SUMMARY
Non-structural protein 1 of influenza A virus, NS1A, is a conserved virulence factor comprised of an N-terminal double-stranded RNA (dsRNA)-binding domain (RBD) and a multifunctional C-terminal effector domain (ED), each of which can independently form symmetric homodimers. Here we apply 19F NMR to NS1A from influenza A/Udorn/307/1972 virus (H3N2) labeled with 5-fluorotryptophan (5-F-Trp), and demonstrate that the 19F signal of Trp187 is a sensitive, direct monitor of the ED helix-helix dimer interface. 19F relaxation dispersion data reveal the presence of conformational dynamics within this functionally important protein-protein interface, whose rate is over three orders of magnitude faster than the kinetics of ED dimerization. 19F NMR also affords direct spectroscopic evidence that Trp187, which mediates intermolecular ED:ED interactions required for cooperative dsRNA binding, is solvent exposed in full-length NS1Aat concentrations below aggregation. These results have important implications for the diverse roles of this NS1A epitope during influenza virus infection.
Keywords: 19F NMR, 5-Fluorotryptophan, Conformational dynamics, Effector domain, Influenza A virus, Non-structural protein 1
INTRODUCTION
Influenza A viruses constitute a genus of enveloped viruses from the Orthomyxoviridae family characterized by a segmented negative-stranded RNA genome encoding up to 14 proteins (Wise et al., 2012) that are either incorporated into the virion particle or expressed in the infected host cell, so-called “structural” and “non-structural” proteins, respectively (Medina and Garcia-Sastre, 2011). The multifunctional non-structural protein 1 of influenza A virus, NS1A, plays an integral role in subverting the innate antiviral response of the host and also in regulating several virus functions (Hale et al., 2008b; Krug and Garcia-Sastre, 2013). This highly conserved hub protein in influenza infection consists of a 73-residue N-terminal double-stranded RNA-binding domain (RBD) tethered by a flexible linker to an effector domain (ED) which binds to a plethora of host cellular proteins, followed by an unstructured C-terminal polypeptide segment. Several structural studies on NS1A RBD (Cheng et al., 2009; Chien et al., 1997; Liu et al., 1997) and ED (Bornholdt and Prasad, 2006; Hale et al., 2008a; Kerry et al., 2011; Xia et al., 2009) domains, as well as full-length NS1A (Bornholdt and Prasad, 2008), have established that both domains adopt independent, unique homodimer structures. The isolated RBD forms a unique six-helical head-to-tail symmetric homodimer featuring an A-form RNA-binding epitope conserved across influenza A and B viruses (Cheng et al., 2009; Yin et al., 2007). The isolated ED adopts a novel α-helix β-crescent fold and also forms a homodimeric structure (Bornholdt and Prasad, 2006; Hale et al., 2008a; Kerry et al., 2011; Xia et al., 2009).
Some controversy concerning the exact nature of the biologically relevant dimer interface of NS1A ED was dispelled by recent biophysical studies conclusively establishing that the ED dimer interface in solution encompasses the C-terminal portion of the long α-helix in each subunit (Aramini et al., 2011; Kerry et al., 2011). The focal point of this `helix-helix' dimer interface is a highly conserved tryptophan residue, Trp187. This ED dimerization epitope interacts with host targets, such as the 30-kDa subunit of cleavage and polyadenylation specificity factor (CPSF30) (Das et al., 2008). Structural studies of NS1A ED in complex with domains from CPSF30 (Das et al., 2008) and the p85β subunit of phosphoinositide 3-kinase (PI3K) (Hale et al., 2010) have also revealed that ED dimer dissociation is a prerequisite for complex formation. Independent of these host protein interactions, this same surface epitope of the ED plays an important role in the mechanism of cooperative dsRNA-binding by full-length NS1A in vitro (Aramini et al., 2011) and in vivo (Ayllon et al., 2012). This promiscuous behavior of ED facilitates the multiple functions of NS1A in a spatial and temporal fashion within the infected host cell (Kerry et al., 2011). Moreover, the ability of NS1A to bind multiple partner proteins suggests underlying conformational plasticity (Nobeli et al., 2009). All of these studies coupled with recent progress in the design of NS1A-based inhibitors (Jablonski et al., 2012) and attenuated viruses (Richt and Garcia-Sastre, 2009), underscore the growing interest in the NS1A protein as a target for the development of novel therapeutics to combat future outbreaks of potentially deadly forms of influenza A virus (Imai et al., 2012).
Since the 1960s, 19F has been recognized as a valuable NMR probe for biological systems due to its numerous favorable properties, including its nuclear spin (I = ½), high natural abundance (100%), extremely high resonance frequency and sensitivity (83% that of 1H), minimal inherent 19F background signals, and the exquisite sensitivity of its chemical shift to changes in local environment (Danielson and Falke, 1996; Gerig, 1994; Kitevski-LeBlanc and Prosser, 2012). Indeed, 19F chemical shift values for a given residue type incorporated into a folded protein can span 5 to 20 ppm (Gerig, 1994). Moreover, because of the comparable atomic radii of hydrogen and fluorine, incorporation of fluorinated amino acids into proteins generally results in relatively minor structural perturbations, although this is highly probe- and protein-dependent (Danielson and Falke, 1996; Kitevski-LeBlanc and Prosser, 2012). As a result of these traits, 19F NMR has been successfully applied to a wide variety of protein structural and dynamics studies, including probing site-specific conformational changes in proteins, protein complexes, and membrane proteins, monitoring ligand binding, and in-cell NMR studies (Didenko et al., 2013; Kitevski-LeBlanc and Prosser, 2012). In particular, the general importance of tryptophan at protein:protein interfaces combined with the relative paucity of this amino acid in proteins makes the combination of 19F NMR and incorporation of fluorinated tryptophan analogs an ideal strategy for directly probing protein interaction surfaces in complexes, including dimers (Liu et al., 2012).
Here we have applied 19F NMR to the study of NS1A from influenza A/Udorn/307/1972 virus (H3N2), Ud NS1A. Full-length Ud NS1A contains a total of four tryptophan residues, one in the RBD (Trp16) and three in the ED (Trp102, Trp187, and Trp203) (Figure 1A). We incorporated 5-fluorotryptophan (5-F-Trp), containing a fluorine substitution at the C5 (IUPAC, Cζ3) position of the in dole ring, into the RBD, ED, and a truncated form of full-length Ud NS1A lacking the C-terminal 22 residues, NS1A′, and assigned the 19F signals corresponding to the four 5-F-Trp residues by site-directed mutagenesis. We demonstrate that the 19F signal for Trp187 can be used to directly monitor the oligomerization state (i.e., monomer and dimer) of the ED. Moreover, the concentration dependence of19F line shapes and relaxation properties of the19F signal for Trp187 demonstrate conformational heterogeneity and μs-to-msmotional dynamics within the ED dimer at this important protein interaction interface. Such interfacial dynamics provide a biophysical mechanism by which the energetics of molecular recognition can be modulated by attenuating the entropy changes associated with dimerization (Huang and Montelione, 2005). Finally, in full-length NS1A, where the N-terminal RBD forms a tight dimer, 19F NMR provides direct spectroscopic evidence that intermolecular ED:ED interactions within the ca. 50 kDa full-length NS1A protein dimer do not occur in solution at sub aggregate protein concentrations (≤ 50 μM). These data support a mechanism of cooperative dsRNA binding in which the surface epitope including Trp187 becomes buried only upon formation of the functionally-important protein-protein interface between dimeric NS1A molecules (Aramini et al., 2011).
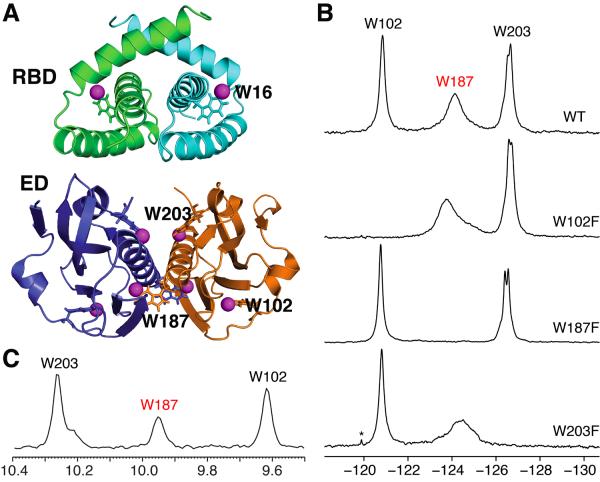
Figure 1.
(A) Locations of the 5-F-Trp residues in the dimer structures of Ud NS1A domains. Top, Ud NS1A RBD (PDB ID, 1NS1) (Chien et al., 1997). Bottom, Ud NS1A ED (PDB ID, 3EE9) (Xia et al., 2009). The fluorine atoms are shown as magenta spheres, and the tryptophan residues within one subunit of each dimer are labeled. Structures were rendered using PyMOL (PyMOL Molecular Graphics System, Version 1.4 Schrödinger, LLC). (B) 19F NMR spectra of 5-F-Trp labeled Ud NS1A ED (530 μM), and its W102F (400 μM), W187F (400 μM), and W203F (200 μM) mutants in high salt pH 8 buffer. Asterisks denote trace amounts of protease inhibitors remaining after purification. Note that the splitting observed for Trp203 under these conditions is more pronounced for the W187F mutant which is a monomer in solution (data not shown). (C) Projection of the indole amide region along the 1H dimension from an 800 MHz 2D 1H-15N TROSY-HSQC spectrum of 400 μM 15N-enriched Ud NS1A ED in pH 6.9 buffer at 27 °C (Aramini et al., 2011).
RESULTS
Effect of 5-F-Trp Incorporation on Ud NS1A ED
Using the protocols described below, we consistently obtained high levels (90%) of biosynthetic 5-F-Trp incorporation into Ud NS1A constructs (Figure S1). To examine the structural effects of 5-F-Trp incorporation, we obtained 1H-15N TROSY-HQSC spectra of 5-F-Trp labeled and un fluorinated Ud NS1A ED (Figure S2A). Aside from the expected absence of the backbone and in dole 15N-1H resonances in fluorinated ED, consistent with the high percent 5-F-Trp incorporation, the similar spectral patterns are indicative of relatively minor structural differences between the proteins. In fact, when mapped onto the crystal structure of Ud NS1A ED (Xia et al., 2009), backbone amide chemical shift perturbations (CSPs) are localized to residues in close proximity to the fluorinated tryptophans (Figure S2B).
Given the established critical role of a tryptophan (Trp187) within the helix-helix dimer interface of NS1A ED (Aramini et al., 2011; Kerry et al., 2011), we next examined the effect of 5-F-Trp labeling on the dimerization dissociation constant, Kd, of Ud NS1A ED by sedimentation equilibrium (Figure S3). The wild type and 5-F-Trp labeled NS1A ED exhibit comparable Kd values in low salt pH 8 buffer (Kd = 12 ± 6 μM and 7 ± 5 μM for wild type and 5-F-Trp NS1A ED, respectively). Taken together with the CSP data above, we conclude that incorporation of 5-F-Trp in Ud NS1A results in only minor biophysical perturbations, making this fluorinated tryptophan analog an appropriate spectroscopic reporter for this protein, particularly its biologically important dimer interface.
Assignment of 5-F-Trp Signals in Ud NS1A ED
The 19F NMR spectrum of 5-F-Trp-labeled Ud NS1A ED exhibits three distinct resonances in a chemical shift window characteristic of 5-F-Trp (δ −120 to −127 ppm), and conservative W→F single-residue mutants facilitate their assignment (Figure 1B). We observe that the 19F resonance corresponding to Trp187 located in the helix-helix dimer interface of NS1A ED (Aramini et al., 2011; Hale et al., 2008a; Xia et al., 2009) is quite broad, a sign of conformational exchange at this site (see below). A broad1H line width is also observed for the Heµ1resonance of Trp187 in native un fluorinated NS1A ED (Figure 1C). In addition, the resonance assigned to Trp203 is split into two resonances under high salt conditions, indicative of two slightly different local environments for the Trp203 side chain (Xia et al., 2009). Interestingly, alanine substitution of individual tryptophans results in small chemical shift changes to the remaining tryptophan resonances, particularly Trp187, indicative of all osteric structural perturbations (Figure S4).
19F NMR to Probe the Dimer-to-Monomer Transition in Ud NS1A ED
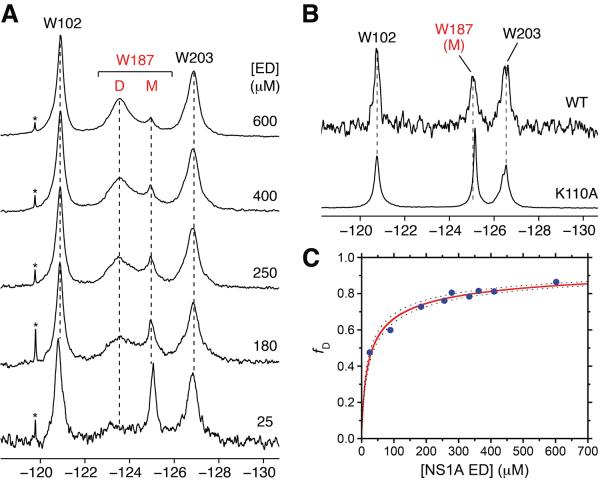
The 19F resonance of Trp187 is highly sensitive to the dimer↔monomer solution equilibrium of Ud NS1A ED. Dilution of 5-F-Trp labeled Ud NS1A ED results in a progressive decrease in the broad resonance for Trp187 and concomitant increase in a second sharper upfield resonance (Figure 2A). This upfield-shifted resonance for Trp187 is also observed for the K110A mutant of Ud NS1A ED (Figure 2B), a mutant of this protein domain that is monomeric (Aramini et al., 2011). Hence, this sharp upfield-shifted resonance corresponds to Trp187 in the exposed monomeric state. Note that the line width of the 19F resonance for Trp187 in the dimer is also significantly broader than those for Trp102 and Trp203 over the entire dilution series (see below). Fitting the concentration dependence of the dimer and monomer states of NS1A ED in slow exchange on the 19F time scale yields a dimer dissociation constant, Kd, of 35 ± 6μM (Figure 2C). This value is comparable to the sedimentation equilibrium results, albeit slightly weaker. This disparity may be attributed to the vastly different protein concentrations required by the two techniques and the poor solubility of NS1A ED at concentrations above ca. 600 μM, which may complicate the estimate of Kd. On the basis of 1D 19F saturation transfer experiments (Figure S5), we estimate exchange rates for ED monomer→dimer and dimer→monomer interconversion under these conditions of kex = 1.2 ± 0.3 and 0.8 ± 0.2 s−1, respectively; that is, with a time constant of 1 s. These rates are, as expected, well below the frequency difference between the Trp187 dimer and monomer 19F resonances (Δν 650 Hz at B0 = 11.7 T). Interestingly, the Kd for the ED homo dimer is highly salt dependent, in that increasing salt significantly weakens the dimerization, resulting in intermediate/fast exchange behavior on the 19F time scale and weaker Kd values by sedimentation equilibrium (Figure S6). This is consistent with the presence of intermolecular salt bridges at the dimer interface in the structure of Ud NS1A ED (e.g., Lys110:Asp189′) (Xia et al., 2009).
Figure 2.
19F NMR is a direct probe of Ud NS1A ED homodimerization. (A) Concentration dependence of the 19F NMR signal of Trp187 within 5-F-Trp labeled Ud NS1A ED in low salt pH 8 buffer. Resonances corresponding to Trp187 in the dimer and monomer states are denoted by `D' and `M', respectively. Asterisks denote trace amounts of residual protease inhibitors after purification. (B) 19F NMR spectra of 5-F-Trp labeled monomeric 25 μM Ud NS1A ED (top) and 500 μM [K110A] NS1A ED (bottom), a known monomeric effector domain mutant (Aramini et al., 2011), in high salt pH 8 buffer. (C) Fit of the fraction of 19F dimer resonance volume as a function of total Ud NS1A ED concentration. Dashed lines represent 95% confidence bounds for the fit.
19F Relaxation to Probe Dynamics at the Dimer Interface of Ud NS1A ED
The slow exchange between the broad dimer and narrow monomer 19F resonances of Trp187 observed under low salt conditions is strongly indicative of conformational exchange within the ED dimer interface, rather than exchange between bound dimer and free monomer states. The line shapes for such a slow/intermediate exchange between monomer and dimer would feature an identical line width for both the bound- and free-state Trp187 19F resonances when the populations are equal, and different line widths throughout the dilution experiment correlated with the fraction of monomer and dimer (Palmer et al., 2001). In contrast, the presence of both broad bound (due to exchange broadening within the dimer) and narrow free (characteristic of the environment in the monomer state) Trp187 19F resonances throughout the entire dilution series (Fig. 2A) is attributed to conformational heterogeneity at the helix-helix interface of the dimer complex.
To test this hypothesis and gain further insights into the conformational dynamics with in the NS1A ED dimer interface, 1D 19F T1, T2, and CPMG relaxation dispersion experiments were performed on 5-F-Trp labeled NS1A ED, at high (dimer only) and intermediate (dimer plus monomer) concentrations, and its monomeric K110A mutant (Table 1; Figure 3A). While T1 values for the various 5-F-Trp residues are relatively insensitive to ED oligomerization state, the 19F T2 of Trp187 is significantly shorter for the dimer resonance compared to the monomer resonance (Table 1), and shorter than the 19F T2 of residues Trp102 and Trp203 in the dimer state. This is consistent with conformational exchange within the interface of the ED dimer, rather than exchange broadening due to inter conversion between bound and free forms (see below), or a simple effect due to the slower overall rotational correlation time of the dimer. For the monomeric mutant, the 19F T2 of Trp187is even longer, indicative off ullsolvent and rotameric accessibility. In addition, 19F CPMG relaxation dispersion results for wild type and [K110A] NS1A ED dramatically demonstrate that the value of R2 for Trp187 in the dimer interface is extremely sensitive to the delay time τcp, in contrast to the transverse relaxation behavior of Trp187 in monomeric ED as well as the other two tryptophan residues, Trp102 and Trp203, in this domain (Figure 3A).
Table 1.
19F T1 and T2 Relaxation Data for 5-F-Trp Residues in Ud NS1A ED, [K110A] NS1A ED, and [C116S,G183C] NS1A EDa
| Sample | Peak | T1 (s) | T2 (ms) |
|---|---|---|---|
| NS1A ED (600 μM) | W102 | 0.58 (0.06) | 4.51 (0.41) |
| W187 dimer | 0.63 (0.06) | 3.34 (0.16) | |
| W203 | 0.61 (0.10) | 5.44 (0.59) | |
| NS1A ED (250 μM) | W102 | 0.49 (0.07) | 5.67 (1.09) |
| W187 dimer | 0.75 (0.26) | 4.0 (1.2) | |
| W187 monomer | 0.71 (0.11) | 11.76 (3.02) | |
| W203 | 0.52 (0.06) | 5.86 (1.29) | |
| [K110A] NS1A ED (200 μM) | W102 | 0.35 (0.04) | 9.83 (2.23) |
| W187 monomer | 0.65 (0.14) | 27.06(5.89) | |
| W203 | 0.42(0.05) | 10.71 (2.14) | |
| [C116S,G183C] NS1A ED (200 μM) | W102 | 0.62 (0.05) | 3.16 (0.32) |
| W187 | 0.65 (0.06) | 4.42 (0.56) | |
| W203 | 0.68 (0.07) | 3.84 (0.55) |
19F relaxation datashown were obtainedat 470.18 MHz in low salt pH 8 bufferand at 20 °C. All T2 data shown correspond to a νCPMG of 2,500 Hz. Standard errors representing 95% confidence bounds for the exponential fits are given in parentheses.
Figure 3.
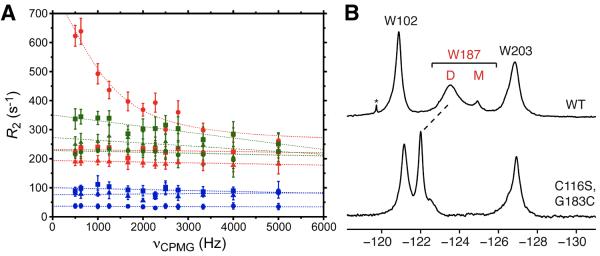
19F NMR CPMG relaxation dispersion data for Trp102 (squares), Trp187 (circles), and Trp203 (triangles) in 5-F-Trp labeled 600 μM Ud NS1A ED (red), 200 μM [K110A] NS1A ED (blue), and 500 μM[C116S,G183C] NS1A ED (disulfide form; no DTT) (green) in low salt pH 8 buffer at 20 °C. (B) 19F NMR spectra of 5-F-Trp labeled Ud NS1A ED (top) and disulfide-bonded 500 μM [C116S,G183C] NS1A ED (bottom), in low salt pH 8 buffer. The downfield shift of the 19F resonance for Trp187 is indicated by a dashed line. The native Cys116 was mutated to a serine in order to prevent a potential mixture of disulfide adducts.
In order to further test this model, we generated and studied a disulfide cross-linked ED dimer that locks the NS1A ED in adimeric form. By mutating Gly183 to a cysteine, we generated a disulfide-bonded dimer that was confirmed by polyacrylamide gel electrophoresis (Figure S7). The locked dimer exhibits a significant reduction in Trp18719F resonance line width, with concomitant down field shift compared to wild type NS1A ED (Figure 3B). Locking down the interfacial dynamics of the NS1A dimer also abolishes the 19F relaxation dispersion effect observed for Trp187 in the wild type protein (Figure 3A). Taken together, these data conclusively demonstrate the presence of slow motion (micro- to millisecond) conformational exchange within the ED helix-helix dimer interface.
Acquiring 19F relaxation dispersion data on the 5-F-Trp labeled NS1A ED dimer at a second magnetic field strength affords a quantitative analysis of the conformational dynamics at the ED dimer interface (Figure 4). As expected, increasing magnetic field leads to an increase in 19F transverse relaxation rate for each of the 5-F-Trp residues in the protein, but again only Trp187 exhibits a relaxation dispersion effect. Fitting the 19F relaxation dispersion curves for Trp187 to a general two-state exchange model (Carver and Richards, 1972) yields a conformational exchange rate, kex, of 6430 ± 1180 s−1. It is important to note that this conformational exchange regime for the ED dimer is over three orders of magnitude faster than the kinetics of dimer↔monomer interconversion determined above, revealing that the latter process has a negligible contribution to the observed line width of the 19F resonance for Trp187.
Figure 4.
Field dependent 19F CPMG relaxation dispersion data for Trp102 (squares), Trp187 (circles), and Trp203 (triangles) of 5-F-Trp labeled Ud NS1A ED dimer (600 μM) in low salt pH 8 buffer. Transverse relaxation rates (R2) as a function of νCPMG acquired at magnetic field strengths of 11.7 T (470.18 MHz) and 14.1 T (564.62 MHz) are shown in red and cyan, respectively. The relaxation dispersion curves for Trp187 at both fields were fit to a two-state exchange model (Carver and Richards, 1972),yielding the following results: kex = 6430 ± 1180 s−1; R20 (470.18 MHz) = 269 ± 18 s−1; R20 (564.62 MHz) = 328 ± 16 s−1; α = 2.3 ± 0.6.
19F NMR of Ud NS1A′
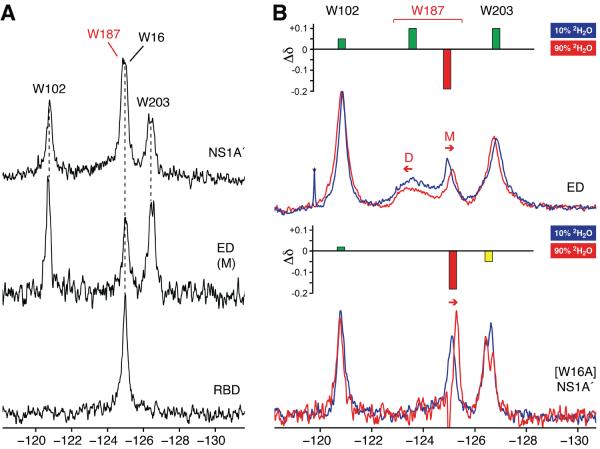
19F NMR was also used to directly probe Trp187 in the ca. 50 kDa Ud NS1A′dimer (Figure 5A). An analogous upfield, solvent-exposed signal for Trp187 is observed in NS1A′ at concentrations up to ca. 50 μM. This signal is degenerate with that of Trp16 in the RBD. In addition, the 19F resonances for both Trp102 and Trp203 in NS1A′ correlate with those in the isolated ED. At higher protein concentrations, significant protein oligomerization via the helix-helix ED interface is observed (Aramini et al., 2011), resulting in protein precipitation. Hence, 19F NMR provides direct spectroscopic evidence that full-length NS1A dimerizes at low concentrations via its RBD, with the surface epitope encompassing Trp187 fully exposed and not involved in either intradimeric or oligomeric ED:ED interactions. These ED:ED interactions occur only between dimers at higher concentrations, and can provide a mechanism for the cooperative binding of full-length NS1A molecules to large dsRNA molecules (Aramini et al., 2011).
Figure 5.
(A) 19F NMR spectra of 5-F-Trp labeled Ud NS1A′ (50 μM), monomeric ED (25 μM), and RBD (200 μM) in high salt pH 8 buffer. (B) Overlay of 19F NMR spectra of 5-F-Trp labeled Ud NS1A ED (top) and [W16A] NS1A′ (bottom) in 10% (blue) and 90% (red)2H2O. Sample conditions: i) 150μM (10% 2H2O) and 300 μM (90% 2H2O) NS1A ED in low salt pH 8 buffer; ii) 35 μM [W16A] NS1A′ in high salt pH 8 buffer. Insets above both spectral overlays depict the change in 19F chemical shift (Δδ) and degree of solvent exposure for each 5-F-Trp colored as follows: green, buried, red, exposed, and yellow, partially exposed. Resonances corresponding to Trp187 in the dimer and monomer states of NS1A ED are denoted by `D' and `M', respectively.
Solvent Exposure of Tryptophans in Ud NS1A from 19F NMR
It is well established that solvent exposure of fluorinated residues can be directly correlated to solvent induced isotope shifts in19F NMR resonances (Kitevski-LeBlanc and Prosser, 2012). For example, free 5-F-Trp in solution exhibits a δ −0.2 ppm upfield shift when the solvent is changed from H2O to 2H2O (Kitevski-LeBlanc and Prosser, 2012; Rule et al., 1987). At intermediate NS1A ED concentrations, where both dimer and monomer are present, exchange into highly deuterated solvent results in a significant upfield shift in the monomer Trp187 signal due to the deuterium isotope effect (Figure 5B; Table 2). In contrast, the Trp187 dimer peak exhibits a small down field shift. These results are consistent with a change in the local environment of the Trp187 side chain from a buried environment in the dimer to a solvent-exposed state in the monomer. The solvent-induced isotope shift data also indicate that Trp102 and Trp203 are fully or partially buried, consistent with the 3D structure of Ud NS1A ED (Aramini et al., 2011; Xia et al., 2009). We observe similar trends for analogous experiments on NS1A ED at high salt (Figure S8). However, the interpretation of solvent induced isotope chemical shift changes under these conditions is complicated by multiple factors; namely, both the monomer/dimer equilibrium, which may be perturbed by changing the solvent, and the deuterium isotope shift itself. In addition, the 19F resonance for Trp187 in the disulfide-bonded [C116S, G183C] NS1A ED dimer exhibits no solvent isotope shift (Table 2), confirming that its side chain is indeed buried within the helix-helix interface of this “locked” dimer.
Table 2.
Solvent Induced Isotope Shifts of 5-F-Trp Residues in Ud NS1A ED, [C116S,G183C] NS1A ED, and [W16A] NS1A′a
| Sample | Peak | δ19F (ppm) 10% 2H2O | δ19F(ppm) 90% 2H2O | Δδ (ppm) | Solvent Exposure |
|---|---|---|---|---|---|
| NS1A ED | W102 | −120.86 | −120.81 | +0.05 | buried |
| W187 monomer | −124.94 | −125.13 | −0.19 | exposed | |
| W187 dimer | −123.6 | −123.5 | +0.1 | buried | |
| W203 | −126.84 | −126.74 | +0.10 | buried | |
| [C116S,G183] NS1A ED | W102 | −121.16 | −121.11 | +0.05 | buried |
| W187 | −121.99 | −122.01 | −0.02 | buried | |
| W203 | −126.92 | −126.93 | −0.01 | buried | |
| [W16A] NS1A′ | W102 | −120.78 | −120.76 | +0.02 | buried |
| W187 | −125.11 | −125.29 | −0.18 | exposed | |
| W203b | −126.50 | −126.55 | −0.05 | partially buried |
Data shown were obtained on the following samples: i) NS1A ED (150 μM in 10% 2H2O; 300μM in 90% 2H2O) in low salt pH 8 buffer; ii) [C116S,G183C] NS1A ED (500 μM in 10% 2H2O; 300μM in 90% 2H2O) in low salt pH 8 buffer;iii) [W16A] NS1A′, 35 μM protein in high salt pH 8 buffer.
Chemical shift values reported for W203 are an average of the overlapped signals observed under these buffer conditions
In the case of the NS1A′ dimer, to avoid confusion due to Trp16 and Trp187 19F resonance overlap we examined the solvent-induced isotope shift effects on a [W16A] NS1A′ mutant. The Trp187 resonance in [W16A] NS1A′exhibits a similar upfield solvent induced isotope shift (Figure 5B; Table 2). These experiments confirm that the upfield shifted Trp187 signals for dilute NS1A ED and the ca. 50 kDa NS1A′dimer correlate with Trp187 in a fully solvent exposed state.
DISCUSSION
In this study we employed 5-F-Trp incorporation and 19F NMR spectroscopy to obtain unique insights into the structure and dynamics of the influenza A virus NS1 (NS1A) protein and its isolated domains. Our results provide direct spectroscopic evidence in solution for the presence of dynamic conformational exchange within the biologically important ED dimer interface, and that this same interface is exposed in both the isolated ED monomer and the full-length protein at concentrations below aggregation. These conclusions have important ramifications for the behavior of this multifunctional protein in influenza A viral infection, as discussed below. Such interfacial dynamics can modulate the energetics of molecular recognition processes (Huang and Montelione, 2005), and are probably more common than is generally believed, particularly for proteins like NS1A which utilize epitope plasticity to allow for promiscuous interactions with multiple protein partners.
The presence of slow exchange between the broad dimer and narrow monomer Trp187 19F resonances under low salt conditions as a function of NS1A ED concentration, along with the dramatic relaxation dispersion properties of the Trp187 dimer 19F signal, reveals that the ED dimer undergoes exchange between two or more conformations in solution in a micro- to millisecond time window. The lack of slow motion dynamics for Trp102 and Trp203 in the ED dimer indicates that such fluctuations are confined to the helix-helix interface. Moreover, this slow motion can be arrested by locking the ED in a dimeric form with an engineered disulfide linkage afforded by mutation of Gly183, which is in close proximity to its symmetry related residue (Gly183′) within the helix-helix interface of the Ud NS1A ED dimer structure (Xia et al., 2009).
Conformational heterogeneity within the ED dimer interface in solution, demonstrated here, is consistent with a recent analysis of several influenza A ED dimer crystal structures in the literature, where it was recognized that the ED protomers adopt a range of relative orientations across various influenza A virus strains and crystal forms (Kerry et al., 2011). Assuming that these crystal structures sample the energy landscape of this critical protein-protein interaction, we propose that our19F solution NMR data may be attributed to a large amplitude rocking motion at the ED dimer interface within a range of interdomain angles consistent with the “snapshots” observed by X-ray crystallography (Figure 6) (Kerry et al., 2011; Xia et al., 2009). There may also be additional contributions to the interfacial dynamics due to multiple side chain conformations of the Trp187 residues that are buried in the interface. However, our 19F relaxation data for the Ud NS1A ED dimer alone do not provide a precise atomic level description of the interconverting conformational sub-states at the helix-helix interface.
Figure 6.

Superimposed crystal structures of NS1A ED showing “snapshots” of the conformational heterogeneity of the helix:helix interface. All dimer structures were oriented to optimally-superimpose the indole ring atoms of Trp187 (shown in grey). (A) View showing the range of movements of Trp187′ in the second protomer as well as the long helix (residues 171 to 188) in both protomers (α1 and α1′). Crystallographic dimers are colored based on the relative angle between Trp187 and Trp187′ indole rings, from smallest (yellow) to largest (blue). (B) Close-up view of the environment that would be sensed by a fluorine atom at the C5 position of Trp187 (magenta sphere). Residues in the second protomer that are consistently within 5 Å of the fluorine position in Trp187 of the first protomer are labeled. Structure coordinates correspond to selected crystallographic dimer units from the NS1A ED literature coordinates for the following influenza A strains and crystal forms: A/Puerto Rico/8/1934 (H1N1), PDB ID, 2GX9 (Bornholdt and Prasad, 2006); A/Udorn/307/1972 (H3N2), PDB IDs, 3EE8 and 3EE9 (Xia et al., 2009); A/California/07/2009 (H1N1), PDB ID, 3M5R; A/Reassortant/IVR108 (Sydney/5/1995 × Puerto Rico/8/1934) (H3N2), PDB ID, 3O9U (Kerry et al., 2011). Structures were rendered using PyMOL.
The 19F relaxation results presented in this work also illustrate the advantage of employing 19F NMR and a strategically placed fluorine probe in the second ring of the indole moiety to directly monitor conformational exchange phenomena at protein:protein interfaces mediated by tryptophan residues compared to, for example, the indole Hε1 resonance, a signal we exploited in earlier studies of the Ud NS1A ED dimer interface (Aramini et al., 2011). In spite of evidence for some exchange broadening of the Trp187 side chain Hε1 resonance compared to the analogous resonances for other tryptophan residues in NS1A ED (Figure 1C), this 1H resonance features a much narrower chemical shift dispersion and a different local environment, rendering it less sensitive to conformational exchange phenomena compared to the fluorinated tryptophan strategy. In the context of the broader 19F protein NMR literature, these dynamical results on the NS1A ED dimer interface constitute a novel application of19F NMR relaxation to directly probe slow motion dynamics within a protein:protein interface The NMR properties of 19F have, have been exploited recently to illuminate slow motion conformational dynamic processes in other types of interactions including protein-ligand interactions (Ahmed et al., 2007), peptide-bicelleinteractions (Buer et al., 2010), changes in dynamics accompanying protein complex formation (Acchione et al., 2012), and the thermodynamic and kinetic aspects of domain swapping (Liu et al., 2012). In addition, 19F CPMG relaxation approaches were recently employed to characterize the conformational exchange behavior of fluorinated Phe residues in the thermal folding intermediate of calmodulin (Kitevski-Leblanc et al., 2013).
The solvent exposure of Trp187 within the ca. 50 kDa NS1A′ dimer is consistent with recent advances in our understanding of the multifunctional behavior of this influenza protein (Aramini et al., 2011; Ayllon et al., 2012; Kerry et al., 2011). The same epitope that mediates ED dimerization also plays a critical role in binding to host proteins such as CPSF30(Das et al., 2008). Moreover, sequestering of dsRNA by NS1A occurs in a cooperative fashion mediated by intermolecular ED:ED interactions leading to oligomerization (Aramini et al., 2011). Mutating the tryptophan at this interface perturbs interferon antagonism by the virus (Ayllon et al., 2012). Recent structural, in vitro, and in vivo studies led to the hypothesis that spatial and temporal regulation of NS1A activity is governed by “helix-open” (i.e., Trp187 exposed) and “helix-closed” (i.e., Trp187 buried) states of NS1A, in which the RBD always adopts a tight dimeric structure while the ED can explorevarious quaternary states (Kerry et al., 2011). Hence, plasticity at this ED:ED epitope facilitates the multiple interactions of NS1A, namely the ability to both mediate NS1A oligomerization during cooperative dsRNA binding and also for specific-interactions with various host proteins.
Finally, the potential correlation between the dynamical properties of the ED dimer interface in NS1A and the multiple functions of this viral protein, proposed here and by others (Kerry et al., 2011), is relevant to our broader understanding of molecular recognition and function in biology. Over the last decade, NMR spectroscopy has played a central role in a burgeoning body of literature that establishes the delicate balance between dynamics, thermodynamics (entropy), and macromolecular activity and binding affinity (Huang and Montelione, 2005; Mittag et al., 2010; Mittermaier and Kay, 2009; Tzeng and Kalodimos, 2011; Wand, 2013). A central theme within this field is the concept that dynamics and entropy underpin conformational selection, where proteins are thought to be primed for activity by adopting a range of pre-existing interconverting conformational sub-states (Boehr et al., 2009) in protein-ligand, protein-protein, and protein-nucleic acid interactions, largely dispelling the historical view of protein structure as being “static”. This has broad implications in many biological processes, ranging from catalysis (Henzler-Wildman and Kern, 2007), to cellular signaling (Smock and Gierasch, 2009), and nucleic acid recognition (Mackereth and Sattler, 2012). Intrinsic internal dynamics and conformational entropy in proteinsand protein-protein interfaces can modulate binding affinities and binding modes (Huang and Montelione, 2005), particularly in systems featuring promiscuous interactions (Nobeli et al., 2009; Perkins et al., 2010; Schreiber and Keating, 2011). In fact, recent elegant methyl relaxation studies revealed that these phenomena are intimately correlated with modulation of molecular recognition by calmodulin (Marlow et al., 2010) and the catabolite activator protein, CAP (Tzeng and Kalodimos, 2012). In our work, we observe exchange broadening on a slower (microsecond) timescale specifically at a protein-protein interface. Therefore, conformational plasticity and residual entropy at the NS1A ED dimer interface, as well as in the flexible and biologically important interdomain linker (Bornholdt and Prasad, 2008; Kerry et al., 2011; Li et al., 2011), may be critical factors that nature exploits to modulate partner binding affinities and allow NS1A to access the variety of structural states required for it to effectively carry out its multiple functions in the host cell.
In summary, we have demonstrated the value of applying 19F NMR to directly probe the oligomerization state and conformational dynamics of the NS1A protein of influenza A virus by monitoring a fluorinated analog of the critical Trp187 at the helix:helix intermolecular interface of the effector domain. This work lays the foundation for future 19F NMR studies of the interactions of influenza NS1A with dsRNA (Aramini et al., 2011; Cheng et al., 2009; Wang et al., 1999), host target proteins, such as the F2F3 tandem zinc finger domains of CPSF30 (Das et al., 2008; Twu et al., 2006), and molecular fragment libraries.
EXPERIMENTAL PROCEDURES
Cloning, Expression, Purification, and Sample Preparation
The following three constructs of influenza A/Udorn/307/1972 (H3N2) NS1A were cloned, expressed, and purified following previous protocols (Acton et al., 2011; Aramini et al., 2011) with minor modifications: NS1A(1–15), NS1A(1–73), and NS1A(85–215), hereafter referred to as Ud NS1A′, RBD, and ED, respectively. Because the C-terminal 22 residues of full-length Ud NS1A are unstructured and lead to insolubility, these residues were not included in our constructs. The three different NS1A constructs were expressed with the following affinity purification tags: i) both the 215-residue NS1A′ and the 73-residue RBD were fused to an N-terminal 6xHis tagged SUMO protein, and ii) the ED was followed by a C-terminal 6xHis tag. All single- and double-residue mutants were made using the Quik Change site-directed mutagenesis kit (Stratagene) along with the appropriate primers (Table S1), and verified by sequencing. The SUMO fusion proteins were cloned into the pSUMO vector (LifeSensors) modified to be ampicillin resistant, whereas the ED constructs were cloned into the pET21_NESG vector (Acton et al., 2011). Protein expressions were carried out in Escherichia coli BL21(DE3)-Gold (Agilent) cells containing the rare tRNA codon-enhanced pMgK plasmid and IPTG-inducible T7 polymerase. Cultures were grown in 2 L baffled flasks at 37 °C in MJ9 minimal medium (Jansson et al., 1996) supplemented with ampicillin and kanamycin. For the production of 15N-labeled proteins 15NH4SO4 (Cambridge Isotope Laboratories) was used as the sole nitrogen source in the MJ9 medium. For 5-F-Trp incorporation, cultures were grown until an A600 of ≈ 0.5 units, cooled on ice with addition of 50 mg/L 5-fluoro-DL-tryptophan (Sigma), followed by incubation at 17 °C for 1 h prior to induction with 1 mM IPTG. Protein expression was carried out overnight at 17 °C with shaking at 225 rpm. Cultures were subsequently centrifuged and decanted, and pellets were stored at −80 °C until purification.
All expressed NS1A protein constructs were purified by immobilized metal ion affinity chromatography (IMAC) followed by size-exclusion chromatography in the final buffer for NMR spectroscopy. Pellets were resuspended in 25 mL of nickel affinity column Binding Buffer [50 mM Tris-HCl pH 7.5, 500 mM NaCl, 40 mM imidazole, 1 mM tris(2-carboxyethyl)phosphine, 0.02% NaN3], followed by sonication, and clarification by centrifugation at 26,000 g for 45 min. After clarification, proteins were purified using an ÄKTAxpress™ system (GE Healthcare) equipped with a 5 mL HisTrap HP affinity column. In the case of NS1A ED and its mutants, samples were immediately loaded onto a HiLoad 26/60 Superdex 75 gel filtration column equilibrated in pH 8 buffer [50 mM Tris-HCl pH 8.0, 30 or 300 mM NaCl, 2.5% (v/v) glycerol, 10 mM DTT]. For the SUMO fusions of NS1A′ and RBD, an aliquot (1:50–100 mass ratio) of yeast SUMO protease Ulp1 containing an N-terminal 6xHis tag expressed and purified in-house (Ii et al., 2007) was added, and the sample was incubated at 4 °C overnight (Panavas et al., 2009). Complete SUMO cleavage was confirmed by SDS-PAGE. Cleaved NS1A′ was then purified using a HiLoad 26/60 Superdex 200 gel filtration column equilibrated in pH 8 buffer. Due to its comparable size and resulting similar retention time, free NS1A RBD could not be separated from the cleaved SUMO tag using size-exclusion chromatography. Consequently, the sample was buffer exchanged with a HiPrep 26/10 desalting column into Rebinding Buffer [50 mM Tris-HCl pH 7.5, 500 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine, 0.02% NaN3], and passed over a 5 mL HisTrap HP affinity column to remove cleaved SUMO tag and SUMO protease. The flow-through containing RBD was finally purified using a HiLoad 26/60 Superdex 75 gel filtration column equilibrated in pH 8 buffer. The purities of all NS1A proteins used in this study were confirmed by SDS-PAGE and MALDI-TOF mass spectrometry. Unless otherwise indicated, samples of 5-F-Trp labeled NS1A constructs for NMR and analytical ultracentrifugation were prepared in low (30 mM NaCl) or high (300 mM NaCl) salt pH 8 buffer containing 10% (v/v) 2H2O and concentrated by ultrafiltration (Amicon, Millipore). The buffer conditions were carefully optimized in order to promote dimerization of the isolated ED (low salt) or enhance the solubility of full-length NS1A (high salt). Samples for deuterium isotope effect NMR experiments were prepared by performing three rounds of 1:10 dilution with pH 8 buffer in 90% 2H2O followed by concentration by ultrafiltration, resulting in a final 2H2O concentration of ≈ 90% (v/v).
Efficiency of 5-F-Trp Incorporation
To assess the efficiency of 5-F-Trp incorporation, aliquots of NS1A ED expressed in various labeling media were diluted in 100 mM Tris-HCl pH 8.0, 10 mM CaCl2, 8 M urea, 10 mM DTT and denatured at 60 °C for 25 min. Proteolysis was carried out by adding 0.1 μg chymotrypsin (Sigma), and incubating the mixture at 37 °C for 2 h. Proteolytic digestion was arrested by addition of 2 μL 5% (v/v) trifluoroacetic acid (TFA), and complete digestion was confirmed by SDS-PAGE. The resulting peptide digest was mixed (1:100) with 10 mg/mL - Cyano-4-hydroxycinnamic acid (CHCA) in 50% acetonitrile, 1% TFA, and 1 μL was plated for reflected MALDI TOF/TOF on an ABI-MDS SCIEX 4800 mass spectrometer. Mass spectrometry spectral data were analyzed using Data Explorer software (Applied Biosystems). Fluorinated peptides were selected using the m/z protease digest prediction tool Protein Prospector (Chalkley et al., 2005) and confirmed with tandem (MS/MS) mass spectrometry. In this study, we consistently observed high levels (≥ 90%) of biosynthetic 5-F-Trp incorporation, hence precluding the use of other approaches (which we also tested; not shown), including the addition of glyphosate to arrest endogenous bacterial expression of aromatic amino acids (Kim et al., 1990) or condensed single protein production (cSPP) technology (Schneider et al., 2009).
Analytical Ultracentrifugation
Sedimentation equilibrium (SEQ) analytical ultracentrifugation experiments were performed at 20 °C using a Beckman XL-I instrument with an AnTi60 rotor. The sample chambers used Epon double sector centerpieces with sapphire windows. Samples of wild type and 5-F-Trp Ud NS1A ED (each 175 μL containing 15 μM total protein) were prepared in low salt pH 8 NMR buffer [50 mM Tris-HCl pH 8.0, 30 mM NaCl, 2.5% (v/v) glycerol, 10 mM DTT, 10% (v/v) 2H2O]. Sedimentation equilibrium scans were recorded after 72 h at three rotor speeds: 15,000, 25,000, and 31,000 rpm. The protein partial specific volume, solvent density and solvent viscosity were calculated using Sednterp 1.09 (Laue et al., 1992). Data analysis was performed using Sedphat 10.40 (Schuck, 2003; Vistica et al., 2004). For each protein, the sedimentation equilibrium profiles were globally fitted to a monomer-dimer equilibrium model using mass conservation and rotor stretch restraints. An F-statistics error mapping approach (Johnson and Straume, 1994) was used to determine the 95% confidence intervals for the homodimerization dissociation constant.
NMR Spectroscopy
One-dimensional (1D) 19F NMR spectroscopy was performed locked and at 20 °C on a Varian INOVA 500 MHz spectrometer equipped with a room temperature 5-mm 1H/19F switchable probe at a frequency of 470.18 MHz. All 19F NMR spectra were acquired using VNMRJ 2.1B and referenced to external neat trichlorofluoromethane, CFCl3. Typical 1D 19F NMR acquisition parameters were as follows: a 20,000 Hz sweep width (42.5 ppm), a 0.35 s acquisition time, a 5 s relaxation delay time, and a 5.0 μs 90° pulse length. 1D 19F NMR spectra were processed with 20 Hz exponential line broadening and displayed using matNMR 3 (van Beek, 2007). Two-dimensional (2D) 1H-15N TROSY-HSQC spectra were acquired locked and at 25 °C on a Bruker AVANCE 800 MHz NMR spectrometer equipped with a 5-mm TXI cryoprobe, referenced to internal 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS), processed with NMRPipe (Delaglio et al., 1995), and displayed using Sparky 3 (Goddard and Kneller, 2006).
Composite 1H and 15N backbone amide chemical shift perturbations, Δδcomp, were computed from assigned 1H-15N TROSY-HSQC spectra of 15N-enriched wild type (Aramini et al., 2011) and 5-F-Trp NS1A ED acquired under identical conditions using Eq. 1 (Farmer et al., 1996):
| (1) |
To obtain the dissociation constant for homodimerization, Kd, of NS1A ED by 19F NMR, values of the fraction of dimer, fD, at each ED concentration were obtained from the volumes of Trp187 dimer and monomer 19F signals determined by Lorentzian line fitting using MATLAB 7.12.0 (MathWorks). Assuming a two-state dimer-to-monomer equilibrium, the Kd is related to fD, as well as the fraction of monomer, fM, and total protein concentration, PT, according to Eq. 2 (Byeon et al., 2003).
| (2) |
Expressing this relationship in terms of fD as a function of PT and Kd yields a quadratic expression Eq. 3, which we employed to fit the concentration dependence of fD to compute Kd by non-linear least squares fitting using MATLAB 7.12.0.
| (3) |
To determine the exchange rate of interconversion between NS1A ED monomer and dimer, a series of 1D 19F saturation transfer experiments were recorded on mixed monomer/dimer samples, with selective irradiation of the monomer or dimer 19F signals using a train of 50 ms Gaussian pulses each separated by a 1 ms delay (Mayer and Meyer, 2001), total saturation times ranging from 0.05 to 2.0 s, and a relaxation delay of 7 s. In the limit of reversible chemical exchange of a nucleus, X, between two non-equivalent sites, A and B, selective saturation of resonance X in site B results in adecay of magnetization of X in site A, MA, as a function of saturation time, t, according to Eq. 4 (Forsén and Hoffman, 1963), where M0A and M∞A are MA at t=0 and t→∞, respectively, and λA is related to the exchange rate, kA, and transverse relaxation rate, R1A, by Eq. 5.
| (4) |
| (5) |
Hence, the exchange rate for a two-site slow exchange process can be calculated from the linear slope in a plot of ln(MA − M∞A) against t.
Longitudinal (T1) and transverse (T2) 19F relaxation time measurements were determined using classic 1D inversion recovery and Carr-Purcell-Meiboom-Gill (CPMG) spin-echo pulse sequences, respectively. 19F inversion recovery experiments were acquired with a series of variable delay times, τ, (i.e., 0.0625, 0.125, 0.25, 0.5, 1.0, 2.0, 4.0, 8.0 s) and a relaxation delay of 7 s. 19F CPMG experiments, comprised of a 90x − [τcp − 180y − τcp]n pulse train, were acquired with a series of spin-echo evolution times, T, optimized for a given τcp and ED construct (i.e., 8 points ranging from T = 1.6 to 128 ms) and a relaxation delay of 5 s. Longitudinal and transverse relaxation times were computed by fitting plots of 19F signal intensity, I, for a given 5-F-Trp residue, determined by Lorentzian line fitting, as a function of variable delay or spin-echo times according to Eqs. 6 and 7, respectively, using MATLAB 7.12.0.
| (6) |
| (7) |
19F CPMG relaxation dispersion experiments, where the transverse relaxation rate, R2, is determined as a function of the delay between 180° pulses (2τcp), were acquired with a series of τcp values (ranging from τcp = 50 to 500 μs) and plotted against CPMG frequency, νCPMG, as defined by Eq. 8.
| (8) |
Additional 19F CPMG relaxation dispersion data on the 5-F-Trp labeled NS1A ED dimer at a second magnetic field strength were acquired at 20 °C on a Bruker AVANCE III HD 600 MHz NMR spectrometer equipped with a 5-mm TCI HCN cryoprobewith 1H coil tunable to 19F at a frequency of 564.62 MHz. A series of pseudo-2D spectra were acquired as a function of T (8 points ranging from 1.0 to 12.5 ms) and νCPMG (ranging from 500 to 5,000 Hz), processed within TopSpin3.2 (Bruker), and analyzed using MATLAB 7.12.0 as described above. The conformational exchange rate, kex, for ED dimer motion was obtained by fitting the field dependent 19F relaxation dispersion data for Trp187 to a general two-state exchange model (Carver and Richards, 1972),using the program GUARDD (Kleckner and Foster, 2012) (http://code.google.com/p/guardd/).
Supplementary Material
HIGHLIGHTS
19F NMR applied to 5-F-Trp labeled Ud NS1A and its isolated RBD and ED domains
19F NMR reveals conformational exchange at ED dimer interface monitored by Trp187
ED dimer conformational exchange is >103 times faster than kinetics of dimerization
19F NMR shows Trp187 is exposed in full length NS1A at subaggregate concentrations
ACKNOWLEDGMENTS
We thank R. Guan, A. Palmer III,J. Prestegard, and P. Rossi for valuable scientific discussions, C. Anklin and D. Baldiserri for assistance with acquiring19F NMR data at a second magnetic field strength in the Applications Laboratory, Bruker BioSpin, Billerica, MA,S. Brill for kindly providing the yeast SUMO protease Ulp1 plasmid used in this work, and B. Amer, K. Conover, K. Field, S. Kim, and C. Schauder for technical support. We also thank an anonymous reviewer for valuable suggestions that helped improve this manuscript. This work was supported by grants from the National Institute of General Medical Sciences Protein Structure Initiative U54-GM094597 (to G.T.M.), National Institutes of Health U01 AI074497 (to G.T.M. and R.M.K.), and National Institutes of Health R01 AI11772 (to R.M.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION Supplemental Information includes eight figures and one table and can be found with this article online at:
REFERENCES
- Acchione M, Lee YC, DeSantis ME, Lipschultz CA, Wlodawer A, Li M, Shanmuganathan A, Walter RL, Smith-Gill S, Barchi JJ., Jr. Specific fluorine labeling of the HyHEL10 antibody affects antigen binding and dynamics. Biochemistry. 2012;51:6017–6027. doi: 10.1021/bi300455t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AH, Loh AP, Jane DE, Oswald RE. Dynamics of the S1S2 glutamate binding domain of GluR2 measured using 19F NMR spectroscopy. J Biol Chem. 2007;282:12773–12784. doi: 10.1074/jbc.M610077200. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, et al. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J Biol Chem. 2011;286:26050–26060. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayllon J, Russell RJ, Garcia-Sastre A, Hale BG. Contribution of NS1 Effector Domain Dimerization to Influenza A Virus Replication and Virulence. J Virol. 2012;86:13095–13098. doi: 10.1128/JVI.02237-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of influenza virus NS1 effector domain. Nature Struct Mol Biol. 2006;13:559–560. doi: 10.1038/nsmb1099. [DOI] [PubMed] [Google Scholar]
- Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer BC, Chugh J, Al-Hashimi HM, Marsh EN. Using fluorine nuclear magnetic resonance to probe the interaction of membrane-active peptides with the lipid bilayer. Biochemistry. 2010;49:5760–5765. doi: 10.1021/bi100605e. [DOI] [PubMed] [Google Scholar]
- Byeon IJ, Louis JM, Gronenborn AM. A protein contortionist: core mutations of GB1 that induce dimerization and domain swapping. J Mol Biol. 2003;333:141–152. doi: 10.1016/s0022-2836(03)00928-8. [DOI] [PubMed] [Google Scholar]
- Carver JP, Richards RE. General 2-site solution for chemical exchange produced dependence of T2 upon Carr-Purcell pulse separation. J Magn Reson. 1972;6:89–96. [Google Scholar]
- Chalkley RJ, Hansen KC, Baldwin MA. Bioinformatic methods to exploit mass spectrometric data for proteomic applications. Methods Enzymol. 2005;402:289–312. doi: 10.1016/S0076-6879(05)02009-4. [DOI] [PubMed] [Google Scholar]
- Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- Chien CY, Tejero R, Huang Y, Zimmerman DE, Rios CB, Krug RM, Montelione GT. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nature Struct Biol. 1997;4:891–895. doi: 10.1038/nsb1197-891. [DOI] [PubMed] [Google Scholar]
- Danielson MA, Falke JJ. Use of 19F NMR to probe protein structure and conformational changes. Annu Rev Biophys Biomol Struct. 1996;25:163–195. doi: 10.1146/annurev.bb.25.060196.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Ma LC, Xiao R, Radvansky B, Aramini J, Zhao L, Marklund J, Kuo RL, Twu KY, Arnold E, et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A. 2008;105:13093–13098. doi: 10.1073/pnas.0805213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Didenko T, Liu JJ, Horst R, Stevens RC, Wuthrich K. Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr Opin Struct Biol. 2013 doi: 10.1016/j.sbi.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr., Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat Struct Biol. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- Forsén S, Hoffman RA. Study of moderately rapid chemical exchange reactions by means of nuclear magnetic double resonance. Journal of Chemical Physics. 1963;39:2892–2901. [Google Scholar]
- Gerig JT. Fluorine NMR of Proteins. Progress NMR Spectroscopy. 1994;26:293–370. [Google Scholar]
- Goddard TD, Kneller DG. Sparky 3. University of California; San Francisco, CA: 2006. [Google Scholar]
- Hale BG, Barclay WS, Randall RE, Russell RJ. Structure of an avian influenza A virus NS1 protein effector domain. Virology. 2008a;378:1–5. doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Hale BG, Kerry PS, Jackson D, Precious BL, Gray A, Killip MJ, Randall RE, Russell RJ. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc Natl Acad Sci U S A. 2010;107:1954–1959. doi: 10.1073/pnas.0910715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008b;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Montelione GT. Structural biology: proteins flex to function. Nature. 2005;438:36–37. doi: 10.1038/438036a. [DOI] [PubMed] [Google Scholar]
- Ii T, Mullen JR, Slagle CE, Brill SJ. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair (Amst) 2007;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski JJ, Basu D, Engel DA, Geysen HM. Design, synthesis, and evaluation of novel small molecule inhibitors of the influenza virus protein NS1. Bioorg Med Chem. 2012;20:487–497. doi: 10.1016/j.bmc.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J Biomol NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- Johnson ML, Straume M. Comments on the analysis of sedimentation equilibrium experiments. In: Schuster TM, Laue TM, editors. Modern Analytical Ultracentrifugation. Birkhäuser; Boston, Massachusetts: 1994. [Google Scholar]
- Kerry PS, Ayllon J, Taylor MA, Hass C, Lewis A, Garcia-Sastre A, Randall RE, Hale BG, Russell RJ. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One. 2011;6:e17946. doi: 10.1371/journal.pone.0017946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HW, Perez JA, Ferguson SJ, Campbell ID. The specific incorporation of labelled aromatic amino acids into proteins through growth of bacteria in the presence of glyphosate. Application to fluorotryptophan labelling to the H(+)-ATPase of Escherichia coli and NMR studies. FEBS Lett. 1990;272:34–36. doi: 10.1016/0014-5793(90)80442-l. [DOI] [PubMed] [Google Scholar]
- Kitevski-Leblanc JL, Hoang J, Thach W, Larda ST, Prosser RS. 19F NMR studies of a desolvated near-native protein folding intermediate. Biochemistry. 2013;52:5780–5789. doi: 10.1021/bi4010057. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc JL, Prosser RS. Current applications of 19F NMR to studies of protein structure and dynamics. Prog Nucl Magn Reson Spectrosc. 2012;62:1–33. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Kleckner IR, Foster MP. GUARDD: user-friendly MATLAB software for rigorous analysis of CPMG RD NMR data. J Biomol NMR. 2012;52:11–22. doi: 10.1007/s10858-011-9589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Garcia-Sastre A. The NS1 protein: a master regulator of host and viral functions. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of Influenza. Wiley-Blackwell; 2013. pp. 114–132. [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, United Kingdom: 1992. pp. 90–125. [Google Scholar]
- Li W, Noah JW, Noah DL. Alanine substitutions within a linker region of the influenza A virus non-structural protein 1 alter its subcellular localization and attenuate virus replication. J Gen Virol. 2011;92:1832–1842. doi: 10.1099/vir.0.031336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lynch PA, Chien CY, Montelione GT, Krug RM, Berman HM. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nature Struct Biol. 1997;4:896–899. doi: 10.1038/nsb1197-896. [DOI] [PubMed] [Google Scholar]
- Liu L, Byeon IJL, Bahar I, Gronenborn AM. Domain Swapping Proceeds via Complete Unfolding: A 19F and 1H NMR Study of the Cyanovirin-N Protein. J Am Chem Soc. 2012;134:4229–4235. doi: 10.1021/ja210118w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackereth CD, Sattler M. Dynamics in multi-domain protein recognition of RNA. Curr Opin Struct Biol. 2012;22:287–296. doi: 10.1016/j.sbi.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. The role of conformational entropy in molecular recognition by calmodulin. Nat Chem Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- Medina RA, Garcia-Sastre A. Influenza A viruses: new research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mittermaier AK, Kay LE. Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci. 2009;34:601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Nobeli I, Favia AD, Thornton JM. Protein promiscuity and its implications for biotechnology. Nat Biotechnol. 2009;27:157–167. doi: 10.1038/nbt1519. [DOI] [PubMed] [Google Scholar]
- Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Panavas T, Sanders C, Butt TR. SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2009;497:303–317. doi: 10.1007/978-1-59745-566-4_20. [DOI] [PubMed] [Google Scholar]
- Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional, and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Richt JA, Garcia-Sastre A. Attenuated influenza virus vaccines with modified NS1 proteins. Curr Top Microbiol Immunol. 2009;333:177–195. doi: 10.1007/978-3-540-92165-3_9. [DOI] [PubMed] [Google Scholar]
- Rule GS, Pratt EA, Simplaceanu V, Ho C. Nuclear magnetic resonance and molecular genetic studies of the membrane-bound D-lactate dehydrogenase of Escherichia coli. Biochemistry. 1987;26:549–556. doi: 10.1021/bi00376a029. [DOI] [PubMed] [Google Scholar]
- Schneider WM, Inouye M, Montelione GT, Roth MJ. Independently inducible system of gene expression for condensed single protein production (cSPP) suitable for high efficiency isotope enrichment. J Struct Funct Genomics. 2009;10:219–225. doi: 10.1007/s10969-009-9067-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21:50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol. 2006;80:3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- van Beek JD. matNMR: a flexible toolbox for processing, analyzing and visualizing magnetic resonance data in Matlab. J Magn Reson. 2007;187:19–26. doi: 10.1016/j.jmr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Wand AJ. The dark energy of proteins comes to light: conformational entropy and its role in protein function revealed by NMR relaxation. Curr Opin Struct Biol. 2013;23:75–81. doi: 10.1016/j.sbi.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Riedel K, Lynch P, Chien CY, Montelione GT, Krug RM. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 1999;5:195–205. doi: 10.1017/s1355838299981621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012;8:e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Monzingo AF, Robertus JD. Structure of NS1A effector domain from the influenza A/Udorn/72 virus. Acta Crystallogr D Biol Crystallogr. 2009;65:11–17. doi: 10.1107/S0907444908032186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Khan JA, Swapna GV, Ertekin A, Krug RM, Tong L, Montelione GT. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J Biol Chem. 2007;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.