Abstract
Introduction
Under normal conditions, the spatial distribution of resting cerebral blood flow and cerebral metabolic rate of glucose are closely related. A relatively new magnetic resonance (MR) technique, pseudo-continuous arterial spin labeling (PCASL), can be used to measure regional brain perfusion. We identified a Parkinson's disease (PD)-related perfusion and metabolic covariance pattern in the same patients using PCASL and FDG-PET imaging and assessed (dis)similarities in the disease-related pattern between perfusion and metabolism in PD patients.
Methods
Nineteen PD patients and seventeen healthy controls underwent [18F]-fluorodeoxyglucose positron emission tomography (FDG-PET) imaging. Of 14 PD patients and all healthy controls PCASL-MRI could be obtained. Data were analyzed using scaled subprofile model/principal component analysis (SSM/PCA).
Results
Unique Parkinson's disease-related perfusion and metabolic covariance patterns were identified using PCASL and FDG-PET in the same patients. The PD-related metabolic covariance brain pattern is in high accordance with previously reports. Also our disease-related perfusion pattern is comparable to the earlier described perfusion pattern. The most marked difference between our perfusion and metabolic patterns is the larger perfusion decrease in cortical regions including the insula.
Conclusion
We identified PD-related perfusion and metabolic brain patterns using PCASL and FDG-PET in the same patients which were comparable with results of existing research. In this respect, PCASL appears to be a promising addition in the early diagnosis of individual parkinsonian patients.
Keywords: Parkinson's disease, FDG-PET imaging, PCASL-MRI, Disease-specific covariance patterns, SSM/PCA method
Highlights
-
•
Unique PD-related perfusion and metabolic covariance patterns were identified.
-
•
PD-related metabolic pattern is highly reproducible compared to earlier reports.
-
•
PCASL is a promising addition in the early diagnosis of parkinsonian patients.
1. Introduction
For many years, nuclear imaging techniques have been used to visualize disease-related changes in brain perfusion and glucose metabolism in neurodegenerative brain diseases.
Sokoloff et al. were the first to report that under physiological steady state conditions, cerebral blood flow (CBF) is coupled to the level of cerebral oxygen (CMRO2) and glucose consumption (Sokoloff, 1977). Leenders et al. measured rCBF and rCMRO2 in patients with Parkinson's disease (PD). They showed an increase of regional blood flow and oxygen metabolism in the basal ganglia of the affected hemisphere in PD patients with predominantly unilateral disease (Leenders, and Wolfson, 1984). In the 1980s, a SPECT tracer, 99Tcm-hexamethylpropyleneamine oxime (99Tcm-HM-PAO), was developed to detect cerebral blood flow with SPECT-imaging. The PET tracer [18F]-fluorodeoxyglucose (FDG) allows the measurement of cerebral metabolic rate of glucose (CMRglc). Regional differences in cerebral glucose metabolism have been reported in parkinsonian syndromes using univariate methods (Teune et al., 2010). Data-driven multivariate methods are increasingly used to examine disease-specific metabolic covariance patterns in parkinsonian syndromes (Ma et al., 2007; Teune et al., 2013). This has improved our understanding of the pathophysiology of these diseases as well as our ability to diagnose patients at an earlier disease stage (Tang et al., 2010a).
Ma et al. reproduced this PD-related metabolic covariance pattern, using H215O PET scanning, indeed suggesting that cerebral blood flow and glucose metabolism are tightly coupled in PD patients (Ma et al., 2007). However, in clinical practice, blood flow measurements with PET are not widely used because it is a demanding and time-consuming procedure, while spatial resolution of SPECT is less optimal. Recently, it has become possible to measure brain perfusion with a relatively new MR technique, pseudo-continuous arterial spin labeling (PCASL). It permits the noninvasive measurement of perfusion with MRI by using a train of radio frequency (RF) pulses and magnetic field gradient pulses to achieve labeling of spins in flowing blood (Dai et al., 2008; van Osch et al., 2009). Ma et al. analyzed the expression of the existing PD-related metabolic covariance pattern in a small number of parkinsonian patients using perfusion MRI (continuous arterial spin labeling) and concluded that perfusion MRI can be used for accurate quantification of disease-related covariance patterns (Ma et al., 2010). Melzer et al. were the first to identify a PD-related perfusion covariance pattern (Melzer et al., 2011).
In this study we identified a PD-related perfusion and metabolic covariance pattern in the same patients using PCASL and FDG-PET imaging and assessed (dis)similarities in the disease-related pattern between perfusion and metabolism in PD patients.
2. Patients & methods
2.1. Subjects
The study was approved by the medical ethics committee of the University Medical Center Groningen. Voluntary written informed consent was obtained from each subject after verbal and written explanation of the study, in accordance with the Declaration of Helsinki. This study aimed to include 17 healthy controls and 20 PD patients in which FDG-PET and different MR image modalities were to be recorded. All healthy controls underwent both FDG-PET and PCASL-MRI. One PD patient did not have an FDG-PET scan. Three PD patients appeared not to be MRI proof (due to dental implants, large artifacts on functional MRI) and in 2 PD patients PCASL was not correctly recorded due to technical issues. In the current analysis we have 17 gender and age-matched healthy controls (HC) (5 females, 12 males; mean age 61.5 years) and 19 PD patients (6 females, 13 males; mean age 63.9 years) but only 14 PD patients (4 females, 10 males; mean age 63.8 years) who underwent both FDG-PET and PCASL. PD patients had to fulfill the UK Brain Bank criteria for PD (Litvan et al., 2003). Healthy controls were not allowed to have first-degree family members with parkinsonism or dementia. All subjects underwent MRI and within 8 weeks an FDG-PET scan and two neuropsychological tests (Mini Mental State Examination (MMSE) and Frontal Assessment Battery (FAB)) and PD patients underwent examination using the Unified Parkinson’s Disease Rating Scale (UPDRS) part 3: motor symptoms. Neuropsychological scores did not differ significantly between groups (mean HC: MMSE 29, FAB 17; mean PD (19): MMSE 29, FAB 16, mean PD (14): MMSE 28, FAB 16) indicating that PD patients were non-demented. PD patients had mild to moderate motor symptoms with Hoehn & Yahr stage: mean (in both 19 and 14 groups) 1 and standard deviation (SD) 1 vs 0.5 and a UPDRS part 3: in both 19 and 14 groups mean 18; SD 7. Antiparkinsonian medication was withheld for at least 12 hours and benzodiazepines were withheld 24 hours before MRI and FDG-PET scanning.
2.2. Image acquisition and preprocessing
MR imaging was performed on a 3 T MRI scanner (Achieva 3 Tesla, Philips Healthcare, Best, The Netherlands) using a standard 8-channel SENSE head coil. Subjects were wearing ear protection and instructed to lie still with their eyes closed, and to avoid falling asleep.
To enable PCASL imaging, the scanner was equipped with locally developed software. Pseudo-continuous labeling was performed by employing a train of Hanning-shaped RF pulses (tip angle 18°, duration 0.5 ms) with an interpulse pause of 0.5 ms in combination with a balanced gradient scheme. PCASL images were obtained in a dynamic mode of 2 × 30 volumes (labeled and control) with an echo time of 14 ms, repetition time (TR) of 4200 ms, 23 axial slices, field of view (FOV) of 240 mm with an 80 × 80 matrix and an isotropic voxel size of 3 × 3 × 6 mm.
All 30 labeled and control volumes were first motion-corrected in SPM8 (Functional Imaging Laboratory, running in Matlab 7.10.0 (R2010a, Mathworks)). Volumes were smoothed with an 8 mm full-width-at-half-maximum isotropic Gaussian kernel (FWHM). Next, volumes were coregistered to the subject's own T1 weighted images. Analogous to Van Dijk et al. (2010) labeled and control images were filtered against a time-course extracted from a ROI in white matter and cerebrospinal fluid (CSF) drawn on a T1 image. Removal of signal from CSF and white matter is motivated by the fact that these regions contain a relatively high proportion of physiological noise (e.g. cardiac and respiratory cycles). Thereafter, labeled perfusion-weighted images were subtracted from control images, creating one mean PCASL image per subject. These steps were performed using Matlab scripts developed in-house.
FDG-PET imaging was performed in a 3D mode using a Siemens Biograph mCT-64 PET scanner. Image acquisition was performed in a resting state with the subject's eyes closed in a dimly lighted room with minimal auditory stimulation. A 6-minute static frame was acquired starting 30 min after the injection of 200 MBq FDG in 4 ml saline.
The FDG-PET images were iteratively reconstructed using the OSEM algorithm with 3 iterations and 24 subsets on a matrix of 400 × 400 and smoothed with 5 mm FWHM. No zoom was applied, resulting in images with an isotropic voxel-size of 2 mm with a specified resolution of 5 mm in the center of the field of view. Scatter and attenuation corrections were applied based on the acquired low dose CT.
A study-specific template of all mean PCASL images was spatially normalized to a standard brain PET template (Montreal Neurological Institute; MNI, supplied in SPM8) and then used to spatially normalize the individual mean PCASL images. An example of the mean PCASL images of seventeen healthy controls is shown in Fig. 1c. FDG-PET images were directly spatially normalized to a PET template and then both were smoothed with 10 mm FWHM.
Fig. 1.
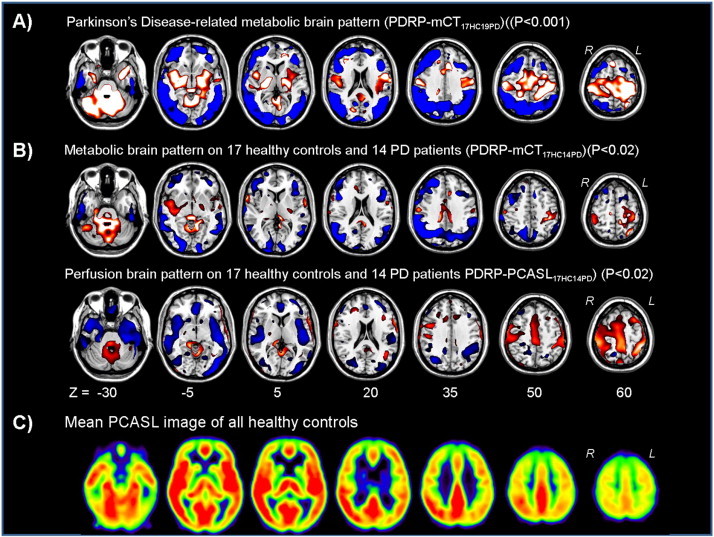
A) (T) Map of the Parkinson's disease (PD)-related metabolic brain patterns by using 17 healthy controls (HC) and 19 PD (PD) patients (PDRPmCT17HC19PD). The map was overlaid on a T1 MR template. Relative metabolic decreases (blue) and increases (red) compared to the control group are thresholded at T = 3.7–6.7 (p = 0.001). Seven transversal slices through the brain are shown.B) (T) Maps of the PD-related metabolic and perfusion covariance brain patterns (PDRP-mCT17HC14PD and PDRP-PCASL17HC14PD) calculated by using the same HC and PD subjects and overlaid on a T1 MR template. Relative metabolic and perfusion decreases (blue) and increases (red) compared to the control group are thresholded at T = 2.5–5.0 (p = 0.02). Seven transversal slices through the brain are shown.C) Example of the mean PCASL images of all healthy controls. Seven transversal slices through the brain are shown. Yellow/red = high perfusion, black/blue = low perfusion.
2.3. Statistical analysis
SSM/PCA was applied using software written in-house, based on the methods of Spetsieris and Eidelberg (2011). The correctness of the in-house written script was verified in an earlier validation study of the already identified PD, MSA and PSP metabolic brain patterns (Teune et al., 2010; Spetsieris and Eidelberg 2011). We performed both the analysis with the Eidelberg software and our own software, which provided identical results. A 35% threshold of the whole-brain intensity maximum was applied to remove out-of-brain voxels which results in a mask of mainly gray matter, followed by a log transformation (Spetsieris and Eidelberg 2011). This gray matter mask was applied to the PCASL images without subsequent log transformation. After removing between-subject and between-region averages, a principal component analysis (PCA) was applied. The components explaining the largest amount of variance were selected. Together these components accounted for at least 50% of the variance. A disease-related metabolic covariance pattern was determined by a linear combination of the selected principal components with the lowest AIC (Akaike information criterion) value in a stepwise regression procedure. Thereafter, a leave-one-out cross validation procedure was performed, providing an average estimate of the disease-related metabolic brain patterns and the error of this average per voxel. By dividing the average estimate by its Standard Error of the Mean, a T-score was obtained. The metabolic PD-related pattern (PDRP-mCT17HC19PD) was thresholded at T = 3.7 (corresponding to p = 0.001) assuming normality of data. The perfusion brain pattern (PDRP-PCASL17HC14PD) was thresholded at T = 2.5 (corresponding to p = 0.02) and overlaid onto a T1 MR template using MRIcron. For comparison purposes using the same HC and PD subjects, a second metabolic brain pattern (PDRP-mCT17HC14PD) was identified using the same threshold as for the perfusion pattern. Furthermore, for PDRP-mCT17HC19PD, a leave-one-out cross validation was performed. For each subject left out, its subject score was calculated using the pattern obtained from the remaining subjects. These subject scores were transformed into z-scores with respect to the healthy control population (z-score with a mean of zero and a standard deviation of one) and displayed in a scatter plot. Receiver-operating-characteristic (ROC) curves were determined for the probability values of PD. Optimum cut-off probability values for classifying individual patients were calculated by identifying an inflection point on the ROC curve that corresponded to the best combination of sensitivity and specificity. Patients were classified as correctly diagnosed if their probability value was higher than the cut-off value. Thereafter, we calculated sensitivity, specificity, and positive and negative predictive values (PPV and NPV respectively).
To compare the present PDRP-mCT17HC19PD using a high resolution mCT camera with our previously published PD-related metabolic brain pattern using an HR+ camera (PDRP-HR18HC20PD) (Teune et al., 2010) z-scores were calculated for the HC and PD subjects scanned on the mCT camera by using the PDRP-HR18HC20PD as a reference and vice versa.
Moreover, subject scores of the PCASL images were calculated and transformed into z-scores with respect to the PCASL healthy control population using the PDRP-mCT17HC19PD as a reference.
3. Results
Spatial covariance analysis was performed on FDG-PET scans and PCASL scans from PD patients. The model with the lowest AIC value and hereafter considered as disease-related was determined by a linear combination of principal components (PCs) 1, and 2 with variance accounted for (VAF) of 27.1% (16.7 and 10.4% respectively) for PDRP-mCT17HC19PD; PCs 1, 2, 4, and 5 (37.7% VAF (18.4, 8.9, 5.6 and 4.8% respectively)) for PDRP-mCT17HC14PD and PCs 3 and 5 (16.1% VAF (9.9 and 6.1% respectively)) for PDRP-PCASL17HC14PD. A leave-one-out cross validation was subsequently performed resulting in disease-related patterns (Fig. 1a,b). The PD-related metabolic covariance pattern (PDRP-mCT17HC19PD) was characterized by relatively decreased cortical metabolic activity in the temporal, posterior parietal, inferior parietal, lateral occipital, and prefrontal association cortices and SMA. Relative increases were seen in the cerebellum and pons, thalamus and pallidum, sensorimotor cortex, limbic association cortex, paracentral lobule and left SMA (Fig. 1a).
The PD-related perfusion covariance pattern (PDRP-PCASL17HC14PD) was characterized by relatively decreased cortical perfusion activity bilaterally in the temporal, insular, posterior parietal, inferior parietal, lateral occipital and prefrontal association cortices. Relative increases were seen in the cerebellum and pons, right thalamus and pallidum, sensorimotor cortex, paracentral lobule and supplementary motor area (SMA) (Fig. 1b). In Fig. 1b the PD-related metabolic brain pattern was shown using the same group as was used to determine the perfusion related pattern (PDRP-mCT17HC14PD). The same regions were visible as in the PDRPPDRP-mCT17HC19PD, although at a lower statistical threshold.
The corresponding z-scores showed an overlap between the patients and healthy controls using PDRP-mCT17HC19PD. A cut-off value of z = 0.31 resulted in a sensitivity of 94.7%, positive predictive value of 75%, specificity of 64.7% and negative predictive value of 91.7% for correct PD classification of an individual patient (Fig. 2a).
Fig. 2.
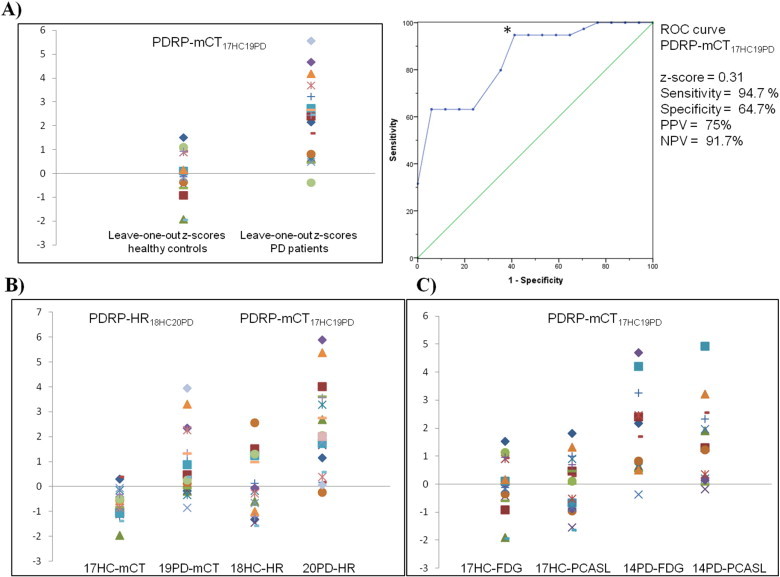
A) Scatter plot and receiver-operating-characteristic (ROC) curve for the Parkinson's disease (PD)-related metabolic brain pattern by using 17 healthy controls (HC) and 19 PD (PD) patients (PDRP-mCT17HC19PD). On the y-axis of the scatter plot the leave-one-out z-scores are displayed and on the x-axis the healthy control group and the PD patient group. * = Inflection point on the ROC curve (z-score); y-axis ROC curve = sensitivity, x-axis ROC curve = 1 − specificity; PPV = positive predictive value; NPV = negative predictive value.B) Scatter plot displaying z-scores for healthy controls and Parkinson's disease patients scanned on the high resolution mCT camera (17HC-mCT and 19PD-mCT (first two columns)) by using the previously published metabolic brain pattern on the HR+ camera (Teune et al., 2013) (PDRP-HR18HC20PD) as a reference. In the third and fourth columns z-scores are displayed for the HC and PD subjects scanned on the HR+ camera (18HC-HR and 20PD-HR) by using PDRP-mCT17HC19PD as a reference.C) Scatter plot displaying z-scores for healthy controls and Parkinson's disease patients and their FDG and PCASL scans (17HC-FDG, 17HC-PCASL, 14PD-FDG and 14PD-PCASL respectively) by using the PD-related metabolic brain pattern (PDRP-mCT17HC19PD) as a reference.
The PDRP-mCT17HC19PD z-scores did not correlate with the UPDRS motor scores of PD patients (Spearman's rho = 0.294 and p = 0.222).
The present PD-related metabolic brain pattern using a high-resolution mCT (PDRP-mCT17HC19PD) showed high reproducibility with our previously published PD-related metabolic brain pattern using an HR+ camera (PDRP-HR18HC20PD) (Fig. 2b). We computed the z-scores of the FDG and PCASL images of 17 healthy controls and 14 PD patients on the PDRP-mCT17HC19PD (Fig. 2c) and performed a correlation analysis. The Pearson correlation coefficient was 0.5 (p = 0.004).
4. Discussion
In this study we identified PD-related perfusion and glucose metabolic brain patterns measured in the same patients. The PD-related metabolic covariance brain pattern is in high accordance with previously described disease-related metabolic brain patterns in different cohorts in PD patients using standard clinical FDG-PET imaging (Ma et al., 2007; Teune et al., 2013). The present PDRP-mCT17HC19PD did not correlate with UPDRS motor scores. This is probably related to the relatively short disease duration of the PD patients in comparison with previous reports with more advanced stage PD patients (Ma et al., 2010; Tang et al., 2010b).
We obtained a disease-specific perfusion brain pattern using PCASL-MR imaging. Our disease-related pattern is comparable to the PD-related perfusion brain pattern described by Melzer et al. characterized by decreased perfusion in the posterior parieto-occipital cortex, and middle frontal gyri and preserved perfusion in the globus pallidus, putamen, anterior cingulate and post/precentral gyri (Melzer et al., 2011). The most characteristic difference between our perfusion brain pattern and previous reports is the decrease of cerebral perfusion in the insular cortex on both sides in our data. However, Helmich et al. have shown that PD patients had decreased connectivity between the posterior putamen and various cortical regions contributing to the corticostriatal loop, including the insula using resting-state functional MRI data (Helmich et al., 2010). Performing PCASL and FDG-PET imaging in the same patients enabled a comparison between both patterns. z-Scores of PCASL datasets onto the metabolic pattern correlated positively (0.5) with the z-scores of FDG datasets onto the metabolic pattern. This value indicates that there is an overlap, however, not complete. As discussed above, there was a larger perfusion decrease in the PD-related pattern in cortical regions including the insula than in the metabolic brain pattern.
Another difference is the less pronounced increased perfusion in the basal ganglia in the perfusion-related pattern compared to the metabolic brain pattern. This is probably related to a lower signal-to-noise ratio. It is known that MRI head coils with multiple receive channels result in better signal to noise ratio in the cortex than in deeper brain structures.
Our data shows that PCASL is a promising technique, which could be developed further as an additional measurement to improve image-based diagnosis of parkinsonian syndromes. Our results need to be replicated in larger samples, particularly in prospective validation samples and especially in PCASL data.
5. Conclusion
We identified PD-related perfusion and metabolic brain patterns using PCASL and FDG-PET in the same patients which were comparable with results of existing research. In this respect, PCASL appears to be a promising addition in the early diagnosis of individual parkinsonian patients.
Disclosure
This research was funded with a grant of “Stichting ParkinsonFonds”.
Acknowledgments
We would like to thank M.F. Masman for the assistance with PCASL image reconstruction and M. Segbers for the assistance with FDG-PET image reconstruction.
References
- Dai W., Garcia D., de Bazelaire C., Alsop D.C. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2008;60:1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmich R.C., Derikx L.C., Bakker M., Scheeringa R., Bloem B.R., Toni I. Spatial remapping of corticostriatal connectivity in Parkinson's disease. Cerebral Cortex (New York, N.Y.: 1991) 2010;20:1175–1186. doi: 10.1093/cercor/bhp178. [DOI] [PubMed] [Google Scholar]
- Leenders K.L., Wolfson L., Jones T. Cerebral blood flow and oxygen metabolism measurement with positron emission tomography in Parkinson's disease. Monographs in Neural Sciences. 1984;11:180–186. doi: 10.1159/000409209. [DOI] [PubMed] [Google Scholar]
- Litvan I., Bhatia K.P., Burn D.J., Goetz C.G., Lang A.E., McKeith I. Movement Disorders Society Scientific Issues Committee report: SIC Task Force appraisal of clinical diagnostic criteria for parkinsonian disorders. Movement Disorders: Official Journal of the Movement Disorder Society. 2003;18:467–486. doi: 10.1002/mds.10459. [DOI] [PubMed] [Google Scholar]
- Ma Y., Huang C., Dyke J.P., Pan H., Alsop D., Feigin A. Parkinson's disease spatial covariance pattern: noninvasive quantification with perfusion MRI. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2010;30:505–509. doi: 10.1038/jcbfm.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Tang C., Spetsieris P.G., Dhawan V., Eidelberg D. Abnormal metabolic network activity in Parkinson's disease: test–retest reproducibility. Journal of Cerebral Blood Flow and Metabolism: Official Journal of the International Society of Cerebral Blood Flow and Metabolism. 2007;27:597–605. doi: 10.1038/sj.jcbfm.9600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer T.R., Watts R., Macaskill M.R., Pearson J.F., Rüeger S., Pitcher T.L. Arterial spin labelling reveals an abnormal cerebral perfusion pattern in Parkinson's disease. Brain: A Journal of Neurology. 2011;134:845–855. doi: 10.1093/brain/awq377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff L. Relation between physiological function and energy metabolism in the central nervous system. Journal of Neurochemistry. 1977;29:13–26. doi: 10.1111/j.1471-4159.1977.tb03919.x. [DOI] [PubMed] [Google Scholar]
- Spetsieris P.G., Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson's disease: methodological issues. NeuroImage. 2011;52:2899–2914. doi: 10.1016/j.neuroimage.2010.10.025. 20969965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.C., Poston K.L., Dhawan V., Eidelberg D. Abnormalities in metabolic network activity precede the onset of motor symptoms in Parkinson's disease. Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 2010;30:1049–1056. doi: 10.1523/JNEUROSCI.4188-09.2010. 20089913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.C., Poston K.L., Eckert T., Feigin A., Frucht S., Gudesblatt M. Differential diagnosis of parkinsonism: a metabolic imaging study using pattern analysis. Lancet Neurology. 2010;9:149–158. doi: 10.1016/S1474-4422(10)70002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teune L.K., Bartels A.L., de Jong B.M., Willemsen A.T., Eshuis S.A., de Vries J.J. Typical cerebral metabolic patterns in neurodegenerative brain diseases. Movement Disorders: Official Journal of the Movement Disorder Society. 2010;25:2395–2404. doi: 10.1002/mds.23291. [DOI] [PubMed] [Google Scholar]
- Teune L.K., Renken R.J., Mudali D., de Jong B.M., Dierckx R.A., Roerdink J.B.T.M. Validation of parkinsonian disease-related metabolic brain patterns. Movement Disorders: Official Journal of the Movement Disorder Society. 2013;28(4):547–551. doi: 10.1002/mds.25361. 23483593 [DOI] [PubMed] [Google Scholar]
- Van Dijk K.R., Hedden T., Venkataraman A., Evans K.C., Lazar S.W., Buckner R.L. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. Journal of Neurophysiology. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Osch M.J., Teeuwisse W.M., van Walderveen M.A., Hendrikse J., Kies D.A., van Buchem M.A. Can arterial spin labeling detect white matter perfusion signal? Magnetic Resonance in Medicine: Official Journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2009;62:165–173. doi: 10.1002/mrm.22002. [DOI] [PubMed] [Google Scholar]


