/− mice results in ablation of allergen- and infection-induced Th2 cell responses.
RESULTS
Bone-Marrow-Derived PDL2+ DCs Have a Distinct Phenotype
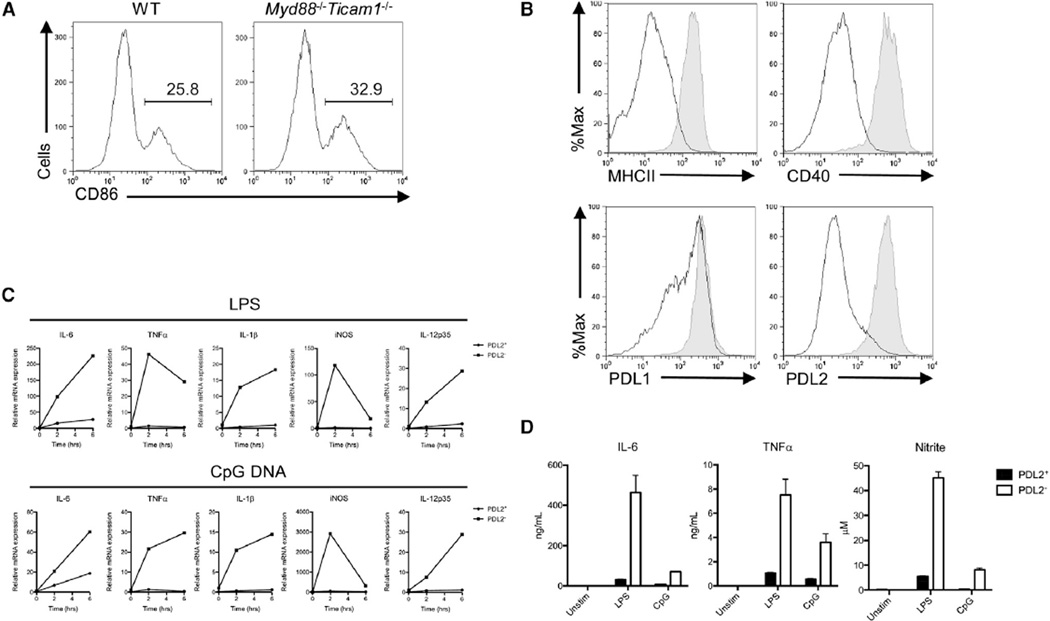
In vitro generated bone-marrow-derived DCs (BMDCs) grown with granulocyte macrophage colony-stimulating factor (GM-CSF) (Inaba et al., 1992), typically exhibit an immature cell surface phenotype characterized by low expression of the costimulatory molecule CD86 (Figure 1A) and major histocompatibility complex MHC class II molecules (Figure 1B). However, it has long been observed that these cultures also contain a smaller fraction of CD11c+ CD86hi MHC-IIhi DCs (~20%−35%), which arise spontaneously in the absence of exogenous inflammatory mediators or microbial products (Jiang et al., 2007) (Figure 1A). In addition, the spontaneously mature DCs expressed high amounts of CD40, programmed death ligand-1 (PDL1) and PDL2 (Figure 1B). We found that PDL2 expression was stably associated with this DC subset (see below), and we will therefore refer to them here as PDL2+ DCs. Interleukin-4 (IL-4) is known to induce PDL2 expression on dendritic cells (Ishiwata et al., 2010). Interestingly, in vitro generation of PDL2+ DCs could be promoted by IL-4 and inhibited by transforming growth factor-β (TGF-β) (see Figure S1 Aavailable online). In addition, the fraction of PDL2+ DCs in bone-marrow cultures could be increased by mechanical disruption of E-cadherin-mediated cell adhesion (Figure S1B), as previously described (Jiang et al., 2007).
Figure 1. Bone-Marrow-Derived PDL2+ Dendritic Cells Are Phenotypically Mature and Hyporesponsive to Proinflammatory Stimuli.
(A) Percentages of CD86hi BMDCs in GM-CSF-supplemented bone-marrow cultures from WT C57BL/6 and Myd88−/− Ticam1−/− mice. Gated on CD11c+ cells.
(B) Cell-surface expression of maturation markers (MHCII, CD40) and B7 family members (PDL1, PDL2) by CD86hi (shaded) and CD86lo (solid line) BMDCs.
(C) Induction of proinflammatory genes (IL-6, TNF-α, IL-1β, iNOS, and IL-12p35) in PDL2+ and PDL2− BMDCs stimulated with LPS or CpG DNA. PDL2+ and PDL2− DC subsets were sorted from GM-CSF-supplemented bone-marrow cultures at day 5 and stimulated with LPS or CpG DNA for indicated times. Data are representative of three independent experiments.
(D) Secretion of proinflammatory cytokines (IL-6, TNF-α) and nitric oxide by PDL2+ and PDL2− BMDCs stimulated with LPS or CpG DNA. Sorted PDL2+ and PDL2− BMDCs were stimulated with LPS or CpG DNA for 24 hr. IL-6 and TNF-α amounts in the supernatant were measured by ELISA, and NO production was measured by the Greiss assay as nitrite concentration. Data are representative of three independent experiments, and bar graphs show mean ± SD.
See also Figure S1.
PDL2+ DCs Are Hyporesponsive to TLR Stimulation
The conventional immature BMDCs are highly sensitive to stimulation with microbial TLR ligands: they undergo maturation and cytokine production necessary for naive T cell activation (Reis e Sousa, 2001). As expected, stimulation of immature PDL2− BMDCs with LPS or CpG DNA resulted in a robust proinflammatory response, as measured by gene expression and secretion of proinflammatory mediators. In contrast, PDL2+ DCs are unresponsive to stimulation with LPS and CpG DNA (Figures 1C and 1D). Unresponsiveness to LPS is likely due to a low amount of TLR4 expression in PDL2+ DCs, whereas unresponsiveness to CpG DNA might be due to reduced endo-cytic activity of PDL2+ DCs (Figure S1C and S1D).
PDL2+ DCs Promote Th2 Responses
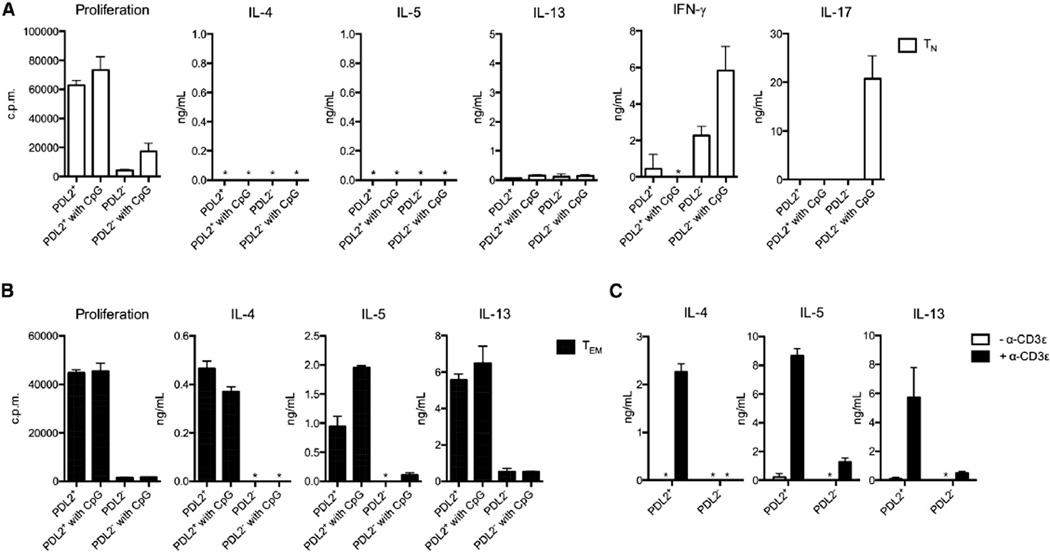
We next compared the ability of bone-marrow-derived PDL2+ DCs and conventional PDL2− DCs to stimulate T cell responses in vitro. Because both DC subsets express similar amounts of toll-like receptor-9 (TLR9), we used CpG DNA for their stimulation. PDL2+ DCs, with or without CpG DNA treatment, induced very robust proliferation of naive (CD62Lhi CD44lo) and effector or memory (CD62Llo CD44hi) CD4+ T cells (Figures 2A and 2B) in the presence of anti-TCR and anti-CD3 stimulation. However, they did not induce differentiation of naive CD4+ T cells into Th1, Th2, or Th17 cell effectors (Figure 2A), nor did they induce expression of Foxp3 in naive CD4+ T cells (data not shown). As expected, upon CpG DNA stimulation, the conventional PDL2− DCs induced Th1 and Th17, but not Th2 cell differentiation of naive T cells (Figure 2A). To determine the antigen specificity of the T cell response, we cultured sorted DCs together with naive OT-II T cells in vitro in the presence of the cognate OVA peptide (amino acids 323–339). Neither PDL2+ nor PDL2− DCs alone could induce Th2 cell differentiation of OT-II T cells, whereas addition of exogenous IL-4 was sufficient to induce Th2 cell differentiation, as expected (Figure S2A). Furthermore, unlike IL-4, addition of epithelial cell-derived cytokines such as IL-25 and IL-33 did not induce Th2 cell differentiation of naive T cells (Figure S2B), while thymic stromal lymphopoietin (TSLP) had a modest effect (Figure S2C). Interestingly, although PDL2+ DCs failed to promote naive T cell differentiation in vitro, they elicited a strong Th2 cell response in effector or memory CD4+ T cells (Figure 2B). This response was T cell mediated, because it was dependent on T cell receptor (TCR) engagement by anti-CD3s antibody (Figure 2C). The Th2 cell response by effector or memory CD4+ T cells did not require PDL2 expression by DCs but was partially dependent on PD1 and OX40 costimulatory molecule expression in T cells (Figures S2D and S2E). Induction of Th2 cell-associated cytokines did not require MyD88 or TRIF adaptor signaling pathways in DCs (Figure S2F).
Figure 2. PDL2+ BMDCs Induce Th2 Responses in Effector or Memory CD4 T Cells.
(A) Proliferation and cytokine production by naive CD4+ T cells (TN) cultured with PDL2+ or PDL2− BMDCs in the presence of soluble purified α-CD3 antibody and CpG DNA, where indicated. Cytokines in the supernatant from cocultures at day 3 or 4 were measured by ELISA. Proliferation was measured by incorporation of [3H]-thymidine during the last 16 hrs of coculture (cpm). *, not detected (same hereafter). Data are representative of at least three independent experiments, and bar graphs show mean ± SD.
(B) Proliferation and cytokine production by effector or memory CD4+ T cells (Tem) stimulated with BMDCs, same as above (A). Data are representative of at least three independent experiments, and bar graphs show mean ± SD.
(C) Dependence on TCR engagement for the effector or memory Th2 cell response induced by PDL2+ BMDCs. Tem cells were cocultured with PDL2+ or PDL2− BMDCs in the presence or absence of α-CD3ε. Data are represented as mean ± SD.
See also Figure S2.
PDL2+ DC Subset Exists In Vivo
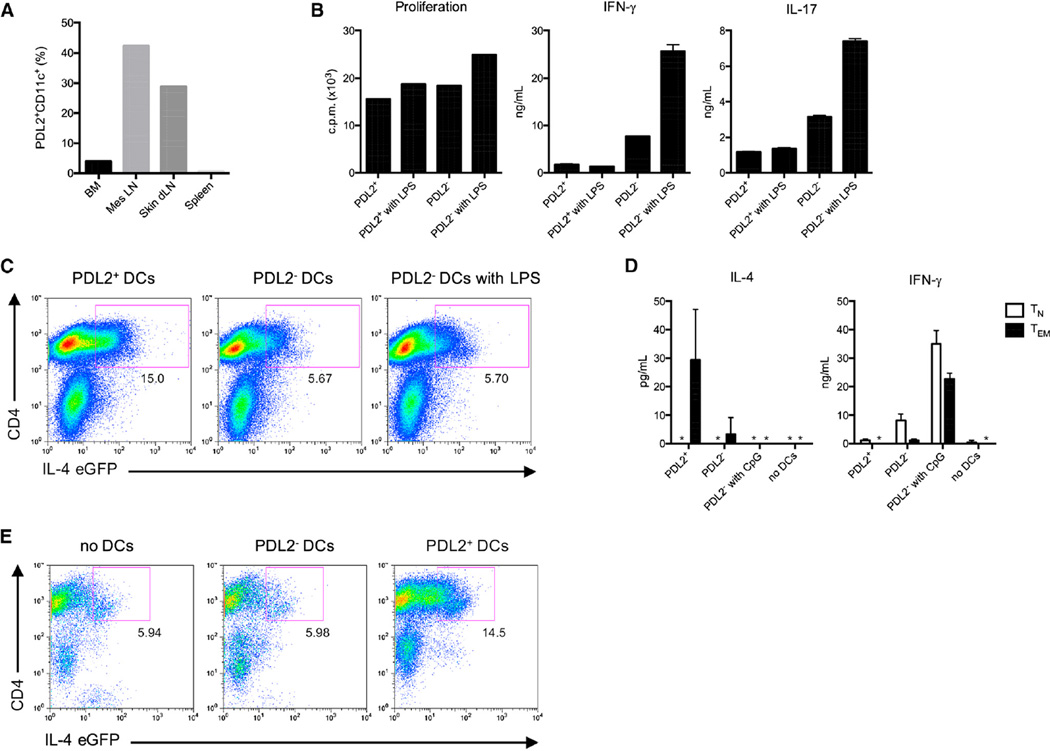
Collectively, these data suggested that bone-marrow-derived PDL2+ DCs might have specialized properties for the regulation of Th2 cell responses. Therefore, we next looked for their in vivo counterparts by using PDL2 expression as a marker. We found a related DC population highly enriched in the mesenteric and skin-draining lymph nodes (dLNs) (Figure 3A). Similar to their bone-marrow-derived counterparts, PDL2+ DCs in the skin dLNs expressed high amounts of CD80, CD86, CD40, and PDL1 (Figure S3A). Moreover, similar to in vitro generated PDL2+ DCs, in vivo PDL2+ DCs had reduced messenger RNA (mRNA) expression of several TLRs (Figure S3B). In the skin dLNs, different DC subsets can be identified based on expression of CD8α and CD205 (Dec205) (Henri et al., 2001). Langer-hans cells (LCs) are CD205hi CD8αint and dermal DCs are CD205int CD8αlo. Gating on CD11c+ PDL2+ DCs, we found that this population contained a mixture of dermal DCs and Langerhans cells, as determined by CD8α and CD205 staining (Figure S3D). Moreover, by staining for epithelial cell adhesion molecule (EpCAM), whichishighly expressedby LCs (Borkowski et al., 1996), we confirmed that a proportion of PDL2+ DCs were LCs (Figure S3E). Dermal DCs can be divided into at least three subsets. The C-type lectin CD301b (Mgl2) has recently been identified as a specific marker for CD207 (Langerin) negative, CD103 negative dermal DCs (Kumamoto et al., 2009). To determine whether cutaneous PDL2+ DCs contained this subset of dermal DCs, we stained these cells for CD301b and found that PDL2+ DCs could be divided into CD301b− and CD301b+ subsets (Figure S3F). Moreover, the PDL2+ CD301b+ cells were CD103 negative, as expected (Figure S3F). Furthermore, the PDL2+ DCs from the skin dLNs expressed CCR7, consistent with them being migratory DCs (Figure S3A). In addition, PDL2+ DCs were able to take up and process a subcutaneously injected fluorescently labeled antigen (DQ-OVA) (Figure S3C), indicating that they have normal endocytic activity in contrast to bone-marrow-derived PDL2+ DCs.
Figure 3. PDL2+ DCs Are Present in Skin-Draining Lymph Nodes and Regulate Effector or Memory Th2 Response.
(A) Percentage of PDL2+ DCs in different lymphoid organs in vivo during homeostasis. BM, bone marrow; Mes LN, mesenteric lymph nodes; skin dLN, skin-draining lymph nodes.
(B) Proliferation and cytokine production by total splenic CD4+T cells stimulated for 3–4 days with PDL2+ or PDL2ࢤ skin dLN DCs, plus or minus LPS stimulation. Data are representative of at least three independent experiments, and bar graphs show mean ± SD.
(C) IL-4-eGFP expression by total CD4+ T cells from 4get mice stimulated with skin dLN PDL2+ or PDL2− DCs. Data are representative of three independent experiments.
(D) IL-4 and IFN-γ secretion by Tn and Tem cells stimulated with skin dLN DCs. Data are representative of three independent experiments, and bar graphs show mean ± SD.
(E) IL-4-eGFP expression by Tem cells from 4get mice stimulated with skin dLN DCs. Data are representative of two independent experiments.
See also Figure S3.
Lymph Node PDL2+ DCs Promote Th2 Cell Response
Similar to their bone-marrow-derived counterparts, PDL2+ DCs from skin dLNs induced proliferation of splenic CD4+ T cells, but unlike PDL2− DCs, they failed to induce interferon-γ (IFN-γ) or interleukin-17 (IL-17) production by T cells (Figure 3B). They also did not induce Foxp3 expression by T cells (Figure S3G). However, PDL2+ DCs from skin dLN did induce IL-4 reporter expression in T cells from 4get mice, in which GFP marks cells with IL-4 gene transcription (Mohrs et al., 2001) (Figure 3C). In addition, similar to in vitro generated DCs, skin dLN PDL2+ DCs induced a Th2 cell cytokine response from effector or memory CD4+ T cells, but did not induce Th2 cell differentiation of naive T cells (Figures 3D and 3E). Collectively, these results suggest that PDL2+ DCs from skin dLNs (specifically, PDL2+ CD301b+ DCs, see below) are in vivo counterparts of bone-marrow-derived PDL2+ DCs and that this DC subset might be specialized for controlling Th2 cell responses.
Differentiation of PDL2+ DCs Is Dependent on IRF4
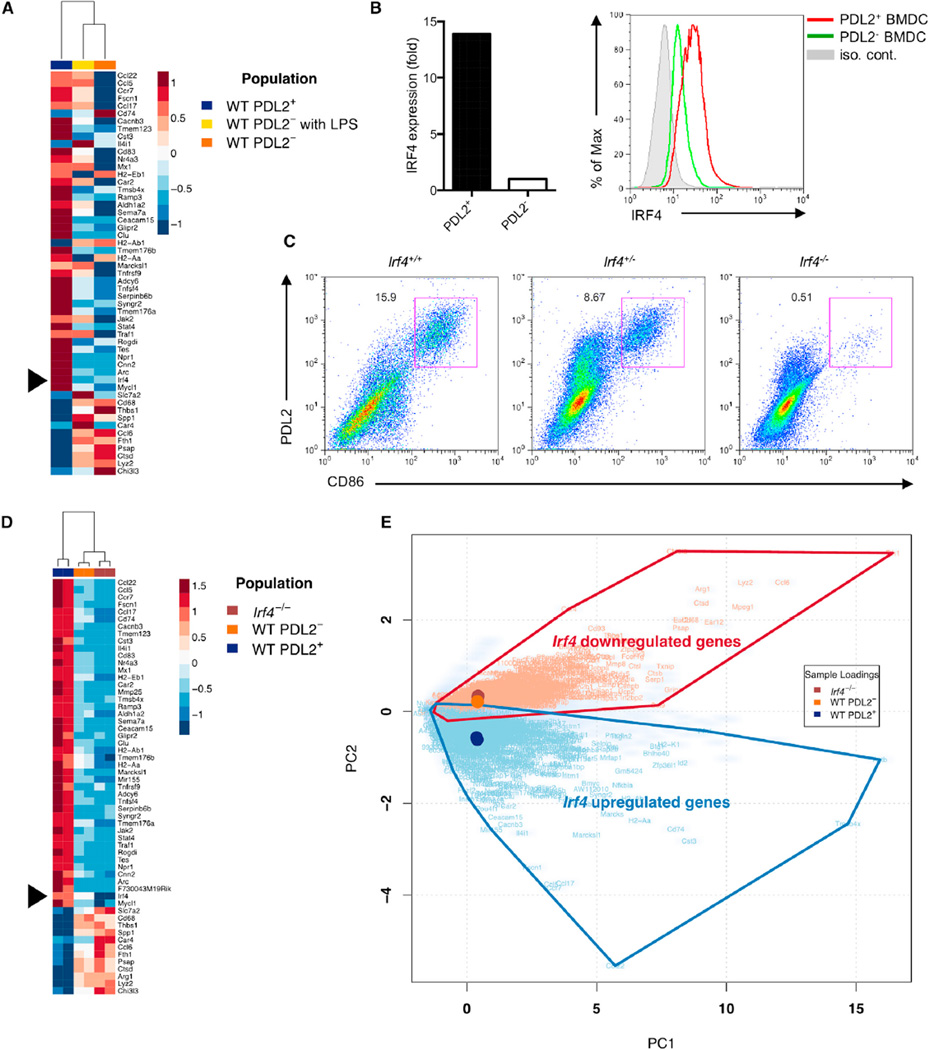
To gain further insight into the unique features of PDL2+ DCs and the mechanism for their Th2 cell regulatory ability, we compared gene-expression profiles of PDL2+ and PDL2− BMDCs by microarray analysis (Figure 4A). Among the genes differentially expressed in these DC populations, the transcription factor IRF4 was preferentially expressed in the PDL2+ DC subset, as validated by mRNA and protein expression (Figure 4B). We therefore investigated whether IRF4 was required for the differentiation of PDL2+ DCs. We generated BMDCs from wild-type (WT) and Irf4-deficient mice (Klein et al., 2006) and found that in the absence of IRF4, the differentiation of PDL2+ DCs was almost completely blocked (Figure 4C). Retroviral reconstitution of IRF4 into IRF4-deficient progenitors restored the differentiation of these cells, as indicated by the presence of PDL2 and CD301b–positive DC subset (Figure S4A).
Figure 4. IRF4 Expression Is Critical for the Development of Bone-Marrow-Derived PDL2+ DCs.
(A) Microarray analysis of log2 transformed expression amounts from three BMDC populations (PDL2+, PDL2−, and LPS stimulated PDL2−). Irf4 is indicated. Genes displayed correspond to those identified as significant by RNA-Seq analysis.
(B) IRF4 mRNA and protein expression in PDL2+ BMDCs was examined by real-time RT-PCR and flow cytometry, respectively. Data are representative of at least three independent experiments.
(C) Percentages of PDL2+ BMDCs in WT, Irf4+/− and Irf4−/− bone-marrow cultures. Data are representative of three independent experiments.
(D and E) RNA-seq analysis of gene expression in three BMDC populations (WT PDL2+, WT PDL2−, and Irf4−/−). Heatmap of square root transformed RPKM values for genes with PC2 scores greater than a designated absolute threshold value of ± 2.38 (D). Biplot derived from PCA analysis of gene summarized RPKM values (cloudy blue) plotted according to two PCs that account for > 80% of total sample variance (E). PC1 describes overall expression and PC2 describes differential expression between populations. Sample loading vectors and the symbols of overlapping, differentially expressed genes (Benjamini-Hochberg adjusted p value < 10−50 by negative binomial testing) are displayed.
See also Figure S4.
RNA-seq analysis confirmed that the expression profile of Irf4−/− BMDCs was similar to WT PDL2− BMDCs and different from that of PDL2+ BMDCs, highlighting an essential role for IRF4 in directing PDL2+ DC development in vitro (Figure 4D and 4E). Principal component analysis (PCA) revealed that a subset of genes that were significantly upregulated in PDL2+ BMDCs and downregulated in the Irf4−/− DC population accounted for the principal biological sample variance not associated with mean gene-expression amounts (Figure 4E). Gene-ontology analysis underscored that, within this subset, genes associated with lymphocyte activation and T cell development were specifically upregulated in PDL2+ BMDCs, whereas those associated with microbial defense responses were downregulated (Figure S4B). Together these results suggest that IRF4 plays a critical role in PDL2+ DC differentiation in vitro.
IRF4 usually cooperates with transcription factors of the Ets or AP-1 family in a cell-type-specific manner through binding to composite DNA motifs to mediate different transcription programs (Ciofani et al., 2012; Glasmacher et al., 2012; Li et al., 2012; Tussiwand et al., 2012). We therefore examined expression of Ets and AP-1 transcription factors in PDL2+ DCs and found that some members of the two families were selectively expressed (Figures S4C and S4D). In particular, the Ets family members SpiB, SpiC and Ehf, as well as the AP-1 family member Batf3, were expressed at higher concentrations in PDL2+ DCs. These transcription factors may thus cooperate with IRF4 to drive the PDL2+ DC-specific gene-expression program, a possibility that will be investigated in future studies.
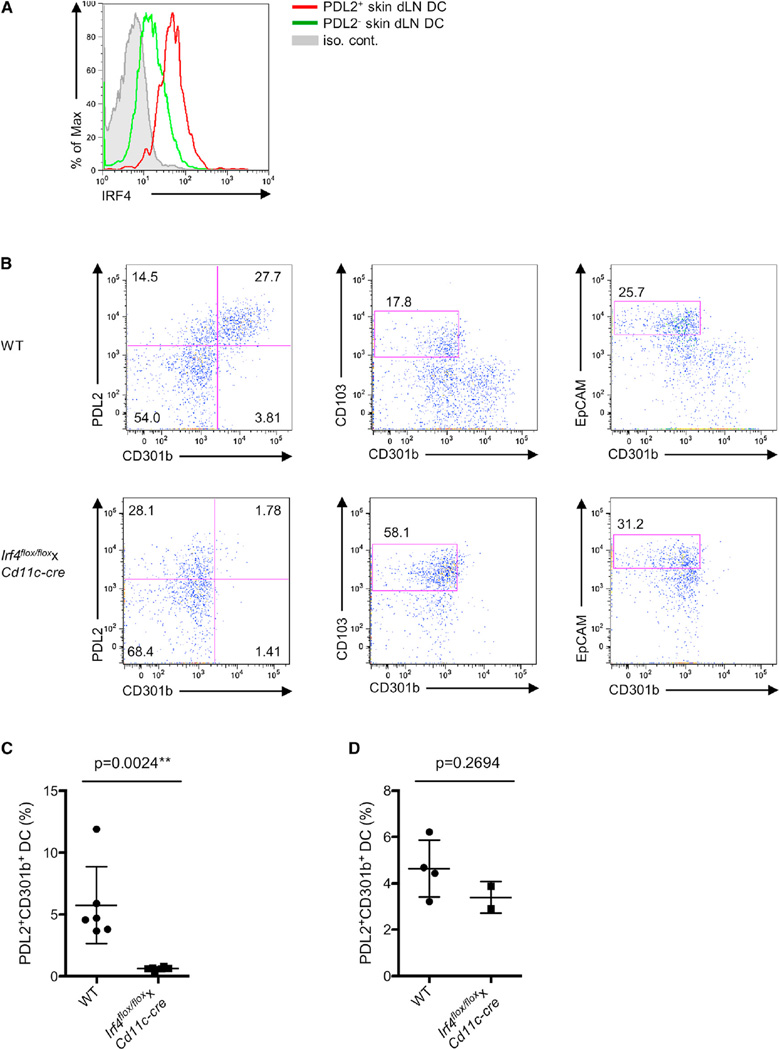
Importantly, IRF4 was also specifically expressed in vivo in PDL2+ DCs in the skin dLNs (Figure 5A). We next crossed Irf4flox/flox mice (Klein et al., 2006) with Cd11c-cre mice, which deletes predominantly in DCs (Caton et al., 2007), to generate mice with Irf4 deleted in DCs (hereafter referred to as Irf4flox/flox xCd11c-cre mice). We found that the PDL2+ CD301b+ DC subset was almost completely eliminated, whereas other DC subsets such as CD103+ dermal DCs and Langerhans cells appeared unaffected in the skin dLNs of Irf4flox/floxxCd11c-cre mice (Figures 5B and 5C). Nevertheless, a normal number of PDL2+ CD301b+ DCs were still detected in the dermis of Irf4flox/floxxCd11c-cre mice (Figure 5D). These data indicate that IRF4 is required for the presence of CD301b+ PDL2+ DCs in skin dLNs, but is not essential for their development in the dermis in vivo.
Figure 5. IRF4 Expression Is Critical for the Presence of PDL2+ CD301b+ DCs in Skin dLNs.
(A) IRF4 protein expression in PDL2+ skin dLN DCs were examined by flow cytometry.
(B) MHCIIhi CD11c+ DCs from skin dLNs of WT and Irf4flox/floxxCd11c-cre mice were examined by flow cytometry for PDL2, CD301b, CD103, and EpCAM expression. The percentages of PDL2+ CD301b+ and PDL2+ CD301b− DCs (left), dermal CD103+ DCs (middle), and EpCAM+ Langerhans cells (right) were shown as indicated. Data are representative of three independent experiments.
(C) Percentages of PDL2+ CD301b+ DCs in the skin dLNs of WT and Irf4flox/floxxCd11c-cre mice. Data are represented as mean ± SD. **p < 0.01.
(D) Percentages of PDL2+ CD301b+ DCs in the ear dermis of WT and Irf4flox/floxxCd11c-cre mice. Data are represented as mean ± SD.
IRF4 in DCs Drives Th2 Responses
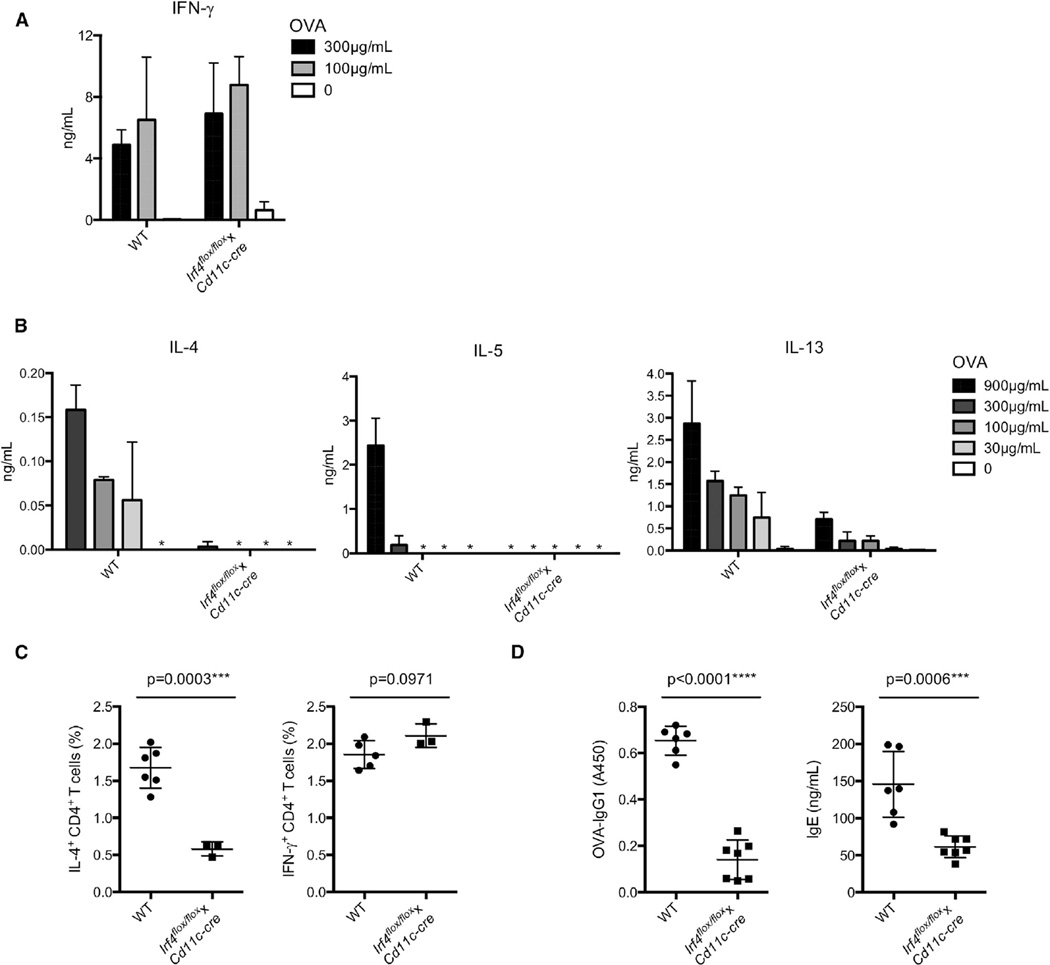
Next, to investigate the role of IRF4-dependent CD301b+ PDL2+ DCs in Th2 cell responses in vivo, Irf4flox/floxxCd11c-cre mice were immunized with OVA by using either LPS or papain as Th1 and Th2 cell inducing adjuvants, respectively. The amount of cytokine production by skin dLN cells was assessed after in vitro restimulation. While the Th1 cell response induced by immunization with OVA plus lipopolysaccharide (LPS) was unimpaired (Figure 6A), the OVA plus papain-induced Th2 cell response, as measured by secretion of IL-4, IL-5, and IL-13, was almost completely ablated in Irf4flox/floxxCd11c-cre mice (Figure 6B). Consistent with this, Irf4flox/floxxCd11c-cre mice had significantly lower percentages of IL-4-producing but not IFN-γ-producing CD4+ T cells after OVA plus papain immunization, suggesting that IRF4 plays a role in DCs in controlling the papain-induced Th2 cell response (Figure 6C). Additionally, Irf4flox/floxxCd11c-cre mice produced significantly lower amounts of immunoglobulin G1 (IgG1) and IgE after immunization with OVA plus papain, suggesting a defect in the Th2 cell-dependent antibody response as well (Figure 6D).
Figure 6. IRF4 Expression in DCs Is Essential for the Papain-Induced Th2 Cell Response.
(A and B) LPS-induced Th1 cell response (A) and papain-induced Th2 cell response (B) in WT and Irf4flox/floxxCd11c-cre mice. WT and Irf4flox/floxxCd11c-cre mice were immunized subcutaneously with OVA plus either LPS or papain as Th1 and Th2 cell-inducing adjuvants, respectively. CD4+ T cells were sorted from inguinal and popliteal LNs at day 6 and stimulated with irradiated splenic APCs and titrating doses of OVA for 4 days. IFN-γ (for LPS response) and IL-4, IL-5, and IL-13 (for papain response) production was measured by ELISA. Data are representative of at least three independent experiments, and bar graphs show mean ± SD.
(C) Percentages of IL-4- or IFN-γ-producing CD4+ T cells in draining inguinal and popliteal LNs in WT and Irf4flox/floxxCd11c-cre mice 6 days after subcutaneous immunization with OVA plus papain. Data are represented as mean ± SD. ***p < 0.001.
(D) OVA-specific IgG1 and total IgE amounts in the serum were measured by ELISA in WT and Irf4flox/floxxCd11c-cre mice at day 21 after subcutaneous immunization with OVA plus papain at day 0 and day 14. Data are represented as mean ± SD. ***p < 0.001, ****p < 0.0001.
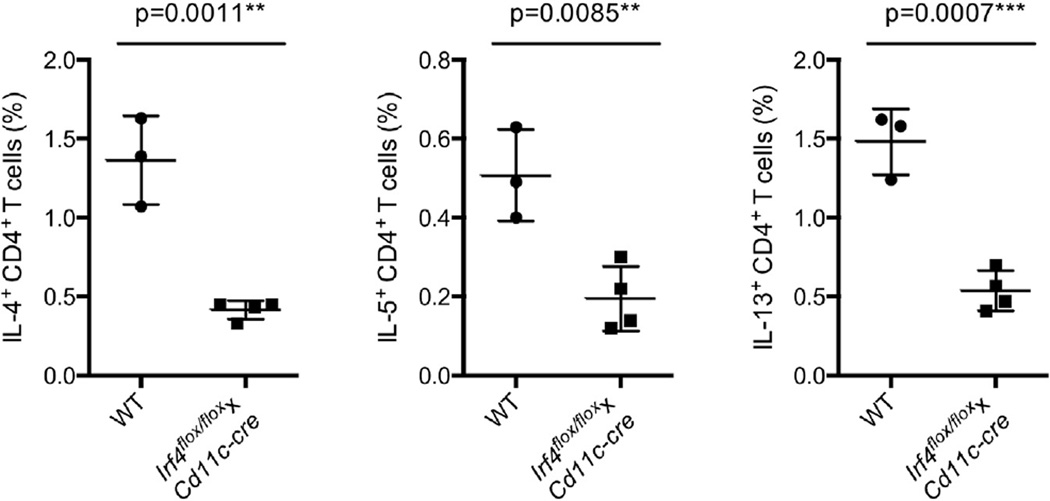
To further examine the role of IRF4 in DCs in controlling Th2 cell responses, we used another commonly used Th2 cell-mediated immunity model—Nippostrongylus brasiliensis infection. In the draining mesenteric lymph nodes 7 days after infection, Irf4flox/floxxCd11c-cre mice had significantly lower numbers of IL-4-, IL-5-, and IL-13-producing CD4+ T cells (Figure 7). A reduced Th2 cell response was also seen in the draining mediastinal LNs of Irf4flox/floxxCd11c-cre mice (data not shown). Collectively, these data indicate that IRF4 expression in DCs is important for regulation of a variety of Th2 cell responses and further suggest that PDL2+ DCs play a specialized role in Th2 cell responses.
Figure 7. IRF4 Expression in DCs Is Essential for Th2 Cell Responses Induced after Infection with Nippostrongylus Brasiliensis.
Percentages of IL-4-, IL-5-, and IL-13-producing CD4+ T cells in draining mesenteric LNs in WT and Irf4flox/floxxCd11c-cre mice 7 days after subcutaneous infection with Nippostrongylus brasiliensis. Data are represented as mean ± SD. **p < 0.01, ***p < 0.001.
DISCUSSION
In contrast to steady progress in characterization of the mechanisms of Th1, Th17, T follicular helper, and Treg cell differentiation, induction of Th2 cell responses remains poorly understood. Although many models have been proposed, the relevant signals and cell types involved in the initiation of Th2 cell responses are still unknown. Likewise, the innate sensor mechanisms responsible for the initiation of Th2 cell responses remain to be elucidated.
Here we investigated functional properties of a DC population characterized by expression of PDL2. These DCs were found in GM-CSF bone-marrow cultures, as well as in mesenteric and skin-draining lymph nodes. Uncharacteristically for DCs, they were unresponsive to stimulation with TLR ligands. Both bone-marrow-derived and dLN PDL2+ DCs strongly induced naive CD4 T cell proliferation, but not differentiation into any of the known T helper lineages. Surprisingly, however, bone-marrow-derived and skin dLN PDL2+ DCs induce robust activation of Th2 effector or memory cells. This finding suggests that restimulation of effector or memory CD4+ T cells might be controlled by distinct DC subsets, although the mechanism for such specificity is currently unclear.
Gene-expression analyses revealed that PDL2+ BMDCs expressed high amounts of the transcription factor IRF4 compared to PDL2– BMDCs. IRF4 has a T cell-intrinsic function in Th2 cell differentiation (Lohoff et al., 2002); it functions in regulatory T cells to control Th2 cell-mediated immune responses (Zheng et al., 2009), it controls plasma cell differentiation and class switch recombination (Klein et al., 2006), and finally it negatively regulates TLR signaling (Negishi et al., 2005) and promotes M2 polarization of macrophages (Satoh et al., 2010). Together, these prior studies point to an important contribution of IRF4-dependent gene expression in Th2 cell-mediated immunity. In addition, IRF4 has been previously implicated in the development of CD11bhi CD8α– conventional DCs in the spleen (Suzuki et al., 2004) and in the regulation of cutaneous DC migration (Bajana et al., 2012). More recently, it was shown that IRF4-dependent CD103+ CD11b+ DCs drive mucosal Th17 cell differentiation in draining mesenteric lymph nodes (Persson et al., 2013; Schlitzer et al., 2013).
We found that PDL2+ CD301b+ DC differentiation in vitro and in vivo was dependent on DC-specific expression of IRF4 and that Irf4flox/floxxCd11c-cre mice were deficient in Th2 cell responses to a protease allergen and a parasitic nematode N. brasiliensis. Consistent with this result, elimination of CD301b+ DCs by an alternative strategy (based on the specificity of CD301b expression) also resulted in ablation of Th2 cell responses (see accompanying paper by Kumamoto et al. 2013). Although the previously reported IRF4-dependent control of DC migration (Bajana et al., 2012) can contribute to this phenotype, we find both a specific defect in the Th2 cell response in Irf4flox/floxxCd11c-cre mice and an extensive IRF4-dependent gene-expression program in PDL2+ CD301b+ DCs.
Although the involvement of DCs in Th2 cell responses has been previously reported (Bell et al., 2013; Hammad et al., 2010; Leon et al., 2012; Phythian-Adams et al., 2010; Plantinga et al., 2013; Steinfelder et al., 2009; Tang et al., 2010), it has been unknown whether there is a specialized subset of DCs performing this function. Although our findings demonstrate the role of IRF4-dependent PDL2+ CD301b+ DCs in Th2 cell responses to protease allergen and parasitic infection, it is possible that Th2 cell responses to other stimuli or in other anatomical locations might be mediated by different DC subsets. Importantly, PDL2+ CD301b+ DCs are unable to induce Th2 cell differentiation of naive T cells in vitro, suggesting that an additional cell type present in vivo but not in the in vitro system might be required. In summary, our data indicate that in addition to Th2 cells, Treg cells, and M2 macrophages, IRF4 plays an essential role in DCs in controlling Th2 cell responses.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6, Tnfrsf4−/−, Irf4flox/flox, OT-II, and 4get mice were purchased from Jackson laboratories. Cd11c-cre (Caton et al., 2007), Pdcd1−/− (Nishimura et al., 1999), Foxp3-GFP.KI (Fontenot et al., 2005), Pdcd1lg2−/− (Nguyen et al., 2002), and Irf4−/− (Klein et al., 2006) mice have been described previously. Animals were 6–8 weeks at time of analyses and were all on a C57BL/6 background. Animals were bred and maintained in a conventional, specific pathogen-free facility at Yale University, and all animal experiments were performed in accordance with regulatory guidelines and standards set by the Institutional Animal Care and Use Committee of Yale University.
Reagents and Antibodies
OVA and LPS were purchased from Sigma Aldrich. Papain was purchased from Calbiochem, EMD Millipore. CpG DNA and OVA peptide (aa 323–339) were generated by the Keck facility. DQ-OVA was purchased from Molecular Probes. Flow cytometry antibodies used include CD86, CD80, CD83, CD11b, CD103, CCR7, CD40, MHCII I-Ab, CD276 (B7-H3), CD11c, CD8a, CD205 (Dec-205), IFN-γ, IL-4, IL-5, and IL-13, all from BD Biosci-ences. CD326 (EpCAM), CD274 (PDL1) and CD273 (PDL2), and IRF4 antibodies were purchased from eBioscience. Recombinant IL-4, IL-25, IL-33, TSLP, and TGF-β were purchased from R&D Systems. CD301b antibody was a generous gift from Drs. Yosuke Kumamoto and Akiko Iwasaki. Retroviral constructs encoding IRF4 was kindly provided by Dr. Alessandra Pernis.
Bone-Marrow DC Cultures
BMDCs were generated from bone-marrow cultures of 1.0 × 106 cells per ml cultured for 5 days in granulocyte-macrophage colony-stimulating factor. BMDC populations were purified by flow cytometry and stimulated with LPS (100 ng/ml), CpG-DNA (3 µM), IL-4 (100 ng/ml), or TGF-β (10 ng/ml).
For endocytosis assays, bone-marrow-derived DCs were incubated on day 5 for 20 min at 4°C or 37°C with OVA-FITC (100 µg/ml), DQ-OVA (100 µg/ml), Dex-FITC (500 µg/ml), and CpG-FITC (3 µM). Cells were collected and thoroughly washed followed by examination by flow cytometry.
In Vivo DC Isolation
dLNs were homogenized with single frosted slides and filtered to obtain a single-cell suspension. Dermal and epidermal sheets were obtained after treatment of the ear skin with Dispase II (2mg/ml) in PBS for 1 hr at 37°C. Dermal sheets were digested in RPMI 1640 medium with 10% FCS, collage-nase type D (6 mg/ml), and hyaluronidase (2 mg/ml) at 37°C for 1hr with frequent pipetting to obtain a single-cell suspension.
Immunizations
Mice were immunized subcutaneously in the rear footpads with either 50 µg papain and 50 µg OVA in PBS or 5 µg LPS and 50 µg OVA emulsified in incomplete Freund’s adjuvant (IFA). Popliteal and inguinal lymph nodes were isolated from mice 6 days later, and CD4+ T cells were purified by magnetic-activated cell sorting. Purified CD4+ T cells (1 × 105) were cultured in U-bottom 96-well tissue culture plates with irradiated splenocytes as antigen-presenting cells (3 × 105) and titrating doses of antigen for approximately 96–120 hr. Supernatants were collected to assess cytokine production by ELISA. For intracellular cytokine staining, popliteal and inguinal lymph nodes were isolated and homogenized from mice at day 6 after immunization. Cells were stimulated in U-bottom 96-well tissue culture plates with 20 ng/ml PMA and 1 µg/ml ionomycin (Sigma) for 2 hrs at 37°C and another 4 hrs at 37°C in the presence of Golgi Plug (BD Biosciences). Cells were then fixed and permeabilized with BD Cytofix/Cytoperm Kit (BD Biosciences) and stained with anti-cytokine antibodies for 1 hr on ice.
Nippostrongylus Brasiliensis Infection
We subcutaneously injected 625 third-stage larvae of N. brasiliensis, purified as described, into mice (Voehringer et al., 2006). Mice were sacrificed at day 7 after infection, and mediastinal and mesenteric lymph nodes were collected. Cells were restimulated in vitro and subjected to intracellular cytokine staining as previously described.
DC and T Cell In Vitro Coculture
Flow cytometry-sorted DCs (2 × 104) were cocultured with either 1 3 105 naive CD4+ T cells (CD44lo CD62Lhi) or 1 3 105 effector or memory CD4+ T cells (CD44hi CD62Llo) purified by flow cytometry in the presence of soluble anti-CD3ε (1 µg/ml) for 72–96 hr. IL-4, IL-25, IL-33, or TSLP (10 ng/ml) was added at the beginning of the culture where indicated. T cell proliferation was assessed by adding [3H]-thymidine for the last 12–16 hrs of the culture. Supernatants were collected to determine cytokine production by ELISA.
Enzyme Linked Immunosorbent Assay (ELISA)
Paired antibodies against IL-4, IL-5, IL-13, IL-6, tumor necrosis factor-α (TNF-α), IFN-γ, and IL-17 were purchased from BD Biosciences to perform ELISAs.
Gene-Expression Analysis
Total RNA from the different DC subsets was isolated with the RNeasy kit (QIAGEN) and prepared for microarray and RNA-seq at the Yale Center for Genome Analysis (YCGA). RNA samples for microarray study were applied to a GeneChip Mouse Genome 430 Array. Scanned CEL files were background corrected, normalized, and summarized by using the oligo (2.12, Biocon-ductor) package. RNA samples for RNA-seq analysis were sequenced on an Ilumina Hi-Seq 2000. Reads were mapped to the mm9 genome reference sequence with the bowtie (0.12.7) algorithm. Read sequences were aligned to the mm9 reference genome and calculated with the easyRNaseq (2.12, Bioconductor) package. Gene ontology for RNA-seq data was performed with the goseq (2.12, Bioconductor) package.
Statistics
Data were analyzed with Student’s t test with Prism4 (GraphPad Software; GraphPad, San Diego, CA). Data are represented as mean ± SD. Normalization, parametric statistical testing, RPKM calculation, and PCA analysis of whole-genome-expression data was performed with the R statistical language (2.15.3).
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank A. Iwasaki, Y. Kumamoto, U. Klein, B. Reizis, L. Chen, and A. Pernis for providing reagents. This study was supported by a grant from the National Institutes of Health (RO1 AI08977, 5RO1 AI055502, and R37 AI046688) and the Howard Hughes Medical Institute.
Footnotes
ACCESSION NUMBERS
The accession number for the microarray data reported in this paper is GSE50898.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.08.028.
REFERENCES
- Bajana S, Roach K, Turner S, Paul J, Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell BD, Kitajima M, Larson RP, Stoklasek TA, Dang K, Sakamoto K, Wagner KU, Reizis B, Hennighausen L, Ziegler SF. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat. Immunol. 2013;14:364–371. doi: 10.1038/ni.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowski TA, Nelson AJ, Farr AG, Udey MC. Expression of gp40, the murine homologue of human epithelial cell adhesion molecule (Ep-CAM), by murine dendritic cells. Eur. J. Immunol. 1996;26:110–114. doi: 10.1002/eji.1830260117. [DOI] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley K, Igyarto BZ, Ortner D, Bobr A, Kashem S, Schenten D, Kaplan DH. Langerhans cells require MyD88-dependent signals for Candida albicans response but not for contact hypersensitivity or migration. J. Immunol. 2012;188:4334–4339. doi: 10.4049/jimmunol.1102759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H, Plantinga M, Deswarte K, Pouliot P, Willart MA, Kool M, Muskens F, Lambrecht BN. Inflammatory dendritic cells-not basophils-are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 2010;207:2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto D, Miller J, Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henri S, Vremec D, Kamath A, Waithman J, Williams S, Benoist C, Burnham K, Saeland S, Handman E, Shortman K. The dendritic cell populations of mouse lymph nodes. J. Immunol. 2001;167:741–748. doi: 10.4049/jimmunol.167.2.741. [DOI] [PubMed] [Google Scholar]
- Henri S, Guilliams M, Poulin LF, Tamoutounour S, Ardouin L, Dalod M, Malissen B. Disentangling the complexity of the skin dendritic cell network. Immunol. Cell Biol. 2010;88:366–375. doi: 10.1038/icb.2010.34. [DOI] [PubMed] [Google Scholar]
- Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, Kaplan DH. Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity. 2011;35:260–272. doi: 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/ macrophage colony-stimulating factor. J. Exp. Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata K, Watanabe N, Guo M, Tomihara K, Brumlik MJ, Yagita H, Pardoll D, Chen L, Shin T. Costimulator B7-DC attenuates strong Th2 responses induced by Nippostrongylus brasiliensis. J. Immunol. 2010;184:2086–2094. doi: 10.4049/jimmunol.0804051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang A, Bloom O, Ono S, Cui W, Unternaehrer J, Jiang S, Whitney JA, Connolly J, Banchereau J, Mellman I. Disruption of E-cadherin-mediated adhesion induces a functionally distinct pathway of dendritic cell maturation. Immunity. 2007;27:610–624. doi: 10.1016/j.immuni.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IL, Kroenke MA, Segal BM. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J. Exp. Med. 2010;207:953–961. doi: 10.1084/jem.20091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Kumamoto Y, Denda-Nagai K, Aida S, Higashi N, Irimura T. MGL2 Dermal dendritic cells are sufficient to initiate contact hypersensitivity in vivo. PLoS ONE. 2009;4:e5619. doi: 10.1371/journal.pone.0005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y, Linehan M, Weinstein JS, Laidlaw BJ, Craft JE, Iwasaki A. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39 doi: 10.1016/j.immuni.2013.08.029. Published online September 26, 2013. http://dx.doi.org/10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon B, Ballesteros-Tato A, Browning JL, Dunn R, Randall TD, Lund FE. Regulation of T(H)2 development by CXCR5+ dendritic cells and lymphotoxin-expressing B cells. Nat. Immunol. 2012;13:681–690. doi: 10.1038/ni.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl. Acad. Sci. USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrs M, Shinkai K, Mohrs K, Locksley RM. Analysis of type 2 immunity in vivo with a bicistronic IL-4 reporter. Immunity. 2001;15:303–311. doi: 10.1016/s1074-7613(01)00186-8. [DOI] [PubMed] [Google Scholar]
- Negishi H, Ohba Y, Yanai H, Takaoka A, Honma K, Yui K, Matsuyama T, Taniguchi T, Honda K. Negative regulation of Toll-like-receptor signaling by IRF-4. Proc. Natl. Acad. Sci. USA. 2005;102:15989–15994. doi: 10.1073/pnas.0508327102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Radhakrishnan S, Ciric B, Tamada K, Shin T, Pardoll DM, Chen L, Rodriguez M, Pease LR. Cross-linking the B7 family molecule B7-DC directly activates immune functions of dendritic cells. J. Exp. Med. 2002;196:1393–1398. doi: 10.1084/jem.20021466. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/s1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 Transcription-Factor-Dependent CD103CD11b Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Phythian-Adams AT, Cook PC, Lundie RJ, Jones LH, Smith KA, Barr TA, Hochweller K, Anderton SM, Hammerling GJ, Maizels RM, MacDonald AS. CD11c depletion severely disrupts Th2 induction and development in vivo. J. Exp. Med. 2010;207:2089–2096. doi: 10.1084/jem.20100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga M, Guilliams M, Vanheerswynghels M, Deswarte K, Branco-Madeira F, Toussaint W, Vanhoutte L, Neyt K, Killeen N, Malissen B, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Artis D. New paradigmsin type 2immunity. Science. 2012;337:431–435. doi: 10.1126/science.1221064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis e Sousa C. Dendritic cells as sensors of infection. Immunity. 2001;14:495–498. doi: 10.1016/s1074-7613(01)00136-4. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfelder S, Andersen JF, Cannons JL, Feng CG, Joshi M, Dwyer D, Caspar P, Schwartzberg PL, Sher A, Jankovic D. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1) J. Exp. Med. 2009;206:1681–1690. doi: 10.1084/jem.20082462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha-dendritic cell development. Proc. Natl. Acad. Sci. USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Tang H, Cao W, Kasturi SP, Ravindran R, Nakaya HI, Kundu K, Murthy N, Kepler TB, Malissen B, Pulendran B. The T helper type 2 response tocysteine proteases requires dendritic cell-basophil cooperation via ROS-mediated signaling. Nat. Immunol. 2010;11:608–617. doi: 10.1038/ni.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D, Reese TA, Huang X, Shinkai K, Locksley RM. Type 2 immunity is controlled by IL-4/IL-13 expression in hematopoietic non-eosinophil cells of the innate immune system. J. Exp. Med. 2006;203:1435–1446. doi: 10.1084/jem.20052448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.