Abstract
The BridgeDb app for Cytoscape allows users to map and annotate identifiers of genes, proteins and metabolites in the context of biological networks. The app greatly simplifies the identifier mapping process in Cytoscape by providing a unified interface to different mapping resources and services. The app also provides a programming interface via Cytoscape Commands that can be utilized for identifier mapping by other Cytoscape apps. In this article we provide a technical guide to the BridgeDb app for mapping identifiers in Cytoscape.
Introduction
Cytoscape 1 is a powerful network visualization tool and platform for data integration and analysis. However, identifier mapping remains a challenge when working with biological data from different sources. We developed the BridgeDb app for Cytoscape to provide utilities for mapping and annotating identifiers in the network context. Built on the BridgeDb open-source framework for identifier mapping 2, the BridgeDb app provides a graphical user interface (GUI) for users and a command interface for other Cytoscape apps to perform identifier mapping in Cytoscape networks. As a result, BridgeDb enables or simplifies the processes of identifier translation, biological entity unification, and functional annotation.
Implementation
The BridgeDb identifier mapping framework ( http://bridgedb.org/) 2 was designed to provide standardized access to gene, protein and metabolite identifier mapping services such as Ensembl BioMart 3, Synergizer 4, PICR 5 and BridgeDb web services and BridgeDb database files. By creating a Java-based abstract layer, BridgeDb enables bioinformatics applications to connect to different mapping resources through the same interface, which greatly alleviates the burden of exploring, maintaining and switching between resources.
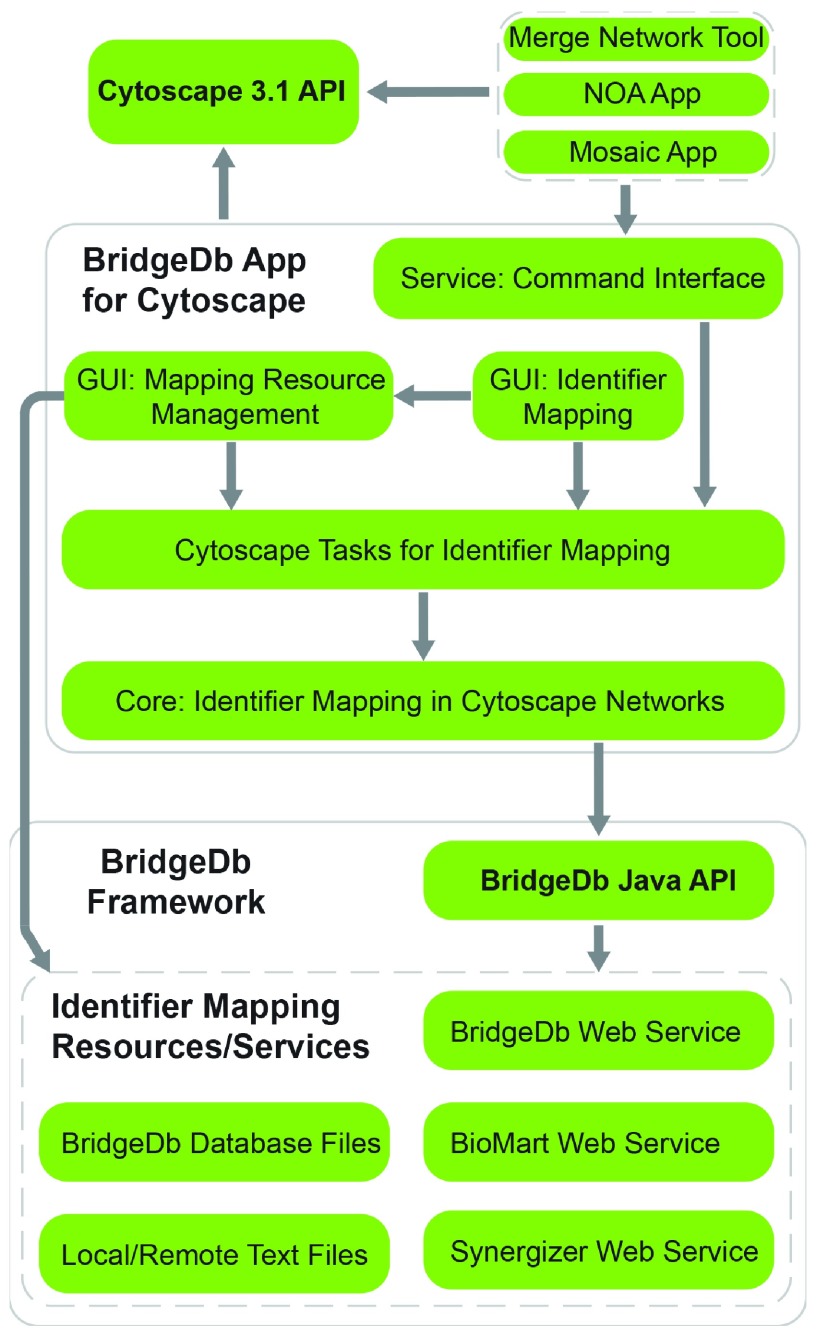
Built upon BridgeDb framework and API, the BridgeDb app for Cytoscape can be used to connect to different mapping resources and map identifiers in Cytoscape netwoks. The BridgeDb app was implemented based on the Cytoscape 3 API. Its predecessor was the CyThesarus plugin for Cytoscape 2. To take advantage of the new OSGi based architecture in Cytoscape 3 and its clearly defined and simplified API, we have rewritten the CyThesaurus plugin into an OSGi bundle app. Figure 1 illustrates the implementation details. The identifier mapping API of BridgeDb framework was wrapped by Cytoscape Task and TaskFactory API, which provide identifier mapping utilities to users through graphical user interfaces for managing mapping resources and performing identifier mapping. The BridgeDb app Tasks were also registered to Cytoscape as command services allowing other apps, such as Mosaic 6 and NOA 7 apps, and the Merge Network tool, to take advantage of BridgeDb app’s identifier mapping capacities.
Figure 1. BridgeDb app implementation.
Results
BridgeDb app user interface
The BridgeDb app provides an intuitive GUI to perform identifier mapping in Cytoscape networks, consisting of two interactive dialogs for resource management and identifier mapping, respectively, which can be opened via the menu item Apps/BridgeDb.
The resource management dialog allows user to add, remove, and select or deselect mapping resources. Once the resources are configured, they will be saved in a global property file and therefore shared among different Cytoscape sessions. Currently BridgeDb app supports local and remote delimited text files, BridgeDb database files, BridgeDb web service, BioMart web service, and Synergizer web service. Web services are easy to access and up to date and therefore are preferable for annotating small to medium-size networks (less than 1,000 nodes). It is highly recommended to download and use the BridgeDb database files for mapping identifiers in large networks in order to save time. Local delimited text files are useful when mapping between non-standard or customized identifiers. To avoid ambiguity, it is recommended to select only one resource unless multiple resources are believed to be complementary to each other. Particularly, resources for different species should not be selected at the same time. Database or text files are also preferable when reproducibility is essential. We will update the app to support more mapping resources as BridgeDb API keeps being developed.
The identifier mapping dialog is the main interface for mapping identifiers stored in the node table in the selected Cytoscape network. The user needs to choose the source identifier types, columns in the node table that contain the source identifiers, target identifier types, and columns to save the target identifiers. When mapping, all specified source identifier types in all selected resources will be queried for matching identifiers. If one source identifier can be mapped onto multiple target identifiers, all target identifiers can be saved as a list in the node table. If a target column exists in the node table, all values in the column will be overwritten with the target identifiers; otherwise, a new column will be created and filled with the target identifiers. Besides a set of supported identifier types (e.g., Entrez Gene ID and UniProt accession), a mapping resource such as BridgeDb database may also have a set of supported attributes (e.g., gene symbol and description).
BridgeDb app command interface
The BridgeDb app also provides a set of Cytoscape commands, which can be used by scripting or by other Cytoscape apps (client apps) to take advantage of the identifier mapping capacities provided by BridgeDb app. Table 1 provides a selected list of commands supported by BridgeDb. Every command has an optional argument ‘appName’ in order to allow every client app to have its own set of identifier mapping resources. Morris et al. ( setsApp 8, published together in the Cytoscape App collection 9) provided an example of how to use commands in client apps.
Table 1. Selected BridgeDb app commands.
Arguments with asterisks (*) are required.
| Command | Arguments | Description |
|---|---|---|
| bridgedb resource add |
classPath=
IDMapper class path*
connString= BridgeDb connection string* displayName= display name appName= name of client Cytoscape app |
Add a mapping resource. appName is the
name of the client Cytoscape app that is using this command. |
| bridgedb resource remove |
connString=
BridgeDb connection string*
appName= name of Cytoscape app |
Remove a mapping resource. |
| bridgedb resource select |
connString=
BridgeDb connection string*
select= true or false appName= name of client Cytoscape app |
Select or deselect an mapping resource. |
| bridgedb id mapping |
network=
Cytoscape network name*
sourceColumn= source column in node table* targetColumn= source ID type* sourceIdType= target column in node table* targetIdType= target ID type* appName= name of client Cytoscape app |
Map identifiers from a column in node
table to another in a Cytoscape network. |
| bridgedb resource config
dialog |
appName= name of client Cytoscape app | Open the resource management/
configuration dialog. |
| appName= name of client Cytoscape app | bridgedb main dialog | Open the main BridgeDb app dialog. |
Example use cases
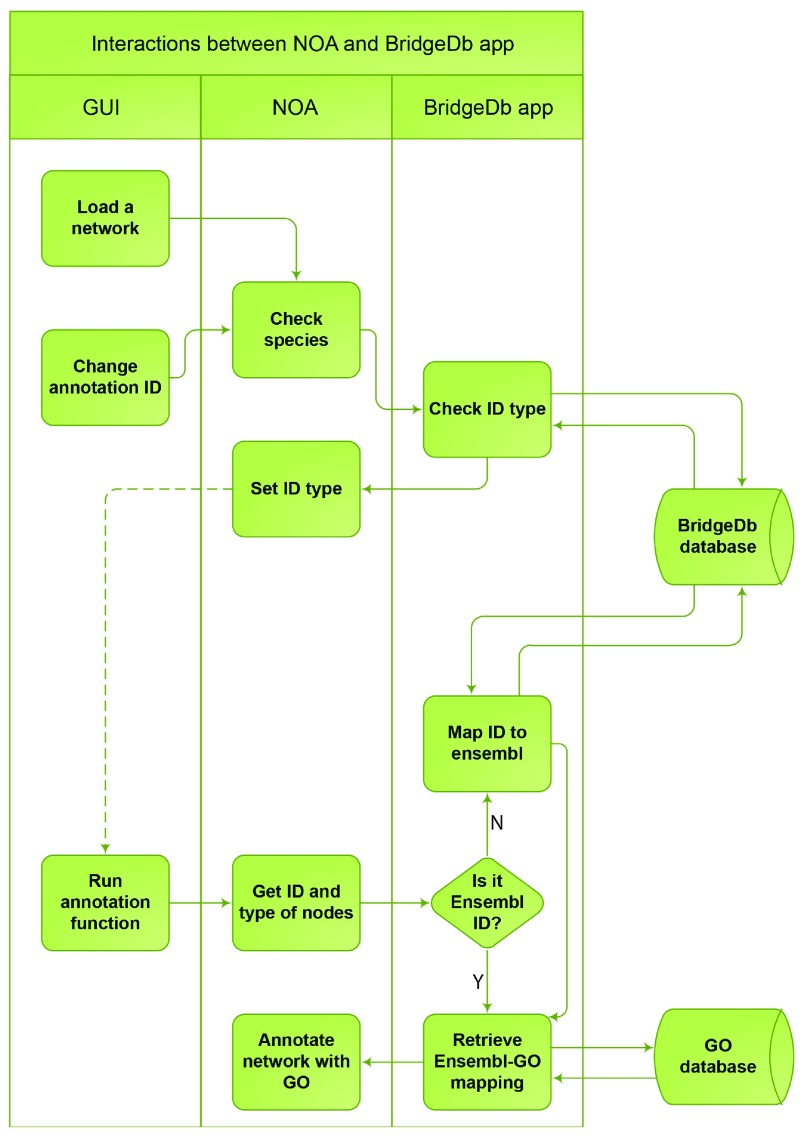
Use cases of BridgeDb app include identifier translation, biological entity unification, and functional annotation in Cytoscape networks. Box 1 provides an example of how to use BridgeDb app to facilitate annotation and integration of networks from public databases. In particular, this example shows the steps to generate a TP53 interaction network by merging networks from different sources using BridgeDb app and Merge Network tool. Figure 2 illustrates the process of the Cytoscape app NOA 7 annotating Cytoscape networks with Gene Ontology by utilizing BridgeDb app’s command services.
Box 1. Example: Using BridgeDb app for merging TP53 interaction networks from different sources.
-
1.
Install and start Cytoscape 3.1.0 or above
-
2.
Select File > Import > Network > Public Databases. . .
-
3.
Enter search condition TP53 AND human, press Search, select database IntAct, and click Import
-
4.
After importing, click No when asking about whether to manually merge networks, and close the import dialog
-
5.
If needed, install BridgeDb from Apps > App Manager
-
6.
Select File > Import > Network > URL. . . > Example:Human Protein-Protein: Rual et al. . . . Nature 2005
-
7.
Use default when asking about setting of the new network
-
8.
Select Apps > BridgeDb > Manager ID Mapping Resources
-
9.
Click Databases, Select database type .bridge
-
10.
Click Download, download Hs_Derby_[date].zip, and unzip the file
-
11.
In Cytoscape, select the unzipped .bridge file
-
12.
Review supported identifier types and click Close
-
13.
Select network rual.sif in Network panel
-
14.
Select Apps > BridgeDb > Map Identifiers
-
15.
Select name as the Source Column in Node Table
-
16.
Select Entrez Gene as Source ID Type(s)
-
17.
Select Uniprot/TrEMBL as Target ID Type
-
18.
Click Insert in the destination ID types table
-
19.
Select Attribute: Symbol as the second Target ID Type
-
20.
Click OK, wait, then click No to close
-
21.
Search for TP53 in the Cytoscape search box
-
22.
Select Select > Nodes > First Neighbors of Selected Nodes > Undirected
-
23.
Select File > New > Network > From selected nodes, all edges
-
24.
Select Tool > Merge > Networks. . .
-
25.
Select the network from IntAct and rual.sif(1)
-
26.
Click Advanced Network Merge
-
27.
Select Matching columns: uniprotkb_accession for IntAct, and Uniprot/TrEMBL for rual.sif(1)
-
28.
Click Merge to get a TP53 interaction network merged from the two sources
Figure 2. NOA app utilizing BridgeDb app services for gene ontology annotation.
Conclusions
In this paper, we presented the BridgeDb app for Cytoscape. By providing a unified interface to various mapping resources, BridgeDb app enables identifier mapping in the network context and therefore greatly eases the process of data integration in Cytoscape.
Software availability
Software available from: http://apps.cytoscape.org/apps/bridgedb
Latest source code: https://github.com/jjgao/bridgedb.cytoscape
Source code as at the time of publication: https://github.com/F1000Research/bridgedb.cytoscape
Archived source code as at the time of publication: http://www.dx.doi.org/10.5281/zenodo.10465 10
License: Lesser GNU Public License 2.1: https://www.gnu.org/licenses/old-licenses/lgpl-2.1.html
Acknowledgements
We thank Scooter Morris for technical support on command interface, Kristina Hanspers for beta testing, and Gary Bader, Mike Smoot, Keiichiro Ono and David States for providing useful comments.
Funding Statement
This work was partially funded by the Google Summer of Code program (JG, CZ, and LZ), the Robertson Foundation (NS), and by NIGMS R01-GM100039 (ARP).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
v1; ref status: indexed
References
- 1.Shannon P, Markiel A, Ozier O, et al. : Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Iersel MP, Pico AR, Kelder T, et al. : The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC bioinformatics. 2010;11(1):5 10.1186/1471-2105-11-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasprzyk A, Keefe D, Smedley D, et al. : EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14(1):160–169 10.1101/gr.1645104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berriz GF, Roth FP: The Synergizer service for translating gene, protein and other biological identifiers. Bioinformatics. 2008;24(19):2272–2273 10.1093/bioinformatics/btn424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Côté RG, Jones P, Martens L, et al. : The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC bioinformatics. 2007;8(1):401 10.1186/1471-2105-8-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang C, Hanspers K, Kuchinsky A, et al. : Mosaic: making biological sense of complex networks. Bioinformatics. 2012;28(14):1943–1944 10.1093/bioinformatics/bts278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Wang J, Hanspers K, et al. : NOA: a cytoscape plugin for network ontology analysis. Bioinformatics. 2013;29(16):2066–2067 10.1093/bioinformatics/btt334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris JH, Wu A, Doncheva NT, et al. : setsapp: Set operations for cytoscape nodes and edges.Manuscript submitted for publication. F1000Research. 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The cytoscape app collection. F1000Research. 2014 [Google Scholar]
- 10.Gao J, Zhang C, van Iersel M, et al. : F1000Research/bridgedb.cytoscape. ZENODO. 2014. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]