Abstract
Antisense oligonucleotides are short nucleic acid sequences designed for use as small-molecule drugs. They recognize and bind to specific messenger RNA (mRNA) or pre-mRNA sequences to create small double-stranded regions of the target mRNA that alter mRNA splicing patterns or inhibit protein translation. Antisense approaches have been actively pursued as a form of molecular medicine for more than 20 years, but only one has been translated to a marketed drug (intraocular human immunodeficiency virus treatment). Two recent advances foreshadow a change in clinical applications of antisense strategies. First is the development of synthetic DNA analogues that show outstanding stability and sequence specificity yet little or no binding to modulator proteins. Second is the publication of impressive preclinical and clinical data using antisense in an exon-skipping strategy to increase dystrophin production in Duchenne muscular dystrophy. As long-standing barriers are successfully circumvented, attention turns toward scale-up of production, long-term toxicity studies, and the challenges to traditional drug regulatory attitudes presented by tightly targeted sequence-specific drugs.
With the advent of recombinant DNA in the 1970s, it was soon realized that bacteria possess a form of regulatory machinery where small RNA transcripts can bind (hybridize) to other target RNAs and inhibit the translation of these targets.1 These antisense RNAs were subsequently recognized as natural translational regulation mechanisms in plants and higher organisms.2 More recently, a specialized form of antisense transcript was found to be a cellular defense mechanism against invading messenger RNAs (mRNAs) (viruses), and this has been harnessed as a popular method to “knock down” specific mRNA transcripts in cultured cell models (short interfering RNAs).3
Attention soon shifted toward development of antisense molecules as a form of small-molecule drug (antisense oligonucleotide [AO]). The approach was intuitive: one needs simply to chemically synthesize short pieces of DNA of about 20 bases, where a specific complementary sequence is designed to hybridize with a desired target mRNA. Such designer AO drugs should show very high specificity and selectivity for binding only the desired target RNA sequence of nucleotides that is predicted by base pairing. Beginning in the mid-1980s, this approach was put to the test in model systems and was shown to work quite well in shutting down the production of the target (undesired) protein.4 Isis Pharmaceuticals, Inc, Carlsbad, California, a company focused on clinical applications of AOs, was incorporated in 1989. Additional companies focusing on AO approaches soon followed.
Despite early promise, uses of AOs as small-molecule drugs have been painfully slow to enter the market and standard of care. Indeed, only a single AO drug has been approved by the Food and Drug Administration (FDA), fomivirsen sodium (Vitravene; Isis Pharmaceuticals, Inc) delivered by intravitreous injection to inhibit cytomegalovirus retinitis in AIDS. Vitravene was approved in 1998 and there have been no subsequent successful approvals in the ensuing 10 years.
What has slowed the progress of AO drugs into the clinical arena, and why may this be changing?
There have been 2 major hurdles: off-target toxic effects and potency or delivery. Regarding toxic effects, most organisms do not take kindly to covert infiltration by foreign DNA or RNA. Indeed, all species have quite effective mechanisms to destroy foreign DNA and RNA as they are more likely than not to be viruses or other undesirable organisms. In addition, many of the clinical trials testing AO drugs have seen evidence of activation of the complement cascade, and this has been a key concern of the FDA. Delivery has also been a consistent problem. Because the target RNAs are always intracellular, it is imperative for the AO drug to achieve intracellular concentrations sufficient to enable it to bind and modulate the target RNA to a significant extent. The fact that AOs typically do not easily cross the lipid bilayers that bound the cell so as to achieve sufficient intracellular potency via systemic (intravenous) delivery has been problematic.
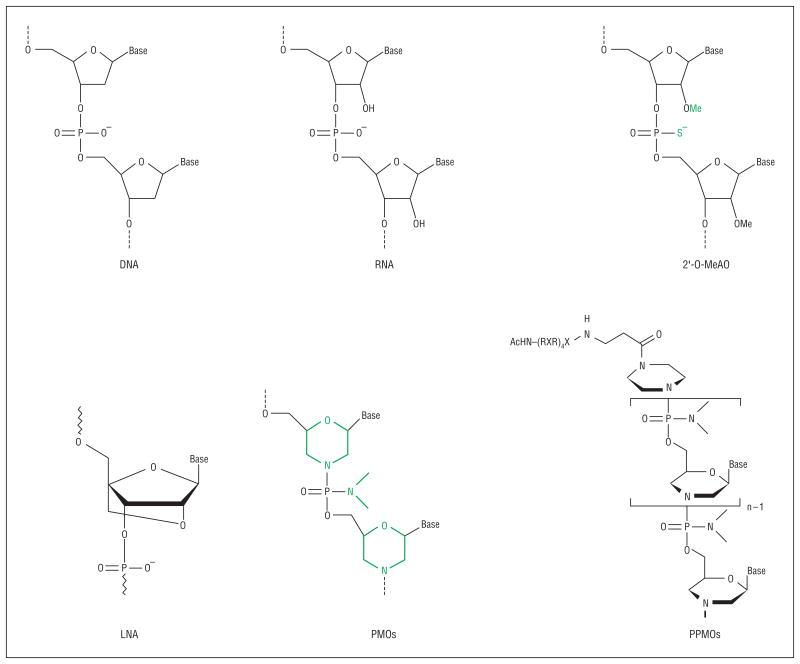
Recent developments are achieving success in overcoming both hurdles. Analogues of nucleic acid have been designed and synthesized in which the ribose backbone of RNA and DNA is replaced with different chemistries (Figure 1). Two are particularly promising: one uses a morpholino backbone (phosphorodiamidate morpholino oligomer [PMO]; AVI BioPharma, Portland, Oregon), and the second uses a locked nucleic acid backbone (Enzon Pharmaceuticals, Inc, Bridgewater, New Jersey). These new backbones are designed to maintain the molecular distance between bases (G, A, T/U, and C), enabling highly sequence-specific base pairing to the target RNA that is stronger in the case of PMO and locked nucleic acid drugs than DNA or RNA AOs. Equally important, these backbones are so dissimilar from the DNA and RNA ribose phosphodiester backbone that they are not recognized by most or any DNA and RNA binding proteins or degrading enzymes, thereby enhancing their stability and avoiding many or all off-target toxic effects.
Figure 1.
Comparison of chemistries used for the exon-skipping approach. Examples of artificially developed antisense oligomers such as 2′-O-methylated antisense oligonucleotides (2′-O-MeAO) (phosphorothioate), locked nucleic acid (LNA), phosphorodiamidate morpholino oligomers (PMOs), and peptide-tagged PMOs (PPMOs) are shown for comparison with DNA and RNA.
The second major barrier has been achieving sufficient intracellular concentrations (delivery). One successful approach is to take advantage of preexisting holes in the plasma membrane of the target cell. Infecting viruses breach the cell membrane during the process of infection and appear to bring along AO drugs in the process. As such, AOs have been quite successful in blocking downstream viral replication within cells, and PMO drugs are showing impressive promise as antiviral antidotes.5 Another preexisting hole is found in muscle cells lacking dystrophin (Duchenne muscular dystrophy [DMD]).6 The unstable plasma membrane of myofibers appears to allow the AO to leak into the cell.7 An additional approach is to modify the AO drugs with cell delivery moieties, chemical adducts that penetrate the cell membrane. One example is the addition of arginine-rich peptides to one end of the AO drug (peptide-tagged PMO) (Figure 1).
From these advances has sprung a resurgence of interest in AO drugs for treatment of genetic disease, cancer, and infectious disease. The purpose of most applications is to knock down a target RNA so that it makes less of the deleterious protein product (eg, tumor growth factor β or hypoxia-inducible factor 1α in cancer cells, viral mRNAs, or dominant gain-of-function toxic proteins in inherited neurological disease). However, the disorder that may be most advanced in such applications is DMD. Here the AOs are used for a quite different objective than for previous applications; explicitly, AOs in DMD are designed to restore function to the target mRNA and protein rather than block it. The remainder of this review focuses on this application.
Rationale and Proof of Principle of Exon-Skipping Therapy
The principle of exon-skipping therapy for dystrophinopathies was initially demonstrated by Dunckley et al8 in cultured mouse muscle cells in vitro. The rationale is as follows. Duchenne muscular dystrophy is caused by mutations of the 79-exon gene (commonly deletions of ≥1 exon). Within the myofiber, the remainder of the gene will be transcribed and spliced together. However, if the triplet codon reading frame of the mRNA is not preserved, the resulting frame shift will lead to the failure of dystrophin protein production. Becker muscular dystrophy (BMD) is a clinically milder and more variable disease in which mutations of the dystrophin gene are commonly such as to preserve the translational open reading frame; thus, after splicing together, the remainder of the gene retains some ability to synthesize the dystrophin protein. The goal of exon-skipping therapies is to force the dysfunctional mRNA with out-of-frame mutations in a patient with DMD to skip (exclude) some additional exons. The loss of additional material directed by AO drugs restores the reading frame, changing a Duchenne out-of-frame transcript to a Becker in-frame transcript. Fortunately, most mutations in the dystrophin gene occur in parts that do not code for functionally essential regions of the protein.
This AO-mediated exon-skipping method has been developed and extensively tested on the dystrophic mdx mouse model of DMD. The mdx mouse harbors a nonsense mutation in exon 23 that prevents translation beyond this point in the transcript. Both local intramuscular injection and systemic delivery of a single AO targeted against exon 23 in the primary transcript excludes this exon from the mRNA, leaving an in-frame transcript that generates dystrophin expression and produces a degree of functional recovery. Intramuscular and systemic injections of AOs for exon splicing of a dog model of DMD have also been demonstrated with a novel cocktail AO strategy (T.Y., S.T., Q.-L.L., T.A.P., A.N., E.P.H., and Masanori Kobayashi, DVM, unpublished data, 2006-2008). The principle is similarly illustrated in humans; van Deutekom et al9 reported single-site intramuscular injections of 2′-O-methyl AO chemistry in 4 boys with DMD, showing evidence of de novo dystrophin production at the injection site.
These data demonstrate that the key hurdles of achieving intracellular delivery and avoiding toxic effects can be cleared. A similar strategy is being explored in other diseases such as myotonia, human immunodeficiency virus, and spinal muscular atrophy.10-12
Hurdles in Bringing Exon Skipping to Standard of Care
Exon skipping using AO drugs has rapidly emerged as the frontline therapeutic approach for DMD. How soon can we expect exon skipping to reach the neuromuscular clinic and standard of care? This approach is breaking new ground and raising challenges not encountered previously in drug development. Different patients have different mutations, and many AO sequences will need to be designed, tested, and FDA approved. Also, current genotype and phenotype data suggest that there may be good in-frame deletions and not-so-good in-frame deletions; simply restoring the reading frame may not be synonymous with restoring dystrophin protein function. The optimization of dystrophin function will likely require deletions of multiple exons, and this will require mixtures of different AOs—new territory for drug development and the FDA. The approach will require regular injections of large amounts of AO drug; what are the long-term toxic effects? Moreover, are the standard toxicity tests appropriate for sequence-specific drugs? Each of these hurdles is discussed briefly in the remainder of this review.
Certain Exon Deletions May Retain More Dystrophin Function than Others
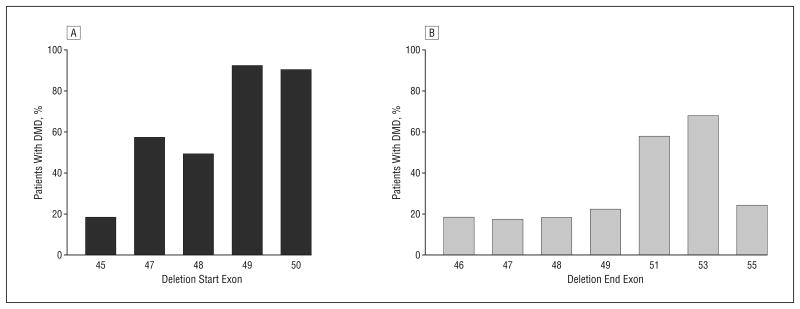
The molecular diagnostics of DMD and BMD frequently refer to the reading frame rule, where out-of-frame deletions are given a DMD diagnosis and in-frame deletions are given a BMD diagnosis. However, as many as 30% of patients with BMD do not adhere to this rule.13 A thorough understanding of reading frames is critical for appropriate design of exon-skipping therapies, both so that the best AO can be given to the patient and so that an optimally functional dystrophin protein is produced as a result of the expected exon skipping. Currently, the best information from which to predict the capabilities of partially deleted dystrophins to rescue the DMD phenotype comes from analysis of the thousands of genotype and phenotype correlations in patients with DMD and BMD that have been published in the literature and on the Internet. We examined all in-frame deletions and determined the proportion of observed cases that showed mild or severe phenotypes. This was gleaned from combined muscular dystrophy databases of 14 countries (from Argentina, Belgium, Brazil, Bulgaria, Canada, China, Denmark, France, India, Italy, Japan, The Netherlands, the United Kingdom, and the United States) at Leiden University (http://www.dmd.nl), excluding diagnoses that did not allow deletion boundaries to be assigned accurately to a specific exon.14 Of all observed in-frame deletion patterns on genomic DNA in the central rod domain hot-spot region (exons 42-57; 28 distinct patterns), 57% (16 of 28 patterns) were associated with DMD rather than BMD. This analysis showed that there are considerable discrepancies between population-based ratios and pattern-based proportions of severe DMD vs mild BMD phenotypes, and interestingly, the ratio of DMD to BMD remarkably varies between specific deletion patterns. For example, in-frame deletions starting or ending around exon 50 or 51 that encode the hinge region were most commonly associated with severe phenotypes (Figure 2) (eg, deletions at exons 47-51, 48-51, and 49-53 are all reported to be associated with a severe DMD phenotype rather than BMD).15,16
Figure 2.
Clinical phenotypes associated with specific start (A) and end (B) sites for in-frame deletions. Percentages of patients with Duchenne muscular dystrophy (DMD) out of patients with DMD or Becker muscular dystrophy with specific start and end exons are shown. Combined muscular dystrophy databases of 14 countries (from Argentina, Belgium, Brazil, Bulgaria, Canada, China, Denmark, France, India, Italy, Japan, The Netherlands, the United Kingdom, and the United States) at Leiden University (http://www.dmd.nl), where diagnoses were performed using multiplex ligation-dependent probe amplification/multiplex amplification and probe hybridization, Southern blotting, or polymerase chain reaction primer sets that allow deletion boundaries to be assigned accurately to a specific exon, are used (deletion start sites: n=288 for exon 45, n=23 for exon 47, n=9 for exon 48, n=12 for exon 49, and n=10 for exon 50; deletion end sites: n=11 for exon 46, n=115 for exon 47, n=95 for exon 48, n=51 for exon 49, n=53 for exon 51, n=40 for exon 53, and n=21 for exon 55).
Two questions arise. First, why do specific patterns of in-frame mutations tend to result in a severe DMD phenotype in contradiction to the reading frame rule? Second, why do different individuals with the same exonic deletion pattern exhibit such different clinical phenotypes? Likely contributory factors include the following: the effect of the specific deletion breakpoints on mRNA splicing efficiency and/or patterns; translation or transcription efficiency after genome rearrangement; and stability or function of the truncated protein structure. The mechanisms controlling accurate splicing of the 79-exon, 2.4 million–base pair dystrophin gene are clearly complex. Introns of the dystrophin gene are highly variable in size, and it is likely that exonic splicing does not take place in an ordered 5′ to 3′ sequence. A complication in interpreting genotype and phenotype correlations is that the deletion in genomic DNA does not always correspond to the material missing from the resulting mRNA. We and others have shown that even in the absence of AOs, a patient may produce 1 or more transcripts that skip additional exons present in the genomic DNA, in effect performing their own private exon skipping.13 Disruption of splice site information (such as an intervening sequence) in some patients with in-frame gene deletions may cause skipping of additional exons at mRNA splicing, thus leading to out-of-frame transcripts from an in-frame genomic DNA deletion as Kesari et al13 have recently described. As Menhart17 has pointed out, it is also likely that quasi-dystrophin variants in the rod domain may show different stability or function because of different types of derangement of spectrinlike repeat domains. Not enough is known about dystrophin structure and function, and the relative importance of the protein sequence within the rod domain remains entirely a matter of speculation. Historically, lack of dystrophin expression has been used as the key criterion for DMD diagnosis. This together with the presence of the DMD clinical picture with such in-frame mutations argues that other confounding variables such as imprecisely defined mutation or aberrant splicing may explain these “exceptions to the reading frame rule.” Thus, it is anticipated that most or all patients with mutations in the central rod domain would benefit from the production of truncated dystrophin.
Paralleling AO Trials: Testing New Exons and Mixtures
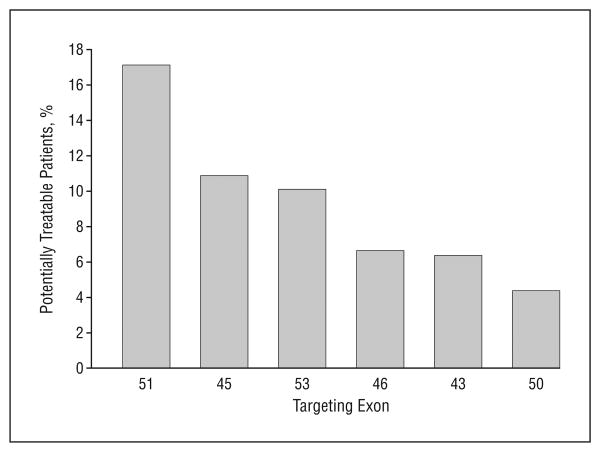
Clinical proof-of-concept trials testing limited intramuscular injection with a 2′-O-methyl AO against exon 51 have been published,9 and similar studies with PMO chemistry are under way in the United Kingdom. Given the many questions concerning the sequence specificity of toxic effects and the large number of AO sequences that will need to be developed as drugs to treat most patients with DMD, it is critical to parallel studies on many more AOs for DMD (Figure 3).
Figure 3.
Targets of exon skipping and population of potentially treatable patients. Percentage of patients with the dystrophin deletion who are potentially treatable by targeting specific exons for Duchenne muscular dystrophy. For example, 17% of patients with Duchenne muscular dystrophy who have the dystrophin deletion can be potentially treated by targeting exon 51 using antisense oligonucleotides.
It should be noted that about 30% of patients with DMD have nondeletion mutations (duplication, nonsense mutations, small rearrangement, or splice site mutations). Most mutations are theoretically amenable to exon skipping; however, there are no hot spots for point mutations, so relatively few patients would be treatable with each targeted exon by comparison with deletion mutations. Moreover, if skipped to remove a nonsense mutation, the exons that are candidates to restore the reading frame in patients with deletions (eg, exons 43, 45, 46, 50, 51, and 53) will require additional deletion of at least 1 further exon to restore the reading frame because these are frame-shifting exons. Thus, only 35% of nonsense mutations are potentially treatable by single-exon targeting, but the combined data of the Leiden DMD mutation database imply that more than 90% could be responsive to multiskipping.14
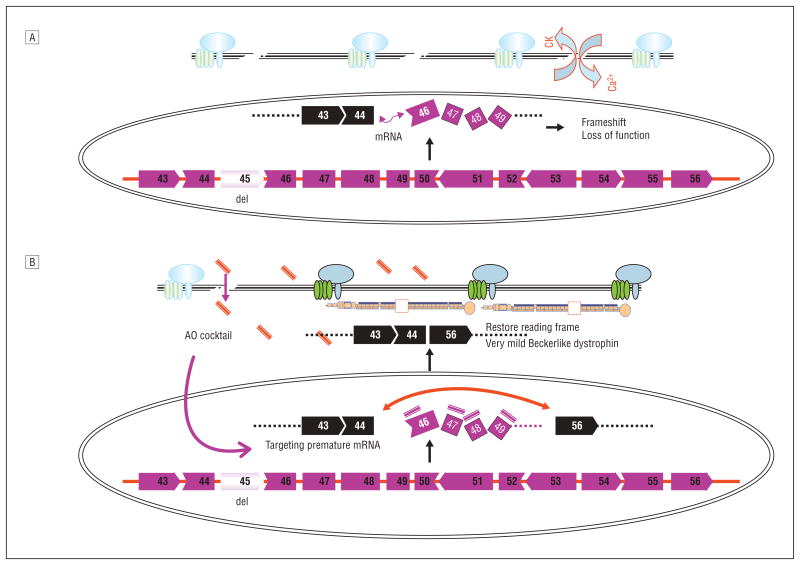
Development of exonic cocktails (mixtures) could resolve a number of problems, including optimization of dystrophin function and covering relatively high proportions of patients with DMD with a single mixture. The mixture approach has clear advantages and disadvantages. As an example, an 11-exon AO cocktail skipping exons 45 through 55 is predicted to result in a particularly mild BMD phenotype (94% of reported patients).14 Encouragingly, this large deletion is regularly associated with clinically milder phenotypes than any of the smaller in-frame deletions within the same range of exons 45 to 55.18 A second advantage is that the cocktail could conceivably be approved as a single drug for most patients with DMD who have dystrophin deletions, independent of their precise deletion, eg, an 11-exon AO cocktail targeting exons 45 through 55 is potentially applicable to more than 60% of patients with a dystrophin deletion (Figure 4).18,19 In total, more than 90% of patients with DMD could potentially be treated by multiskipping, whereas single-exon skipping could treat around half of the patients with dystrophin deletions and point mutations. Systemic studies in the large dog model of DMD have been done using a 3-exon PMO cocktail, and this has clearly been shown to be efficacious by multiple clinical, imaging, histological, and biochemical or molecular end points (T.Y., S.T., Q.-L.L., T.A.P., A.N., E.P.H., and Masanori Kobayashi, DVM, unpublished data, 2006-2008).
Figure 4.
Mechanism of multiexon skipping of exons 45 through 55 to rescue 60% of patients with Duchenne muscular dystrophy with dystrophin deletions A, More than 60% of deletion mutations of the dystrophin gene occur within the hot-spot range of exons 45 through 55 (exon 45 is deleted in this schematic [del]) in Duchenne muscular dystrophy muscles. The messenger RNA (mRNA) of remaining exons is spliced together but the reading frame is disrupted, resulting in failure of the production of functional dystrophin protein. CK indicates creatine kinase; Ca2+, calcium ions. B, An antisense oligonucleotide (AO) cocktail targeting exons 45 through 55 likely enters the Duchenne muscular dystrophy muscle through its leaky membranes, then binds to the dystrophin mRNA in a sequence-specific manner. The AOs block the splicing machinery and prevent inclusion of all exons between exons 45 and 55. Skipping these exons restores the reading frame of mRNA, allowing production of quasi-dystrophin containing exons 1 through 44 and exons 56 through 79, which is not normal but likely retains considerable function as evidenced by patients with clinically milder Becker muscular dystrophy with identical partial dystrophin.
A disadvantage of the cocktail approach is the addition of novel hurdles for FDA or regulatory approval. Current FDA regulations require each component of a drug mixture to undergo toxicological and clinical testing and then require the mixture to similarly undergo toxicological and clinical testing. In the context of an ideal 11-exon AO cocktail, the regulatory barriers become truly intimidating. In addition, the 11-exon cocktail PMO approach would lead to delivery of some AOs that may not have a target in a specific patient (eg, the patient already has a deletion of ≥1 exon in the AO mix). Thus, some parts of the mixture will have no possible potential molecular or clinical benefit to individual patients. This would again be uncharted territory for the FDA. While clinical development of the 11-exon mixture is likely ambitious at present, it will be important to initiate toxicological and clinical trials of exon mixtures for subsets of patients who cannot be treated with a single AO. Also, for future trials on multiskipping such as with exons 45 through 55, we should have as many AOs in hand as possible because they can be used as part of multiskipping AOs.
Personalized Medicine and the FDA: Are Existing Guidelines Appropriate?
Personalized medicine has many definitions, but most share the concept of optimizing a treatment for a particular patient. Designing and using AO drugs targeted for a patient's specific gene mutation would seem to fit well within this rubric. As such, the promising AO exon-skipping approach may bring neuromuscular disease to the frontline in development of drugs for personalized medicine. It is important to examine the existing FDA guidelines for drug development and reinterpret these guidelines in the context of AO and DMD. For example, the drug development pipeline includes phase 1 studies of the drug in healthy volunteers. However, successful on-target exon skipping of the dystrophin gene in healthy volunteers would give them DMD, a clear adverse effect that is entirely irrelevant to toxic effects in the target patient population (boys with DMD). Toxicity tests are currently done in animal models (typically 2 species), but one of the major concerns regarding toxic effects of AO drugs is binding to off-target RNAs. For example, if an AO drug designed for exon skipping of the dystrophin mRNA also binds to the closely related utrophin mRNA, then exon skipping of utrophin might occur and could result in off-target adverse effects. The utrophin sequence of mice or rats is different from the utrophin sequence of humans, so the standard rodent toxicity tests may not accurately assess off-target toxic effects of AOs for human use.
Perhaps the largest challenge facing implementation of exon-skipping therapy for DMD is in developing new approaches to toxicity testing and clinical trial regulatory procedures that are relevant and appropriate for sequence-specific drugs. The pharmaceutical industry often quotes a price tag of $500 million to bring any new drug to the market. Given the discussion earlier, implementation of AO drugs in DMD will require many exon-specific drugs. If the $500 million is assessed for each individual AO sequence, then both time and money become insuperable barriers to helping the existing generation of boys with DMD. The silver lining in this cloud is the lack of any detectable toxic effects with PMO AO drugs to date. If multiple AOs all show a lack of long-term toxic effects, then there is hope that specific AO drugs could be approved with more limited toxicological and phase 1 testing.
A practical resolution of this problem is to consider each component of the potential toxic effects of these highly targeted drugs individually. Tests of the generic toxic effects of morpholinos at the doses at which they are likely to be functionally effective could be conducted quite straightforwardly with either a scrambled or arbitrary sequence of a particular molecular weight. It is the notion of individual sequence-specific toxic effects that raises problems. The argument that any specific sequence may have off-target effects (eg, binding to utrophin transcripts) cannot be properly tested in other species because they may have different potential off-target sequences as compared with those in humans. This carries the dire implication that a lack of sequence-specific toxic effects in a test species can provide no assurance, indeed no information at all, as to the sequence's safety in humans. Tests in healthy human volunteers are also problematic. Ethical issues arise from the possible generation of a pathogenic frameshift in healthy muscle by successful suppression of the targeted exon. Moreover, the lack of innate pathological abnormalities in healthy human muscle would stifle access of the AO to its intended intramuscular target while at the same time providing a different spectrum of potential off-target molecules (eg, utrophin transcripts). A further complication arises from the individualistic nature of the entire rationale, for it precludes the possibility of learning from experience; the probability that any given sequence may be toxic is independent of the number of safe experiences with other sequences. In effect, for safety, we can test for sequence-specific toxic effects only in human volunteers with DMD by progressive dose escalation. Only in this way would the reagents have access to their intended targets as well as any unintended targets in a physiological context that is inappropriately modeled both in other species and in healthy human volunteers.
Acknowledgments
Funding/Support: This work was supported by the Foundation to Eradicate Duchenne, Jane Foundation, the Muscular Dystrophy Association, and a collaborative grant from the National Institutes of Health Wellstone Muscular Dystrophy Research Centers (http://www.wellstone-dc.org).
Footnotes
Author Contributions: Study concept and design: Yokota, Takeda, Partridge, and Hoffman. Acquisition of data: Yokota and Lu. Analysis and interpretation of data: Yokota, Nakamura, and Hoffman. Drafting of the manuscript: Yokota, Partridge, Nakamura, and Hoffman. Critical revision of the manuscript for important intellectual content: Takeda, Lu, and Partridge. Obtained funding: Hoffman. Administrative, technical, and material support: Lu. Study supervision: Yokota, Takeda, Partridge, Nakamura, and Hoffman.
Financial Disclosure: None reported.
Contributor Information
Dr Toshifumi Yokota, Research Center for Genetic Medicine, Children's National Medical Center, Washington, DC.
Dr Shin'ichi Takeda, Department of Molecular Medicine, National Institutes for Neuroscience, Tokyo, Japan.
Dr Qi-Long Lu, McColl-Lockwood Laboratory for Muscular Dystrophy Research, Neuromuscular/ALS Center, Carolinas Medical Center, Charlotte, North Carolina.
Dr Terence A. Partridge, Research Center for Genetic Medicine, Children's National Medical Center, Washington, DC.
Dr Akinori Nakamura, Department of Molecular Medicine, National Institutes for Neuroscience, Tokyo, Japan.
Dr Eric P. Hoffman, Research Center for Genetic Medicine, Children's National Medical Center, Washington, DC.
References
- 1.Spiegelman WG, Reichardt LF, Yaniv M, Heinemann SF, Kaiser AD, Eisen H. Bidirectional transcription and the regulation of phage lambda repressor synthesis. Proc Natl Acad Sci U S A. 1972;69(11):3156–3160. doi: 10.1073/pnas.69.11.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar M, Carmichael GG. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev. 1998;62(4):1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418(6896):430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 4.Kim SK, Wold BJ. Stable reduction of thymidine kinase activity in cells expressing high levels of anti-sense RNA. Cell. 1985;42(1):129–138. doi: 10.1016/s0092-8674(85)80108-2. [DOI] [PubMed] [Google Scholar]
- 5.Warfield KL, Swenson DL, Olinger GG, et al. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2006;2(1):e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman EP, Brown RH, Jr, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman EP. Skipping toward personalized molecular medicine. N Engl J Med. 2007;357(26):2719–2722. doi: 10.1056/NEJMe0707795. [DOI] [PubMed] [Google Scholar]
- 8.Dunckley MG, Villiet P, Eperon IC, Dickson G. Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet. 1998;7(7):1083–1090. doi: 10.1093/hmg/7.7.1083. [DOI] [PubMed] [Google Scholar]
- 9.van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 10.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117(12):3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asparuhova MB, Marti G, Liu S, Serhan F, Trono D, Schumperli D. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J Gene Med. 2007;9(5):323–334. doi: 10.1002/jgm.1027. [DOI] [PubMed] [Google Scholar]
- 13.Kesari A, Pirra LN, Bremadesam L, et al. Integrated DNA, cDNA, and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum Mutat. 2008;29(5):728–737. doi: 10.1002/humu.20722. [DOI] [PubMed] [Google Scholar]
- 14.Yokota T, Duddy W, Partridge T. Optimizing exon skipping therapies for DMD. Acta Myol. 2007;26(3):179–184. [PMC free article] [PubMed] [Google Scholar]
- 15.Covone AE, Lerone M, Romeo G. Genotype-phenotype correlation and germline mosaicism in DMD/BMD patients with deletions of the dystrophin gene. Hum Genet. 1991;87(3):353–360. doi: 10.1007/BF00200919. [DOI] [PubMed] [Google Scholar]
- 16.Talkop UA, Klaassen T, Piirsoo A, et al. Duchenne and Becker muscular dystrophies: an Estonian experience. Brain Dev. 1999;21(4):244–247. doi: 10.1016/s0387-7604(99)00016-9. [DOI] [PubMed] [Google Scholar]
- 17.Menhart N. Hybrid spectrin type repeats produced by exon-skipping in dystrophin. Biochim Biophys Acta. 2006;1764(6):993–999. doi: 10.1016/j.bbapap.2006.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Béroud C, Tuffery-Giraud S, Matsuo M, et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat. 2007;28(2):196–202. doi: 10.1002/humu.20428. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura A, Yoshida K, Fukushima K, et al. Follow-up of three patients with a large in-frame deletion of exons 45-55 in the Duchenne muscular dystrophy (DMD) gene. J Clin Neurosci. 2008;15(7):757–763. doi: 10.1016/j.jocn.2006.12.012. [DOI] [PubMed] [Google Scholar]