Abstract
The protein biochemistry supporting autophagosome growth on the cup-like isolation membrane is likely different from the biochemistry on the closed and maturing autophagosome. Thus, the highly curved rim of the cup may serve as a functionally-required surface for transiently-associated components of the early-acting autophagic machinery. Here we demonstrate that the E2-like enzyme, Atg3, facilitates LC3/GABARAPlipidation only on membranes exhibiting local lipid-packing defects. This activity requires an amino-terminal amphipathic helix similar to motifs found on proteins targeting highly curved intracellular membranes. By tuning the hydrophobicity of this motif, we can promote or inhibit lipidation in vitro and in rescue experiments in Atg3 knockout cells, implying a physiologic role for this stress detection. The need for extensive lipid-packing defects suggests that Atg3 is designed to work at highly-curved membranes perhaps including the limiting edge of the growing phagophore.
Keywords: LC3, GABARAP-L1, Atg3, Curvature
The macroautophagy degradative pathway delivers material into the lysosome through the formation of the autophagosome, a double-membraned vesicle that encapsulates portions of cytoplasm. During autophagosome growth, over thirty unique proteins associate with the autophagosomal membrane in a temporally-defined manner1. Among the final autophagosome-specific proteins to bind the organelle is the Atg8 family of ubiquitin-like proteins, which in mammals, consists of the LC3 and GABARAP subfamilies.Atg8/LC3membrane association is mediated by the covalent attachment of Atg8/LC3 to phosphatidylethanolamine (PE)2, 3. The resulting lipidated protein then drives autophagosome maturation steps including cargo capture, growth and closure of the organelle4, 5, and lysosome targeting or recognition (for review6).
When lipidation is compromised by knockout or mutation of upstream factors, autophagosome maturation is largely aborted7, 8 (see also lipidation-independent autophagy9) and curved cup-like intermediates termed isolation membranes (IMs) accumulate10. Electron tomography reveals that the ends of these cups are likely highly curved; the membranes on either side of the cisterna are within a few nanometers of one another and thus the curvature approximates something smaller than a 25nm vesicle11. Because IMs accumulate specifically when lipidation is blocked, we have begun to explore whether the strident curvature associated with this structure plays a role in the lipidation reaction.
Here we demonstrate that LC3/GABARAPlipidation requires local membrane stress associated with lipid-packing defects as occurs with high molar proportions of conical lipids or strident membrane curvature. This stress is sensed by the lipidation enzyme, Atg3. A major component of the sensing motif is an amino-terminal amphipathic helix with an amino acid organization distinct from helices on other curvature-sensing proteins, including the early autophagy factor Barkor12, 13. This sensing allows LC3/GABARAPlipidation to proceed efficiently even on membranes of simple lipid composition and low densities of the lipidation substrate, PE, provided the underlying membrane exhibits sufficient curvature. In short, the lipidation machinery is engineered to work best on membrane structures analogous to the strongly deformed rim of the growing autophagosome14.
Results
Reconstitution of the lipidation reaction
Covalent attachment of PE to Atg8 is mediated by a ubiquitin-like chain of enzymatic steps involving the E1-like Atg7 and the E2-like Atg3 (Figure 1a). These reactions can be reconstituted in vitro with recombinant enzymes, synthetic liposomes and ATP15. To explore the effects of membrane lipid composition and structure on the efficiency of these reactions, we reconstituted the lipidation of the mammalian Atg8 proteins, LC3B, GABARAP-L1 (GL1 also called GEC1 or Atg8L), and GABARAP-L2 (GL2 also called GATE-16) as described previously16-18. In each case, lipidation requires Atg3, Atg7, lipid and ATP and results in a gel-shift of the Atg8 homologue to a faster mobility (Figure 1b).
Figure 1. In vitro reconstitution of the lipidation reaction.

A) Cartoon of the ubiquitin-like lipidation reaction of autophagy. B) Conjugation of LC3 and two other mammalian homologs (GL1 and GL2) with PE was performed essentially as described previously18. Complete reactions contain 1.5 μM Atg7, 2.5 μM Atg3, 8-12μM LC3/GL1/GL2, 2 mM lipid (30nm liposomes composed of 10 mole% bl-PI, 55 mole% DOPE, 0.15 mole% Rh-DOPE and 34.85 mole% POPC), 1mM DTT and 1mM ATP and were run at 30°C for 90 min. Each reaction was run on a 12% SDS-PAGE gel and visualized by coomassie blue stain. The mobility of the lipidated protein is faster than the corresponding non-lipidated forms. Intermediates in the reaction are also detectable, including the Atg7-GRL1 conjugate, the Atg3-GRL1 conjugate, and a modified form of GRL1, GRL2 and LC3 that is labeled simply as GRL1/GRL2/LC3-intermediate and is likely the adenylated form of each LC3-related protein. C) The lipidation reactions are highly dependent upon the molar percentage of DOPE in the liposomes. Reactions are as in B, but run on 400 nm liposomes. Liposomes are composed of the indicated amount of DOPE, 10 mole% bl-PI, 0.15 mole% Rh-DOPE and POPC. D) Completed reactions in C were subjected to density gradient flotation to isolate only the lipidated material. Floated samples were run on SDS-PAGE and imaged by coomassie. Uncropped versions of all gels are included in the Supplemental Figure 7.
We also resolve several intermediates in the reaction. Ordinarily, LC3/GABARAP becomes covalently associated with Atg7 and then Atg3 via thioester bonds prior to being transferred onto PE. In mixtures missing one or more components, the reaction aborts prematurely, and trace amounts of these labile protein complexes accumulate (Figure 1b and 19,16). Our gel conditions also resolve an additional intermediate that is likely the activated adenylate of LC3/GABARAP, previously detected only by mass spectrometry20. Its formation requires ATP and Atg7, but not Atg3 or liposomes, it forms faster than the fully-lipidated product and is eventually completely consumed (Supplemental Figure 1). Thus, we can detect the production of each intermediate and the fully-lipidated product.
LC3/GABARAP lipidation is sensitive to membrane curvature
Lipidation is much more efficient on liposomes in which DOPE constitutes a high molar percentage of the phospholipid pool (e.g. 15, 16, 21and figures 1c & 1d). As PE is the lipidation target, this could suggest that one or more components of the reaction have a low affinity for PE. However, the efficiency of the reaction drops precipitously from 55 mole% DOPE to 30 mole% DOPE, suggesting that simple affinity for individual phospholipids is not the sole determinant. Alternatively, the high density of DOPE could alter the membrane architecture and thereby influence the reaction. To this end, it is notable that DOPE is a cone-shaped lipid. When cones are packed at high density into planar two-dimensional arrays, local defects and/or membrane stress develop (i.e. 22-25and Figure 2a). To test whether these lipid-packing constraints are important during lipidation, we used two approaches to modify the lipid composition. First, we “fill-the-gaps” by introducing semi-soluble inverted cone-shaped lipids. With 30 mole% DOPE sonicated liposomes, 80% or more of the GL1 is converted to GL1-PE (Figure 2b). When the reaction is titrated withstearoyl-CoA, a single acyl-chain derivative of Coenzyme A with a molecular volume that approximates an inverted cone, lipidation efficiency falls off. At high stearoyl-CoA concentrations, the reaction is very nearly dead.
Figure 2. Lipidation requires local membrane defects.
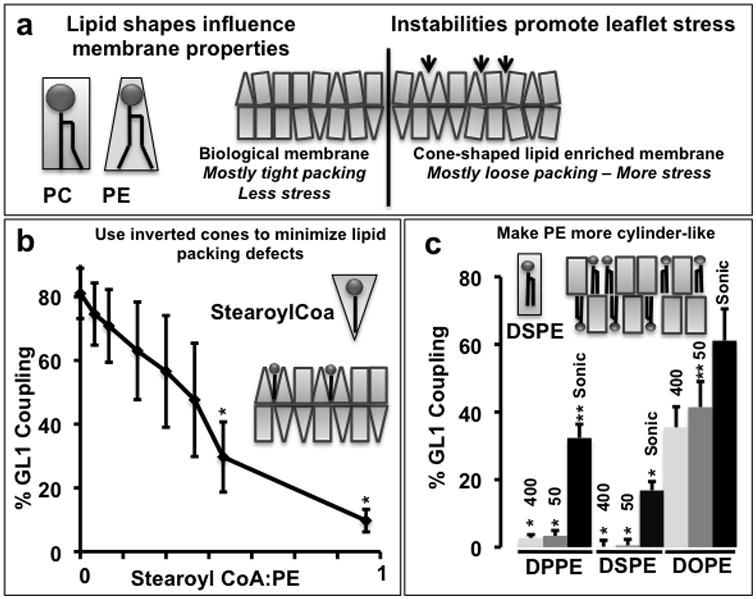
A) Cartoon depicting incompatibility of idealized cone-shaped lipids with planar assemblies. The spontaneous curvature in a lipid monolayer scales qualitatively with the idealized shape of the lipid48 where the idealized volume can be approximated as a cylinder (PC) or a cone (PE). When membrane curvature deviates significantly from the molecular spontaneous radius of curvature for an individual lipid, packing defects arise (i.e. 24, 25) forcing lipids to adopt sub-optimal configurations, producing local monolayer stress (see 22). Resolution of the local stress associated with these incompatibilities can include changes in lipid structure, a reorganization of lipid composition or the insertion of non-lipidic molecules including proteins. B) Filling the defects with inverted cone-shaped lipids inhibits lipidation. In vitro coupling reactions of GL1 were run on sonicated liposomes containing 30 mole% DOPE (2mM total lipid) as in figure 1. To test whether inverted cones influenced the reaction, increasing amounts of Stearoyl CoA was added immediately before initiating the reaction with ATP. Quantification is of n=3 samples, error bars represent standard deviation. p-values represent a comparison to samples without stearyoyl-CoA. **: p<0.1; *:p<0.05. C) Altering the shape of PE to reduce local membrane defects inhibits the lipidation reaction. In vitro coupling reactions of GL1 were run on liposomes of three different sizes (400nm and 50 nm extruded liposomes and sonicated liposomes) and of three different lipid compositions. Lipid compositions include 30 mole% DPPE, DSPE or DOPE, 10% bl-PI, and POPC. Total lipid = 2mM. Quantification is of n=3 samples, error bars represent standard deviation. The extent of lipidation in B and C is plotted as a percentage of total GL1 as determined by densitometry. p-values represent comparison to most active liposomes (sonicated DOPE). **: p<0.1; *:p<0.05.
Alternatively, we can alter the shape of PE itself to favor planar assemblies26 and thus limit the local stress. Here we replaced the unsaturated 18 carbon acyl chains of DOPE with fully saturated 16 or 18 carbon acyl chains (DPPE and DSPE respectively), such that the lipids have a cylindrical rather than conical molecular volume, while keeping the total surface density of reactive PE headgroups the same (Figure 2c). On extruded liposomes, the lipidation reaction is completely dead when either of the cylindrical lipid forms of PE is used despite the high local concentration of substrate head groups. Thus, the membrane environment is an important regulator of protein lipidation.
In contrast, cylindrical PE remains reactive when liposomes are prepared by sonication rather than extrusion (Figure 2c). Sonication produces small liposomes near their curvature limit. The packing of lipids onto a stridently convex membrane is more susceptible to lipid packing defects (Figure 3a). To test whether the lipidation reaction efficiency varies with the curvature of the underlying membrane, we generated liposomes of different diameters by extrusion through membranes of different pore sizes27. Smaller liposomes have a more highly curved surface than larger liposomes (Table I). As before, GL1 is completely converted to the lipidated form when liposomes having 55 mole% DOPE are used (Figure 3b – top). Further, because of the high density of cone-shaped lipids, conversion is independent of the size of the liposome. In contrast, when the DOPE mole% is lowered to 30%, lipidation becomes strongly dependent upon the curvature of the membrane. Under these conditions, liposomes extruded through 800 nm filters are incompatible with lipidationwhereas50 nm liposomes are nearly as efficient as the high DOPE samples.
Figure 3. The lipidation reaction is membrane curvature dependent.
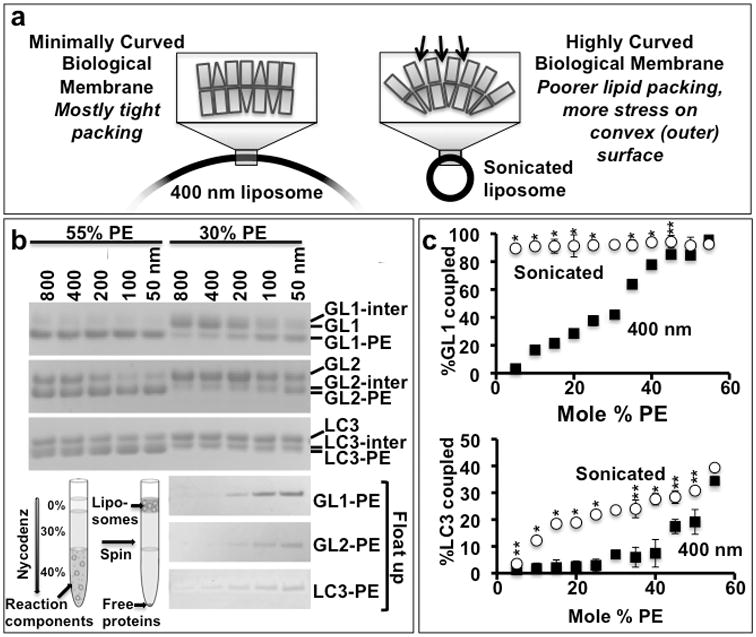
A) Membrane defects resembling those accumulating with high concentrations of PE are prevalent in vivo at sites of high curvature and can be recapitulated in vitro by forming liposomes of different diameters. B) To test the impact of curvature on lipidation, we compared liposomes with unphysiologically high surface densities of PE (55 mole %) as have been used in other Atg8 or LC3 lipidation publications (e.g. 4,21,49), with liposomes having the highest densities of PE observed in mammalian organelle membranes (30 mole %). At physiologically relevant concentrations of DOPE (30 mole %), lipidation becomes membrane curvature dependent. In vitro coupling reactions were performed as in Figure 1, except that liposomes were made by extrusion through membranes with different pore diameters (as indicated). Actual diameters of these liposomes were determined by dynamic light scattering (Table I). B-Top) The lipidation of GL1, GL2, and LC3 proceeds efficiently and with relatively little curvature dependence upon liposomes composed of 55% PE. However, when the PE is reduced to 30%, flat liposomes are no longer suitable for lipidation, while highly curved liposomes (100 nm diameter or less) continue to couple effectively. B-bottom) To confirm that this curvature dependence reveals the lipidated product and not the intermediate, the 30% reactions were each run on nycodenz flotation gradients. Only the lipidated product is recovered at the top of the gradient. C) The lipidation reaction is largely insensitive to PE concentrations when run on highly curved liposomes. Coupling of GL1 and LC3 to PE was assessed on liposomes of two sizes (400 nm – black squares and sonicated – red circles) and varying DOPE mole%. The extent of lipidation is determined as in figure 2. (n=2 for 30 and 55% PE, n=4 for all the other points, error bars represent standard deviation). p-values were determined comparing sonicated to 400nm samples at each PE concentration.**: p<0.1; *:p<0.01. Uncropped versions of all gels are included in the Supplemental Figure 7.
The inherent lipidation efficiencies of GL2 and LC3B are much lower than for GL1. Thus, lipidation of these two homologues is already weakly dependent upon curvature even at high DOPE. On liposomes with more physiological DOPE concentrations, GL2-PE and LC3-PE formation appears to be almost completely limited to highly curved surfaces (Figure 3B– top). However, the bands of the reaction intermediate and the lipidated product are too close to allow quantitative analysis of the reaction. To circumvent this, we subjected the finished reaction to nycodenz density flotation (as in 18). This additional step revealed a sharp sensitivity of all lipidation reactions to liposome size (Figure 3B– bottom).
GL1 coupling is strongly PE concentration-dependent when molecularly flat liposomes are used (Figure 3c-top, squares). However, when these same liposomes are sonicated to produce high surface curvatures, coupling is maximal at as little as 5 mole% DOPE (Figure 3c-top, circles). LC3B coupling is markedly less efficient (Figure 3c-bottom). On flat membranes, there is almost no lipidation until the PE surface density exceeds 40 mole% (Figure 3c-bottom, squares), while LC3B lipidation approaches its maximum efficiency at as little as 15 mole% PE when those liposomes are sonicated (circles). Thus, lipidation efficiency is sensitive to curvature stress and lipid-dependent packing defects. This suggests the involvement of a mechanism that is influenced by both, but in principle the two alterations to membrane structure could influence the reaction in different ways. In either case, these data imply that a uniquely high density of PE is unnecessary on the growing autophagosome provided regions of high curvature are available to support the biochemistry.
Atg3 is a membrane curvature sensor
In samples containing flat membranes or lacking liposomes altogether, both the Atg3-GL1 adduct and the adenylated form of GL1 are still formed (Figure 4a). Thus the transfer from Atg3 to PE is the membrane curvature sensitive step, and Atg3 is likely a component of the curvature sensor. To test if Atg3 binds membranes in a curvature-dependent manner, we mixed Atg3 with liposomes of different diameters and subjected the mixtures to a non-equilibrium binding assay using density gradient flotation. As with lipidation, Atg3 membrane association is curvature insensitive when the surface density of DOPE is exceptionally high (Figure 4b, c), but is strongly curvature-dependent when DOPE is reduced to 30 mole%. Even liposomes lacking any PE can support Atg3 binding if the curvature is sufficiently strident, indicating that Atg3 harbors a curvature-sensitive membrane-binding motif that is independent of any direct interaction with PE.
Figure 4. Atg3 is a membrane curvature sensor.
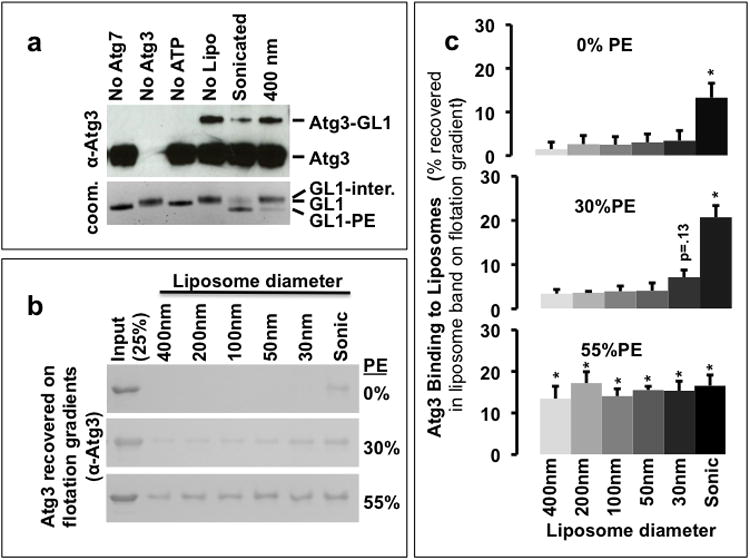
A) The accumulation of the Atg3-GL1 conjugate depends upon the extent of membrane curvature. In vitro GL1 coupling reactions were run as in figure 1. The reactions were visualized by immunoblot against Atg3 (top) and by coomassie staining of GRL1 (bottom). The accumulation of Atg3-GL1 is inversely related to the formation of the lipidated GRL1 suggesting that the membrane curvature-dependent step may involve Atg3 recognition of the membrane surface. B) Atg3 binding to liposomes. Atg3 (10 μM) was incubated at 30°C for 90 min with 0, 30, or 55 mole% DOPE containing liposomes (5 mM lipids) of varying sizes (extrusion membrane dimensions are shown on the figure, actual final sizes were determined by dynamic light scattering (Table I)). The liposome associated Atg3 was recovered by nycodenz density gradient centrifugation and analysed by SDS PAGE. C) Quantification of liposome-associated Atg3. Densitometric plot of the amount of Atg3 recovered in the top fraction in flotation assays as in B. Error bars indicate SEM from three independent experiments. p-values calculated as compared to putative negative control (0% PE, 400 nm). *:p< 0.05. Uncropped versions of all gels are included in the Supplemental Figure 7.
Atg3 targets curved membranes via an amphipathic helix
There are two major paradigms for sensing curvature28. BAR domains rely upon membrane curvature itself (the organization of the headgroups into a non-planar assembly), while amphipathic helices recognize and insert into bilayers exhibiting packing defects as on curved surfaces. Our results in figures 2 and 3 suggest that Atg3 acts via membrane insertion, consistent with an amphipathic helix driven curvature sensor.
According to the hydrophobicity and helix predicting program, Heliquest29, the first 26 amino acids of Atg3 present a high hydrophobic moment (0.398), consistent with an amphipathic distribution of amino acids around a helix. This putative helix includes a hydrophobic face composed of small hydrophobic residues like valine and leucine (figure 5a), bordered on either side by a cationic lysine, likely demarcating the surface of the bilayer. The opposite face is replete with small polar and anionic amino acids. The lack of strongly hydrophobic amino acids in the presumed bilayer-attachment face suggests that this sequence will only weakly partition into stable bilayers, but may interact more strongly with membranes having defects. This distribution is the hallmark for a curvature sensor.
Figure 5. The N-terminus of Atg3 is a curvature-sensing amphipathic helix.
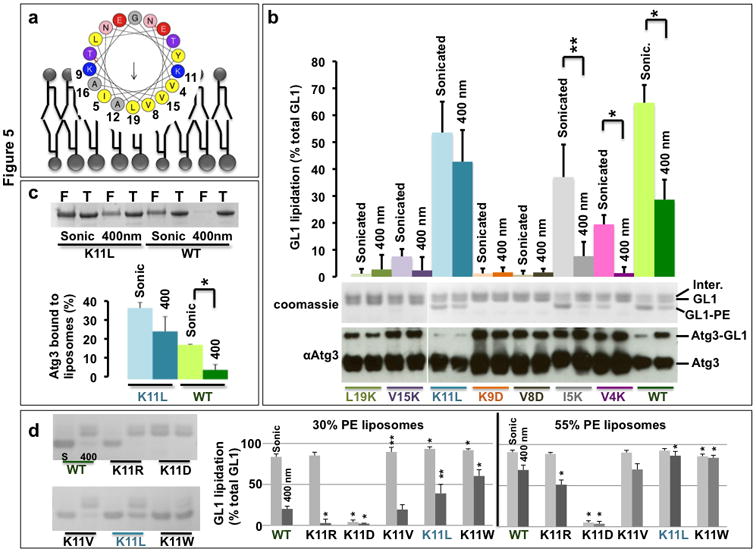
A) Helical wheel representation of the predicted N-terminal amphipathic helix (aa 4-26) of mouse Atg3 (wheel generated in Heliquest29). B) Analysis of reaction efficiency for point mutants of the amphipathic helix. GL1 lipidation reactions were run on 400nm and sonicated liposomes (30 mole% DOPE) with the indicated Atg3 mutants. The extent of GL1-PE formation (top gel) was determined from densitometry and is plotted as percent of total GL1 (bar graph). The ability to form the Atg3-GL1 conjugate was also assessed for each mutant (bottom immunoblot). Note that this conjugate forms in every case, indicating that no mutant inhibited a step upstream in the reaction. Further, this conjugate specifically accumulates in reactions where lipidation is impaired. C) Curvature dependence of K11L mutant binding to liposomes compared to wild-type Atg3. The binding reaction was performed as in Figure 4b on 30DOPE% liposomes. F = Float-up; T = Total Input. D) K11 represents a tunable node through which curvature dependence of the lipidation reaction can be controlled. Lipidation assays were run as in B. Total GL1 lipidation was assessed from reactions with wildtype Atg3 or mutant forms of Atg3 in which position K11 was changed as indicated. Lipidation efficiency on low PE/low curvature liposomes (30% PE, 400 nm) increases with increasing hydrophobicity of the new amino acid, while liposomes with high PE (55%) or high curvature (sonicated) are good substrates for all active forms of Atg3. D-left) coomassie gels of individual lipidation reactions. D-right) densitometry of coomassie-stained gels revealed the ratio of GL1-PE to the sum of all three GL1 species as in figure 2. Light grey bars are sonicated liposomes; dark grey bars are 400 nm extruded liposomes. S = Sonicated liposomes; 400 = 400nm liposomes. For b, c, p-values were calculated between sonicated and 400 nm samples of the same mutant. For d, p-values represent comparison to WT protein on the same liposome size and composition. For all panels, n=3 independent experiments, error bars represent standard deviation and**: p< 0.05; *:p< 0.01. Uncropped gels in Supplemental Figure 7.
To determine whether this helix contributes to curvature-sensing, we generated putative loss-of-function mutants in the hydrophobic face and membrane-delimiting lysines and tested these mutants in lipidation reactions (Figure 5b coomassie gel and bar graph). The mutants fall into three classes (Supplemental Figure 2). 1). Introduction of charged residues into the core of the hydrophobic face destroys the amphipathic character and eliminates lipidation activity (V8D, V15K, L19K). 2). Introduction of lysines near the edges of the hydrophobic face reduces the amount of hydrophobic surface area available to partition into a bilayer (V4K, I5K), making these proteins more reliant upon membrane defects for efficient lipidation. 3). Flipping the charge on a membrane-delimiting lysine from cationic to anionic eliminates lipidation (K9D or K11D). Importantly, each mutant can still form the Atg3-GRL1 intermediate (Figure 5b, western blot) demonstrating that the amphipathic face of the helix is only involved in membrane recognition.
Helices targeting unique curvatures occupy a narrow range of membrane-partitioning energies. While reducing hydrophobicity abrogates membrane binding, marginal increases in hydrophobicity will necessarily reduce or eliminate the curvature-sensing quality of such a helix. To explore this possibility, we mutated the membrane-delimiting lysine at position K11 to leucine. This extends the hydrophobic face to include both the new L11 residue and the neighboring very hydrophobic Y18 (Figure 5a). With stronger hydrophobicity, this helix requires fewer defects for membrane partitioning and thus Atg3 (K11L) already functions well on 400nm surfaces (Figure 5b and 5c). Furthermore, we can modulate the curvature-sensitivity by replacing K11 with increasingly hydrophobic amino acids (V, L, and W - Figure 5d). Thus, the amino-terminal 26 amino acids ordinarily function as a weak amphipathic helix to dictate membrane curvature-sensing and are tunable by subtle modifications of the sequence.
In the absence of Atg3, autophagosome formation aborts at the IM due to a failure in LC3/GABARAP lipidation7. Thus in MEFs derived from Atg3 knockout (Atg3-/-) mice, lipidated LC3 cannot be detected by immunoblot analysis or immunofluorescence (IF). In addition, Atg16-positive IMs accumulate even under basal (unstarved) conditions. These phenotypes are reversible by reintroduction of wildtype Atg37. To establish whether the curvature-sensing motif in Atg3 is required in vivo, we performed rescue experiments in which Atg3-/- MEFs were transduced with wildtype Atg3 (WT) or with each Atg3 variant tested in vitro. Although similar viral titres were used, Atg3 protein expression levels differed for different mutations (Supplemental Figure 3, Figure 6a). Atg3 I5K and K11D failed to express despite secondary attempts at transient overexpression, and thus only the remaining nine mutants were analyzed for their behavior in vivo. Consistent with results obtained in vitro, all mutants that support lipidation on liposomes (Figure 5) rescued lipidation in vivo (Figure 6a). LC3 and GL2 lipidation was robust upon introduction of Atg3 WT, V4K, K11L, K11R, K11V and K11W. GABARAP lipidation was also rescued by active Atg3 mutants, but was only detectable following lysosomal inhibition as reported previously30. LC3-II levels were further increased upon treatment of cells with Bafilomycin A1 (Supplemental Figure 3), indicating that the lipidated LC3 is being targeted to and digested in the lysosome consistent with the formation and maturation of an autophagosome. Introduction of lipidation competent mutants also promoted the formation of LC3-positive immunofluorescent puncta to wildtype levels, while other mutants were without effect (Figure 6b and 6c).
Figure 6. An intact amphipathic helix is necessary for Atg3 function in vivo.
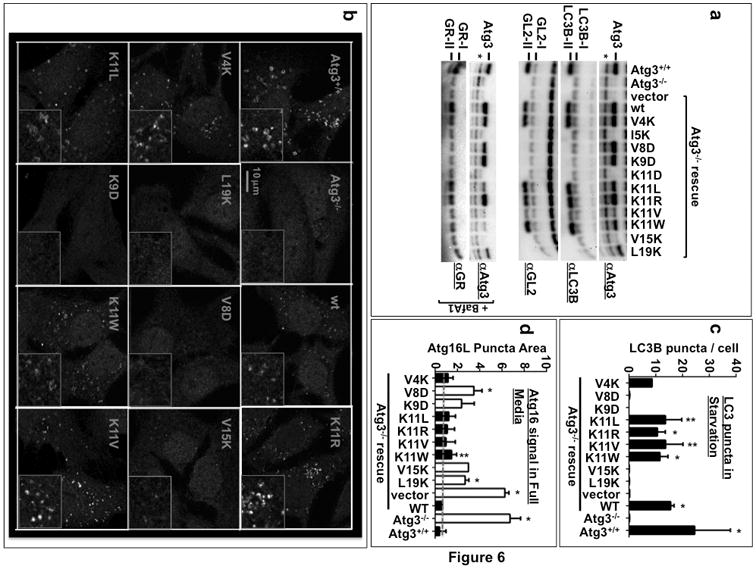
Atg3 knockout MEFs (Atg3 -/-) are incapable of forming LC3-II and do not accumulate LC3 puncta. Instead, they accumulate autophagic intermediates that are positive for Atg16. To test if the amphipathic helix is critical for in vivo function, Atg3 with wild-type or mutant forms of the amphipathic helix was introduced into Atg3 -/- MEFs by lentiviral infection and rescue of the three phenotypes was tested. A) Immunoblot analysis of LC3-II, GL2-II and GABARAP-II formation. GABARAP lipidation is only significantly observed in the presence of Bafilomycin A1 (BafA1: 100 nM). Asterisk indicates a non-specific band recognized by the Atg3 antibody. B) Cells were infected as in A and transduced cells were selected with puromycin for 1-2 days before starvation and immunolabeling with the anti-LC3B antibody. Representative images are shown in B. C) The number of LC3 puncta per cell was counted manually from 22-96 cells per replicate. Raw data can be found in Supplemental Figure 5. p-values represent difference from vector negative control. n = 2-5 independent experiments; the precise n values and number of cells per experiment are listed in the Methods. D) Cells as in B were kept in full media before being subjected to fixation and immunolabeling with the anti-Atg16L antibody. Atg16L puncta area as a fraction of total cellular area was obtained as described in Methods. This ratio was plotted as arbitrary units. Grey dashed line indicates the value for Atg3-/- cells rescued with wild-type Atg3. n = 2 (for V15K) or 3 (for all others) independent experiments; the precise n values and number of fields visualised per experiment are listed in the Methods. Raw data can be found in Supplemental Figure 4. Atg3 variants that support lipidation of LC3 family proteins in vitro (Figure 5) are indicated with black bars, while those that do not support lipidation are indicated with white bars. p-values represent difference from wild-type rescue.**:p<0.1; *: p<0.05. Uncropped versions of all gels are included in the Supplemental Figure 7.
Under basal conditions, Atg3-/- cells accumulate Atg16L-positive IM structures while Atg3+/+ cells have very few7. As previously described7, reintroduction of Atg3 WT into knockout cells fully rescues the Atg16L phenotype, returning puncta levels to numbers similar to wildtype cells. Surprisingly reintroduction of all Atg3 mutants leads to at least some rescue of the knockout phenotype (Figure 6d). Lipidation-competent mutants reduced Atg16L puncta to levels indistinguishable from rescue with wildtype protein expression, indicative of a full rescue response. Lipidation-incompetent mutants however also support a partial rescue, as evidenced by Atg16L puncta numbers well below the vector backbone control but consistently higher than Atg3 WT levels. Other than puncta number, no change in puncta structure or intensity is observed, suggesting that this partial rescue is related to a reduction in the steady-state level of Atg16-membrane structures (Supplemental Figure 4). Importantly, there is no significant variation amongst samples when autophagy is stimulated by starvation, indicating that expression of the Atg3 mutants has not caused a general suppression of the early steps in autophagosome formation (Supplemental Figures 4, 5).
Atg16L is part of a complex including Atg5 and Atg12 that together function as a putative E3 ligase to facilitate Atg3 lipidation activity31-33, perhaps in part by recruiting Atg3 to membranes. For example, Atg16L is ordinarily found on the IM, but when its localization is artificially switched to ectopic sites like the plasma membrane, Atg3-mediated lipidation is also switched to this new site34. To test how Atg3 helix mutants function without Atg16/Atg5-12 mediated membrane targeting, we introduced several Atg3 mutants into Atg5 KO MEFs35. These cells are incapable of LC3 lipidation, but the phenotype is reversible when wildtype Atg5 is reintroduced36. In contrast, no Atg3 mutants could support lipidation in Atg5 nulls, including those that were most effective in vitro, K11W and K11L (Supplemental Figure 6). Thus in vivo, Atg3 activity still requires a functional E3-like ligase and the increased membrane affinity of Atg3 mutants like K11W does not promote Atg5-independent lipidation within the cell. Indeed, Atg16/Atg5-12 likely includes functions beyond membrane targeting, directly activating Atg3 by reconfiguring the active site of Atg332. Atg3 may also regulate Atg16/Atg5-12 function as1) expression levels of Atg5 are sensitive to Atg3 levels7, 2) Atg3 and cargo molecules compete with Atg16/Atg5-12 for access to Atg8-PE37and3) soluble Atg3/Atg16L1/Atg5-12assemblies are detectable in overexpression experiments (e.g. Supplemental Figure 6). Unraveling how Atg3 mutants might influence Atg5 function and intracellular localization will be an important future direction.
Discussion
A curvature-sensing function for Atg3 is consistent with several studies. First, the high proportion of cone-shaped lipids required to lipidate LC3 in vitro16 implies a general advantage for the lipidation machinery on membranes with significant exposure to the hydrophobic core as would be expected at local areas of high curvature13. As high concentrations of cone-shaped lipids are also needed in vitro for yeast and plant autophagy proteins, we expect that this mode of membrane recognition is a general feature of macroautophagy15, 38. Second, the first seven amino acids of yeast Atg3 are already implicated in lipid-binding and show preference for PE-enriched membranes39. How this PE-sensing is accomplished was not determined, but we would now suggest that it arises from the participation of these amino acids in a hydrophobicity- or curvature-sensing amphipathic helix.
Amphipathic helices rely on combinations of hydrophobic interactions with the membrane interior and electrostatic interactions with the polar lipid headgroups. When either of these interactions is strong, membrane binding is efficient. However, at intermediate affinities, binding becomes dependent upon membrane defects like those arising from highly curved surfaces. Because the membrane charge density is higher for membranes late in the secretory cascade or at the plasma membrane, a strong dependence upon electrostatics suggests a plasma membrane directed role for curvature-sensing amphipathic helices, as has been observed for α-synuclein40. In contrast, helices with low charge density are more often associated with highly curved ER or golgi membranes. These include proteins that harbor an ALPS motif like ArfGAP113, 41, and the early-autophagosome protein Barkor12. Theamino-terminal helix of Atg3 has a charge density of zero and a mildly hydrophobic membrane interaction face, akin to many examples of ALPS domains, but it relies upon charged residues positioned at the water-lipid interface, and thus this helix has characteristics of both classes of curvature-sensing motifs. This in-between helical structure may indicate that Atg3 is designed to function at membranes that are compositionally distinct from Barkor targets, either because Atg3 acts later and therefore on a more mature autophagosomal membrane or because Atg3 is also needed at other sites including some membranes with a decidedly more plasma membrane/endocytic composition as occurs in LC3-associated phagocytosis42.
What kind of curvature might Atg3 target in vivo? Unfortunately, we have thus far been unable to visualize Atg3 on membranes in MEFs. Previous studies on proteins harboring an ALPS domain demonstrated that curvature-sensing only becomes robust when the membrane radius of curvature is less than 50 nm13. Atg3 is similar, with dramatic improvements in lipidation efficiency even as the liposome diameter changes from 50 nm extruded liposomes to sonicated liposomes. From combinations of dynamic light scattering and cryo-electron microscopy we know that the 50 nm extrusion process actually produce liposomes of around 90 nm in diameter while sonicated samples are smaller (Table 1), suggesting that both Atg3 binding and LC3/GABARAPlipidation are most efficient on very small liposomes. Intriguingly, tomography of stalled omegasomes, sites of autophagosome initiation in mammals, implies that the leading edge of a growing autophagosome exhibits a very strident curvature11. In individual micrographs of the omegasome, the two bilayers of the autophagosomal membrane are often not resolvable, indicating that the distance across the lumen of these growing structures may be as little as 10 nm. These structures accumulate specifically under conditions where LC3 lipidation is inhibited, including in Atg3 knockout cells, and are thus a likely cellular target for Atg3. By targeting short-lived curved membranes, the cell could achieve both spatial and temporal specificity in the lipidation reaction. Spatially, LC3 or GABARAP would be present at the leading edge of the autophagosome and thus be enriched at sites of cargo encapsulation or autophagosome closure, two activities these proteins are proposed to control6. Temporally, Atg3 would target growing IMs that carry a highly curved rim, but not fully-formed closed autophagosomes. As Atg8 proteins must be removed from the outer membrane prior to the final maturation of the organelle43, continued lipidation on the autophagosome itself would be nonproductive. Likewise, this mechanism would limit the non-specific coupling of LC3 to other membranes that do not exhibit the same strident curvature, complementing Atg4-mediated delipidation at non-autophagosomal membranes44, to promote general fidelity in the LC3-lipidation system. More broadly, Atg3 is just one of several autophagy proteins targeting the growing IM. Many of these proteins (but probably not all, e.g. 45) are released from the autophagosome shortly after or concurrent with its maturation to a closed vesicle46. Thus, perhaps many proteins in the pathway will share a preference for curved bilayers analogous to the behaviors of Atg3 (described here) and Barkor12 and suggested to be essential for Atg1747.
Supplementary Material
Acknowledgments
Atg3 KO MEFs were a gift from Masaaki Komatsu, (Tokyo Metropolitan Organization for Medical Research, Japan) and Atg5 KO MEFs from Noboru Mizishuma (Tokyo University). This work was funded by grants from the NIH (GM100930 - TM) and (NS063973 - TM and AY) and the ERC (advanced grant 268888 – BA) and by Yale fellowships to GP (BioSTEP) and TM (Anderson). We also thank Anjali Jotwani and Brian Mugo for help with electrophoresis experiments, Erdem Karatekin for help with dynamic light scattering and Frederic Pincet for helpful comments.
S.Nath conceived, designed and carried out liposome experiments, analysed the data and assisted with a draft of the manuscript. J.D. conceived, designed and performed cell culture experiments and assisted with statistical analyses. V.S. conceived, designed and performed cell culture experiments and assisted with statistical analyses. G.P. made the initial discovery of curvature sensing in a liposomal system and carried out experiments with lipid composition tests. W.M.F. conceived, designed and carried out experiments testing activity in Atg5 KO cell lines. S.Nag developed protein expression strategies and generated many of the tools supporting both the liposomal and cell culture studies. J.B. analysed imaging data. A.Y. conceived and designed experiments involved in Atg5 KO rescues and assisted with the final manuscript. B.A. conceived and designed experiments on liposome curvature studies, assisted with data analysis and assisted with the manuscript. T.J.M. conceived the project, designed experiments, carried out liposome experiments and data analysis and wrote the manuscript.
Footnotes
There are no competing financial interests for any of the authors involved in this study.
Citations
- 1.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 3.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. Journal of cell science. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 4.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. The EMBO journal. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sou YS, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishida Y, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. The Journal of cell biology. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS letters. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi-Nishino M, et al. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2009;6:301–303. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 15.Ichimura Y, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. The Journal of biological chemistry. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 16.Sou YS, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. The Journal of biological chemistry. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 17.Tanida I, Sou YS, Minematsu-Ikeguchi N, Ueno T, Kominami E. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. The FEBS journal. 2006;273:2553–2562. doi: 10.1111/j.1742-4658.2006.05260.x. [DOI] [PubMed] [Google Scholar]
- 18.Choy A, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science (New York, NY) 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh-oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. The Journal of biological chemistry. 2008;283:21847–21852. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- 20.Brownell JE, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 23.Hatzakis NS, et al. How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol. 2009;5:835–841. doi: 10.1038/nchembio.213. [DOI] [PubMed] [Google Scholar]
- 24.Vamparys L, et al. Conical Lipids in Flat Bilayers Induce Packing Defects Similar to that Induced by Positive Curvature. Biophys J. 2013;104:585–593. doi: 10.1016/j.bpj.2012.11.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanni S, et al. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys J. 2013;104:575–584. doi: 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamal MM, Mills D, Grzybek M, Howard J. Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:22245–22250. doi: 10.1073/pnas.0907354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hope MJ, Bally MB, Webb G, Cullis PR. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 28.Antonny B. Mechanisms of Membrane Curvature Sensing. Annu Rev Biochem. 2010 [Google Scholar]
- 29.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 30.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. The Journal of biological chemistry. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 32.Sakoh-Nakatogawa M, et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 33.Noda NN, Fujioka Y, Hanada T, Ohsumi Y, Inagaki F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013;14:206–211. doi: 10.1038/embor.2012.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 36.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS letters. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 38.Fujioka Y, et al. In vitro reconstitution of plant Atg8 and Atg12 conjugation systems essential for autophagy. The Journal of biological chemistry. 2008;283:1921–1928. doi: 10.1074/jbc.M706214200. [DOI] [PubMed] [Google Scholar]
- 39.Hanada T, Satomi Y, Takao T, Ohsumi Y. The amino-terminal region of Atg3 is essential for association with phosphatidylethanolamine in Atg8 lipidation. FEBS letters. 2009;583:1078–1083. doi: 10.1016/j.febslet.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Pranke IM, et al. alpha-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. The Journal of cell biology. 2011;194:89–103. doi: 10.1083/jcb.201011118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. The EMBO journal. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 43.Yu ZQ, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8 doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8:177–186. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- 45.Proikas-Cezanne T, Robenek H. Freeze-fracture replica immunolabelling reveals human WIPI-1 and WIPI-2 as membrane proteins of autophagosomes. J Cell Mol Med. 2011;15:2007–2010. doi: 10.1111/j.1582-4934.2011.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 47.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Helfrich W. Elastic properties of lipid bilayers: theory and possible experiments. Z Naturforsch C. 1973;28:693–703. doi: 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- 49.Jotwani A, Richerson DN, Motta I, Julca-Zevallos O, Melia TJ. Approaches to the study of Atg8-mediated membrane dynamics in vitro. Methods Cell Biol. 2012;108:93–116. doi: 10.1016/B978-0-12-386487-1.00005-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


