Abstract
The WAVE regulatory complex (WRC) is a 400-KDa heteropentameric protein assembly that plays a central role in controlling actin cytoskeletal dynamics in many cellular processes. The WRC acts by integrating diverse cellular cues and stimulating the actin nucleating activity of the Arp2/3 complex at membranes. Biochemical and biophysical studies of the underlying mechanisms of these processes require large amounts of purified WRC. Recent success in recombinant expression, reconstitution, purification and crystallization of the WRC has greatly advanced our understanding of the inhibition, activation and membrane recruitment mechanisms of this complex. But many important questions remain to be answered. Here we summarize and update the methods developed in our laboratory, which allow reliable and flexible production of tens of milligrams of recombinant WRC of crystallographic quality, sufficient for many biochemical and structural studies.
Keywords: actin, WAVE, WRC, WASP, reconstitution, protein complex
1. Introduction
Actin nucleation, the formation of a new actin filament from actin monomers, is a crucial and rate-limiting step in actin cytoskeletal dynamics (Pollard & Cooper, 2009). The Arp2/3 complex is a central actin nucleator that binds to the side of existing filaments to promote new filament growth as a branch, creating a complex cortical actin network beneath membranes. The Arp2/3 complex has low basal activity in actin nucleation and requires stimulation by so-called nucleation promoting factors (NPFs). Members of the ubiquitous WASP (Wiskott-Aldrich syndrome protein) family constitute the largest and best understood group of NPFs (Campellone & Welch, 2010; Padrick & Rosen, 2010; Rotty, Wu, & Bear, 2013). WASP family proteins are defined by a conserved C-terminal VCA (Verprolin-homology, Central, Acidic) sequence that is sufficient to stimulate the Arp2/3 complex. Members of the WASP family include WASP/N-WASP, WAVE1/2/3 (WASP-family verprolin homologous protein 1, 2 or 3), and the more recently discovered WASH (WASP and SCAR homolog), WHAMM (WASP homolog associated with actin, membranes, and microtubules) and JMY (junction-mediating and regulatory protein) (Pollitt & Insall, 2009; Takenawa & Suetsugu, 2007).
Many WASP family members are basally inhibited and, in response to activation signals, release the VCA to stimulate actin assembly. WASP and N-WASP are inhibited in cis through intra-molecular contacts between the VCA and an N-terminal GTPase binding domain (GBD)(A. S. Kim, Kakalis, Abdul-Manan, Liu, & Rosen, 2000; Miki, Sasaki, Takai, & Takenawa, 1998; Prehoda, Scott, Mullins, & Lim, 2000; Rohatgi, et al., 1999). In contrast, the WAVE proteins are inhibited in trans by incorporation into a ∼ 400-kDa heteropentameric protein assembly, referred to as the WAVE regulatory complex (WRC). The WRC consists of five proteins (Fig. 1A), Sra1/Cyfip1 (or the ortholog PIR121/Cyfip2), Nap1/Hem2/Kette (or the ortholog Hem1), Abi2 (or the orthologs Abi1 and Abi3), HSPC300/Brick1 and WAVE1/SCAR (or the orthologs WAVE2 and WAVE3) (Eden, Rohatgi, Podtelejnikov, Mann, & Kirschner, 2002). Different orthologs of each component seem exchangeable, allowing assembly of different WRC isoforms (Stovold, Millard, & Machesky, 2005). Within the WRC, the VCA is sequestered in trans through intra-complex interactions (Z. Chen, et al., 2010) (Fig. 1A).
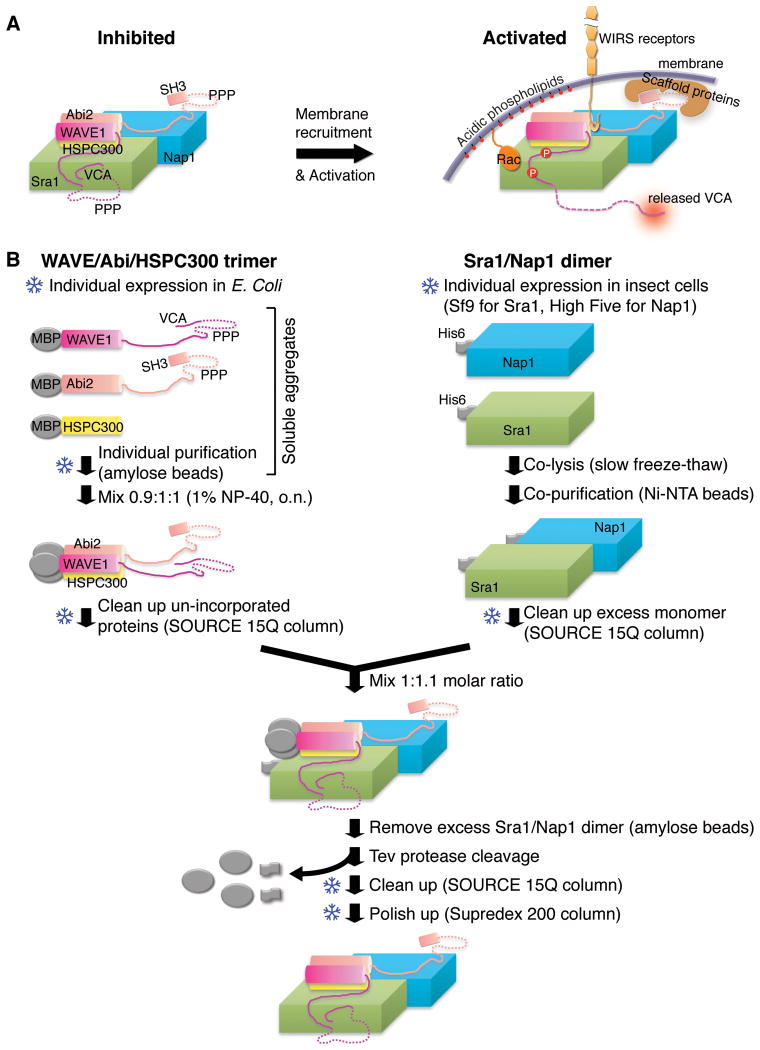
Figure 1.
Activation mechanism and purification strategy of the WRC. (A) Schematic of WRC inhibition, activation and membrane recruitment. Dotted lines indicate unstructured sequences. (B) Schematic of WRC in vitro reconstitution. Snowflake symbols indicate steps after which samples can be frozen and stored for future use.
To function, the inhibited WRC needs to be both recruited to and activated at the membrane by diverse signaling molecules as illustrated in Fig. 1A. These include small GTPases (Rac and Arf), acidic phospholipids (phosphatidylinositol (3,4,5)-trisphosphate, PIP3), kinases (Abl, Cdk5 and ERK2), scaffolding proteins (IRSp53, Toca1 and WRP) (Z. Chen, et al., 2010; Fricke, et al., 2009; Koronakis, et al., 2011; Mendoza, 2013; Miki, Yamaguchi, Suetsugu, & Takenawa, 2000; Oikawa, et al., 2004; Soderling, et al., 2007; Takenawa & Suetsugu, 2007; Westphal, Soderling, Alto, Langeberg, & Scott, 2000), and the recently identified WIRS (WRC interacting receptor sequence)-containing family consisting of a large number of membrane receptors (B. Chen, et al., 2013) These ligands link the WRC to many cellular processes (adhesion, migration, division, fusion etc.) across diverse biological systems, including embryogenesis, neuron morphogenesis and plasticity, immune cell activation and chemotaxis, and cancer invasion and metastasis (Pollitt & Insall, 2009; Takenawa & Suetsugu, 2007).
Mechanistic biochemical and biophysical studies of WRC/ligand interactions require access to purified WRC. Over the last decade, three major strategies have been developed to generate such material. The first involves purification from natural sources, including animal brains, blood or cultured cells (Eden, et al., 2002; Gautreau, et al., 2004; Y. Kim, et al., 2006; Lebensohn & Kirschner, 2009; Weiner, et al., 2006). This method allowed the discovery of the WRC and produces materials preserving native post-translational modifications. As described in the same issue (Hume, Humphreys, & Koronakis, xxx), Koronakis and colleagues recently further developed a new strategy to purify the native WRC from porcine brain extract by using phospholipid bilayer coated silica microbeads, which led to identification of a new WRC activator, Arf (Koronakis, et al., 2011). The above purifications cannot be readily scaled up, and do not allow genetic modification of the WRC components for structure/function studies. The second method is in vivo reconstitution, involving (co-)expression of one or multiple affinity tagged WRC subunits in cultured mammalian or insect cells (Derivery, Lombard, Loew, & Gautreau, 2009; Ismail, Padrick, Chen, Umetani, & Rosen, 2009; Mendoza, et al., 2011). The recombinant WRC is assembled while expressed in cells and is purified using the affinity tags. This method had produced the WRC of sufficient quantity and purity for rigorous biochemical assays, which led to the final reconciliation of debates about whether the WRC is intrinsically inhibited. Here we focus on the third method, in vitro reconstitution, developed and optimized in our laboratory over the last 10 years (B. Chen, et al., 2013; Z. Chen, et al., 2010; Ismail, et al., 2009)(Fig. 1B). This method improves the yield (up to tens of milligrams), the purity (yielding crystal structures of the WRC), and readily allows engineering of the complex to answer mechanistic questions. Through this method, we have been able to accomplish multiple structure-function studies of the WRC (B. Chen, et al., 2013; Z. Chen, et al., 2010; Ismail, et al., 2009; Padrick, et al., 2008).
2. Overview of the in vitro reconstitution method
Generation of recombinant WRC presents substantial challenges. First, expression of the WRC subunits in bacteria either gives poor yield or produces aggregated or truncated proteins. Second, co-expression and purification from eukaryotic hosts suffers from high cost and low yield. Third, inhibition of WAVE depends on a fully assembled WRC. Partially assembled subcomplexes containing WAVE, but missing other components, have high basal activity that obscures biochemical assays. This was actually a problem during our early preparations of the WRC generated by co-expressing all five components in insect cells. Such partial assemblies coincidentally co-purified with the WRC pentamer, giving variable activities to the final material. Only after the contaminants were ultimately removed through an added cation exchange step were we able to obtain consistently inhibited WRC preparations (Ismail, et al., 2009). Similarly, aggregation of the WRC also gives anomalous activation (Lebensohn & Kirschner, 2009). Thus, purifications must rigorously exclude all subcomplexes and aggregates. These complications likely account for previous conflicting observations regarding the basal activity of the WRC (Derivery, et al., 2009; Eden, et al., 2002; Innocenti, et al., 2004; Ismail, et al., 2009; Y. Kim, et al., 2006; Lebensohn & Kirschner, 2009). Below we describe our strategy, which addresses these various difficulties.
The WRC can be viewed as two sub-complexes: a trimer of WAVE, Abi and HSPC300 held together by an N-terminal four-helix bundle, and a dimer formed by the two large structurally homologous proteins, Sra1 and Nap1 (Z. Chen, et al., 2010; Ismail, et al., 2009) (Fig. 1). The in vitro reconstitution follows these logistics: WAVE/Abi/HSPC300 trimer + Sra1/Nap1 dimer = WRC (Fig. 1B).
WAVE/Abi/HSPC300 trimer. Initially, soluble, but aggregated maltose-binding protein (MBP)-fusions of the proteins are separately expressed and purified from E. coli. The three proteins are then mixed in the presence of detergent and incubated for several hours. This allows aggregates to spontaneously disassemble and re-assemble into a WAVE/Abi/HSPC300 trimer. Further purification removes the detergent and un-incorporated protein or sub-complexes.
Sra1/Nap1 dimer. His6-tagged Sra1 and Nap1 are separately expressed in insect cells. The cells are co-lysed allowing formation of the Sra1/Nap1 heterodimer. This heterodimer is then purified and frozen.
WRC pentamer. The heteropentamer is assembled from the purified WAVE/Abi/HSPC300 trimer and the Sra1/Nap1 dimer, followed by purification to remove the excess dimer and affinity tags after protease cleavage.
It is noteworthy that all the affinity tags were fused to the N-termini of the WRC subunits; this produces the desired proteins and preserves the activity while not disturbing the inhibited state of the WRC. We have not explored placing tags at alternative positions for most subunits. However, for the full length Abi and WAVE, bacterial expression sometimes gives significant amounts of truncated products, which can be difficult to separate using conventional chromatography (note that this was not a problem in our previous approach, where all subunits were co-expressed in insect cells, but yields were much lower in that case)(Ismail, et al., 2009). To remove these products, we have recently added an additional C-terminal or internal tag to be used in a second affinity purification step. For the full length Abi, we directly add a His6 tag to the C-terminus. For the full length WAVE, since a C-terminal tag next to the VCA substantially decreases the activity of the WRC in in vitro assays, we insert a His6 or StrepII tag to an internal region between the proline-rich sequence and the VCA (B. Chen, et al., 2013).
The modular nature of this method provides excellent versatility. First, through this method we have been able to produce a variety of WRC isoforms by incorporating different constructs (different truncations or tags), orthologs (Abi1, 2, WAVE1, 2, 3) and homologs (human, Drosophila). Second, we have found that individual subunits or sub-complexes can be frozen, allowing quick combinatorial assembly of different WRC mutants. Third, this method allows for modification of individual subunits prior to reconstitution, suitable for metabolic labeling or chemical modification of specific subunits. For example, the Sra1/Nap1 dimer was selenomethionine labeled to improve x-ray diffraction quality and to solve the phase problem for crystallographic structure determination of the WRC (Z. Chen, et al., 2010).
3. Key reagents
Lysis buffer: 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA and 5 mM β-mercaptoethanol (BME)
MBP-Elution buffer: 20 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% (w/v) maltose, 10% (w/v) glycerol, 2 mM DTT
QA buffer: 20 mM Tris-HCl pH 8.0, 1 mM EDTA and 5 mM BME
QB buffer: QA buffer + 1 M NaCl
WRC-Lysis buffer: 20 mM Tris-HCl pH 8.0, 20 mM imidazole pH 8.0, 200 mM NaCl, 20% (w/v) glycerol and 5 mM BME (no EDTA, as this will block subsequent binding to Ni-NTA affinity resin)
WRC-Ni-Elution buffer: 20 mM Tris-HCl pH 8.0, 300 mM imidazole pH 8.0, 100 mM NaCl, 20% (w/v) glycerol and 5 mM BME (no EDTA)
WRC-MBP-Elution buffer: 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 1% (w/v) maltose, 20% (w/v) glycerol, 1 mM EDTA and 5 mM BME
WRC-QA buffer: 20 mM Tris-HCl pH 8.0, 20% (w/v) glycerol, 1 mM EDTA and 5 mM BME
WRC-QB buffer: WRC-QA buffer + 1 M NaCl
WRC-GF buffer: 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 12% (w/v) glycerol, 1 mM DTT
WRC-100KMEI20G buffer: 10 mM Imidazole pH 7.0, 100 mM KCl, 20% (w/v) glycerol, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT
Protease inhibitors (PI): 2 μg/mL Leupeptin, 2 μg/mL Antipain and 2 mM Benzamidine, diluted from 500x individual stock solutions in water
Ampicillin (Amp): 100 μg/mL, diluted from 1000x stock solution in water
Gentamycin (Gen): 20 μg/mL, diluted from 1000x stock solution in water
We note that the inclusion of glycerol (10 – 20% w/v) in many of the buffers is essential to maintain stability of the WRC. Without glycerol, the complex precipitates over time and shows highly variable activity.
4. Reconstitution of the WAVE/Abi/HSPC300 trimer
4.1 Expression and purification of individual trimer components
WAVE, Abi and HSPC300 are individually expressed and purified from E. coli. We routinely use N-terminal MBP fusion proteins to improve solubility and to facilitate affinity purification. Although MBP has an intermediate affinity for amylose beads, this is substantially improved by multiple MBP tags on the proteins (e.g. multiple copies on the soluble aggregates of individual proteins, or three copies on the reconstituted trimer or WRC).
Below we give a detailed protocol (Fig. 1B) that has been used to purify many different WAVE/Abi/HSPC300 trimers. In the case of full-length WAVE1, 2, 3 or Abi 1, 2, we fuse a His6-tag to or near the protein C-terminus and add a nickel affinity purification step to remove the truncated proteins (B. Chen, et al., 2013). In some cases, we use di-MBP-HSPC300 (with two tandem MBP proteins at the N-terminus) to facilitate pull-down assays (B. Chen, et al., 2013).
4.1.1 Expression constructs for trimer proteins
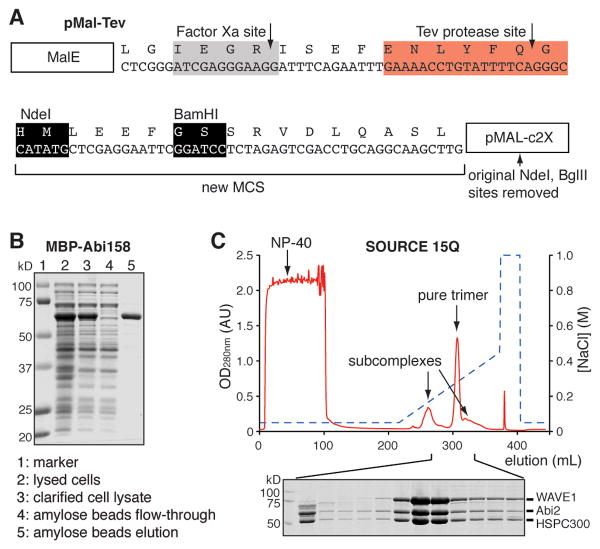
All constructs were cloned into a vector derived from pMAL-c2X (New England Biolabs). This vector has a Tev protease cleavage site introduced between the MBP and the protein of interest as shown in Fig. 2A. Here we use the following constructs as examples: WAVE217 for human WAVE1 (1-217)-(GGS)6-(485-559), in which the proline-rich region (PPP in Fig. 1) is replaced by a (Gly-Gly-Ser)6 linker; Abi158 for human Abi2 (1-158), in which the proline-rich region and the C-terminal SH3 domain are removed; and full length human HSPC300.
Figure 2.
Production of the WAVE/Abi/HSPC300 trimer. (A) Sequence of the pMal-Tev vector used for all trimer constructs. (B) SDS-PAGE gel showing affinity purification of MBP-Abi158 by amylose beads. Other trimer proteins are purified similarly. (C) Anion exchange chromatography of the reconstituted WAVE217/Abi158/HSPC300 trimer. Coomassie brilliant blue stained SDS-PAGE analysis of the indicated fractions is shown below.
4.1.2 Expression of individual proteins in E. coli
We generally use two host strains. We use BL21 (DE3) T1 R cells (New England Biolabs) for MBP-tagged Abi158, HSPC300 or shorter constructs of WAVE. We use ArcticExpress™ (DE3) RIL cells (ArcticExpress cells for short, Stratagene) for MBP-tagged WAVE217 or longer constructs of WAVE and Abi. Protocols are different for the two strains.
Day 1: Transform cells using standard procedures. Incubate plates at 37 °C overnight.
-
Day 2: Use a single colony to inoculate 6 mL of LB/Amp medium in a 15 mL culture tube. Grow this starter culture at 37 °C, 250 rpm overnight.
For ArcticExpress cells, separately inoculate 2 – 3 starter cultures (LB/Amp/Gen, Gen selects for the chaperonin-expressing plasmid).
-
Day 3: Spin down starter culture (2,500 g, 10 minutes, 4°C), resuspend the cell pellets in 1 – 2 L of fresh medium in 4 L flasks supplemented with Amp and 0.2% (w/v) glucose (from sterile 40% w/v stock).
For BL21 (DE3) cells, use LB medium. Grow the culture at 37 °C, 250 rpm until the OD600nm reaches 0.6 – 0.8. Add 1 mM IPTG to induce expression at 18 °C overnight (16 hours).
For ArcticExpress cells, use TB (terrific broth) and baffled flasks. Grow the culture at 37 °C, 250 rpm until the OD600nm reaches 0.4 – 0.6. Then change the temperature to 30 °C until the OD600nm reaches 1.5 – 2. Transfer the flasks to ice water. After 30 min, add 1 mM IPTG to induce expression at 10 °C, 250 rpm for 24 hr.
Day 4: Pellet the cells by centrifugation in 1 L bottles at 1,600 g for 20 min. Resuspend in 30 mL of Lysis buffer per liter of culture (supplemented with PI). Cells may be stored at -80 °C.
4.1.3 Purification of individual proteins by amylose beads
Perform all purification steps at 4 °C. Thaw the frozen cell suspension in tap water for ∼ 10 min. Add PMSF to 1 mM and immediately lyse the cells by sonication (on ice water) or extrusion (we use an Avestin EmulsiFlex-C5 microfluidizer). Clarify the lysed cells by centrifugation at 48,000 g for 30 min at 4 °C. Add the supernatant to 10 mL of amylose beads (New England Biolabs) in an Econo-Column® glass column (2.5 × 10 cm, Bio-Rad) and mix by nutation for 30 min. Drain the lysate through the column, wash the beads three times with 30 mL of Lysis buffer. Elute bound proteins by resuspending the beads in 20 mL of MBP-Elution buffer, waiting 5 minutes, and collecting the eluted proteins by draining through the column. Repeat with an additional 30 mL of MBP-Elution buffer and pool the two eluates. Examine protein purity by SDS-PAGE (Fig. 2B). Measure protein concentration by OD280nm using extinction coefficients calculated by ExPASy (Gasteiger, et al., 2003). Proteins can be flash frozen and stored at -80 °C for at least two years.
4.2 Reconstitution of the WAVE/Abi/HSPC300 trimer
Most individually purified WAVE, Abi and HSPC300 proteins are soluble aggregates. When mixed, trimer assembly from these aggregates is very slow, but inclusion of 1-2% (w/v) NP-40 substantially accelerates assembly. Once the trimer has formed, high-resolution anion exchange (in some cases both cation and anion) is used to remove the detergent and un-incorporated proteins or subcomplexes. Below is a representative protocol, but modifications can be made to adapt to more difficult situations. For instance, choice of detergents or chaotropic agents may vary: although all human WAVE/Abi/HSPC300 trimers can be efficiently assembled with NP-40, a Drosophila WAVE/Abi/HSPC300 trimer could only be assembled with 1% (w/v) CHAPS or OG (Octyl-beta-glucopyranoside), but not NP-40. Thus, this protocol provides a useful template for trimer assembly, especially for proteins from other species.
4.2.1 Reconstitution of trimer
Mix purified WAVE217, Abi158 and HSPC300 in a 0.9:1:1 molar ratio. Add 2 mM DTT, PI and 1% (w/v) NP-40 (now available as Igepal® CA-630, Sigma). Incubate the mixture at 4 °C for 12-48 hours.
4.2.2 Purification of trimer
Dilute the trimer mixture with an equal volume of QA buffer. Centrifuge at 2,500 g for 10 min. Apply supernatant to an 8 mL, 10 mm wide, SOURCE 15Q (GE Healthcare) column using a chromatography system capable of linear gradients at back pressures of 2-3 MPa (e.g. ÄKTA FPLC or similar) (Fig. 2C). After loading, wash the column with 8 column volumes (CV) of QA buffer, and elute the bound proteins using a gradient of 5 – 35% QB buffer developed over 20 CV. The WAVE/Abi/HSPC300 trimer elutes as the major peak, flanked by minor peaks containing partial assemblies (Fig. 2C). In some difficult situations (i.e. WRCs containing full length WAVE or Abi), separation of trimer from partial assemblies is improved at pH 7.0 and/or requires an additional cation exchange step (SOURCE 15S, GE Healthcare). Examine trimer purity by SDS-PAGE (Fig. 2C). Measure protein concentration by OD280nm. Add glycerol to the pooled fractions to 10 % (w/v). If not immediately used, the timer can be flash frozen in liquid nitrogen and kept at -80 °C for at least 2 years.
5. Reconstitution of the Sra1/Nap1 dimer
Sra1 and Nap1 are expressed individually in insect cells, and the Sra1/Nap1 dimer is readily formed upon mixing and co-lysing the cells. A similar strategy can be used to produce complexes with PIR121, or Drosophila Sra and Nap. Variations to consider include separate vs. co-expression, and choice of insect cells.
One precaution to take when handling the Sra1/Nap1 dimer (or individual proteins, and to a lesser extent, the WRC) is that the dimer tends to denature on air/water interfaces, forming a sticky protein film. Therefore, it is important to avoid rough pipetting and foaming (i.e. that created by continued mixing with beads in the presence of air). We also routinely include 20% (w/v) glycerol and 100-200 mM NaCl during purification to lessen these problems.
5.1 Constructs for dimer proteins
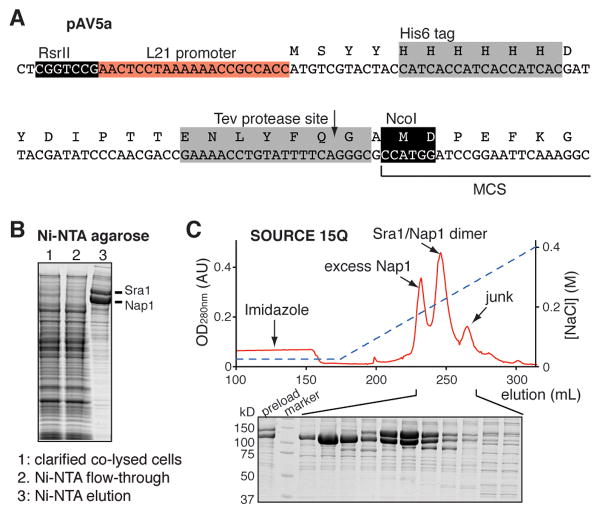
Constructs were cloned into the pAV5a vector, which is derived from pFastBacHTa (Invitrogen) modified by insertion of an L21 promoter sequence from lobster tropomyosin before the start codon (Sano, Maeda, Oki, & Maeda, 2002)(Fig. 3A).
Figure 3.
Production of the Sra1/Nap1 dimer. (A) Modification of the pFastBacHTa vector to enhance expression of dimer proteins by inserting an L21 sequence. (B) Coomassie brilliant blue stained SDS-PAGE analysis following the co-purification of His6-tagged Sra1 and His6-tagged Nap1 by Ni-NTA beads from co-lysed cells. In this case, Nap1 is in excess. (C) Chromatogram of SOURCE 15Q anion exchange purification of the Sra1/Nap1 dimer. Coomassie brilliant blue stained SDS-PAGE analysis of indicated fractions is shown below.
5.2 Insect cell maintenance and baculovirus production
We refer readers to the 2010 Invitrogen Bac-to-Bac and 2002 Invitrogen Growth and Maintenance of Insect Cell Lines manuals for details of baculovirus production and insect cell maintenance, respectively. Below we describe parameters specific to Sra1 and Nap1.
We use Sf9 cells to produce baculoviruses for both Sra1 and Nap1. To produce P1 virus, production and transfection of viral bacmids are performed according to supplier protocols. We examine the cells 72 - 96 hr post-transfection and collect the virus when cells begin to dissociate from the culture dish into the medium. P2 and P3 viruses are prepared as described in the Invitrogen manual. All viruses are stored at 4 °C before use. Instead of determining virus titers, we usually estimate the potency of virus by evaluating protein expression level and percentage of dead cells (normally aiming at ∼20%) after 72 hr of small-scale infection. As protein expression levels decline with virus age, we make new P1 viruses every 6 months to 1 year. We use Sf-900™ II SFM medium (Invitrogen) to maintain and expand the Sf9 cells as suspension culture at 27 °C, shaking at 125 rpm. We use ESF 921 medium (Expression Systems) for High Five ™ cells, which are maintained as stationary culture in T75 flasks at 27 °C in a humidified incubator, but are grown similarly to the Sf9 cells as suspension culture upon expansion for large-scale protein expression. All cultures are supplemented with the Antibiotic-Antimycotic solution (Invitrogen) at 5 ml per L.
5.3 Infection and protein expression
For the human Nap1/Sra1 heterodimer, we have found that separate expression of Sra1 in Sf9 cells and Nap1 in High Five™ cells gives the best overall yield by a significant margin over other combinations (e.g. both proteins together in either Sf9 or High Five™ cells). To express Sra1 or Nap1 proteins, culture 900 mL of desired insect cells at 27°C, 125 rpm in a 2-Liter Delong culture flask (Bellco) until they reach 2×106 cells/ml. Infect the cells with 1.8 mL of P2 or P3 baculovirus suspensions (500-fold dilution). After infection for 65–70 hr, harvest the cells by centrifugation at 1,600 g for 30 min. Resuspend the cells in 30 mL of WRC-Lysis buffer per flask of culture (supplemented with PI), transfer to freezer, and store at -80 °C until needed.
5.4 Purification of Sra1/Nap1 dimer
Estimate protein expression level for each batch of infection by coomassie brilliant blue stained SDS-PAGE. Mix cells to give roughly a 1:1 Sra1:Nap1 ratio. If the viruses are of similar age, this typically requires three parts Sra1-expressing cells with one part Nap1-expressing cells. The protocol below is written for cells from 6×0.9 L Sra1 culture mixed with 2×0.9 L Nap1 culture.
Lyse the cells by slow thawing in wet ice for 1 hr. Pool all cells and add PMSF to 1 mM. Centrifuge the co-lysed cells at 48,000 g for 45 min. Load the supernatant to 20 mL of Ni-NTA agarose beads (Qiagen) in an Econo-Column® glass column (5 × 10 cm, Bio-Rad) by gravity flow. Wash the beads with 3×60 mL of the WRC-Lysis buffer. Elute the bound proteins with 100 mL of the WRC-Ni-Elution buffer (Fig. 3B). The Ni-NTA agarose beads may be re-generated and re-used for subsequent preparations.
Dilute the eluate with an equal volume of the WRC-QA buffer. Centrifuge at 2,500 g for 10 min, and apply the supernatant to 8-mL of SOURCE 15Q packed in a 10 mm wide column using a system capable of performing linear gradients at > 3 MPa (e.g. an ÄKTA FPLC system or equivalent) (Fig. 3C). After loading, wash with 2 CV of 95% WRC-QA/5% WRC-QB buffer, and elute the bound proteins by a gradient of 5 – 35% WRC-QB buffer developed over 20 CV. The first peak is usually excess monomers (Sra1 or Nap1), and the second peak is the Sra1/Nap1 dimer (Fig. 3C). Assess dimer purity by SDS-PAGE (Fig. 3C). Measure protein concentration by OD280nm. If not immediately used, the dimer can be flash frozen and kept at -80 °C for at least 2 years.
6. Reconstitution of the WRC pentamer
The pentameric WRC spontaneously assembles from the WAVE/Abi/HSPC300 trimer and the Sra1/Nap1 dimer subcomplexes (Fig. 1B). The WAVE, Abi, and HSPC300 proteins still bear MBP fusions, allowing capture of the WRC on amylose beads and removal of unincorporated Nap1/Sra1 dimer (which is added in excess) (Fig. 4A). The WRC is further purified by one anion exchange column and a gel filtration step.
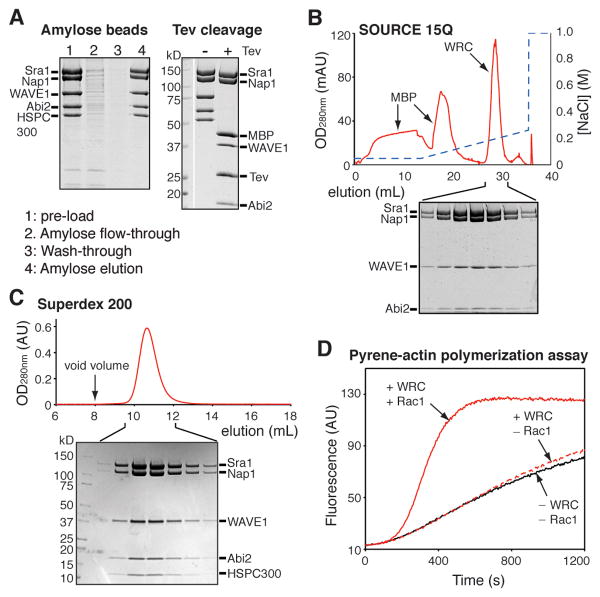
Figure 4.
Reconstitution of the WRC pentamer. (A) Purification of the reconstituted WRC (WAVE217/Abi158/HSPC300/Sra1/Nap1) by amylose beads, followed by Tev protease cleavage. (B-C) UV traces of anion exchange and size exclusion chromatography steps of the WRC purification. Coomassie brilliant blue stained SDS-PAGE analysis of indicated fractions are shown for each chromatogram. In (A-B), untagged HSPC300 (which is smaller than 10 kDa) has been run off of the bottom of both gels. (D) Results from a representative pyrene-actin polymerization assay. All reactions contain 4 μM actin (5% pyrene labeled) and 10 nM Arp2/3 complex. Reconstituted WRC (WAVE217/Abi158/HSPC300/Sra1/Nap1) was added at 100 nM concentration, along with 15 μM GMPNPN-loaded Rac1 as indicated.
6.1 Reconstitution of the WRC
Mix the WAVE/Abi/HSPC300 trimer and the Sra1/Nap1 dimer subcomplexes at a 1:1.1 molar ratio. Note that it is critical to use excess dimer to eliminate unincorporated, free trimer; as described above, the trimer has high activity and can be difficult to separate from the full WRC, and its presence even in small amounts can impart anomalous activity to WRC preparations. After incubation for 1 hr, absorb the WRC to 5 mL of amylose beads in an Econo-Column® glass column (2.5 × 10 cm, Bio-Rad) by rotary mixing for 30 min. Use of gentle agitation to keep the amylose beads suspended is acceptable at this point as the WRC is less prone to denaturation on air-water interfaces than is the Sra1/Nap1 dimer. The bound WRC is purified by gravity flow. After three washes with 15 mL of WRC-Lysis buffer, the bound WRC is eluted with 25 mL of the WRC-MBP-Elution buffer (Fig. 4A).
6.2 Tev cleavage and final purification of the WRC
The MBP and His6 tags are removed using Tev protease cleavage after the amylose bead purification. Add 2 mM DTT and 200 μL of 3 mg/mL purified Tev protease to the WRC eluted from the amylose beads, and incubate at 22 °C for 8 hr. Confirm complete cleavage by SDS-PAGE before continuing (Fig. 4A).
After Tev treatment, the untagged WRC is purified using the SOURCE 15Q anion exchange protocol described in section 5.4 (Fig. 4B). Pool fractions containing pure WRC and concentrate the complex using an Amicon Ultra-15 centrifugal filter unit, MWCO 30 kDa (Millipore) at 2,500 g, 4 °C to reduce the sample volume to 10 mL (or 1 mL if purifying less then 10 mg of protein). Remove any aggregates by centrifugation. Apply the supernatant to a HiLoad 26/600 Superdex 200 column (or a 24-mL Superdex 200 10/300 GL column if loading less than 10 mg of protein) (GE Healthcare), equilibrated with the WRC-GF buffer (for general purposes) or the WRC-100KMEI20G buffer (for actin polymerization assays) (Fig. 4C). Measure protein concentration by OD280nm. Concentrate the purified WRC if desired; at least 10 mg/mL is generally achievable without producing protein precipitation. Aliquot, flash freeze, and store the WRC at -80 °C. WRC can be stored in this fashion for at least one year. We routinely use the Arp2/3-mediated pyrene-actin assembly assay to evaluate the quality of the reconstituted WRC (B. Chen, et al., 2013; Z. Chen, et al., 2010; Ismail, et al., 2009). As shown in Figure 4D, the purified WRC should exhibit marginal basal activity and be readily activated by GMPPNP-bound Rac1 (Fig. 4D).
Acknowledgments
We thank Ayman Ismail, Zhucheng Chen, Da Jia and Junko Umetani for their contributions to this project. B.C. was supported by a fellowship from the American Heart Association. M.K.R. was supported by the NIH (R01-GM056322), the Welch Foundation (I-1544), and the Howard Hughes Medical Institute.
References
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chen Z, Brinkmann K, Pak CW, Liao Y, Shi S, et al. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell. 2013 doi: 10.1016/j.cell.2013.11.048. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468(7323):533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009;66(10):777–790. doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418(6899):790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Fricke R, Gohl C, Dharmalingam E, Grevelhorster A, Zahedi B, Harden N, et al. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE. Curr Biol. 2009;19(17):1429–1437. doi: 10.1016/j.cub.2009.07.058. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Ho HY, Li J, Steen H, Gygi SP, Kirschner MW. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci U S A. 2004;101(13):4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat Cell Biol. 2004;6(4):319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16(5):561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404(6774):151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, et al. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature. 2006;442(7104):814–817. doi: 10.1038/nature04976. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Hume PJ, Humphreys D, Liu T, Horning O, Jensen ON, et al. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc Natl Acad Sci U S A. 2011;108(35):14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36(3):512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev Biol. 2013;24(4):272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, et al. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell. 2011;41(6):661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391(6662):93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 2000;408(6813):732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, et al. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6(5):420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32(3):426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK. Physical mechanisms of signal integration by WASP family proteins. Annu Rev Biochem. 2010;79:707–735. doi: 10.1146/annurev.biochem.77.060407.135452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326(5957):1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J Cell Sci. 2009;122(Pt 15):2575–2578. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290(5492):801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, et al. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol. 2013;14(1):7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Sano K, Maeda K, Oki M, Maeda Y. Enhancement of protein expression in insect cells by a lobster tropomyosin cDNA leader sequence. FEBS Lett. 2002;532(1-2):143–146. doi: 10.1016/s0014-5793(02)03659-1. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, et al. A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci. 2007;27(2):355–365. doi: 10.1523/JNEUROSCI.3209-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovold CF, Millard TH, Machesky LM. Inclusion of Scar/WAVE3 in a similar complex to Scar/WAVE1 and 2. BMC Cell Biol. 2005;6(1):11. doi: 10.1186/1471-2121-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8(1):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, et al. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;4(2):e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD. Scar/WAVE-1, a Wiskott-Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 2000;19(17):4589–4600. doi: 10.1093/emboj/19.17.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]