Abstract
Fibroblast growth factors (FGFs) and their receptors (FGFRs) play significant roles in vertebrate organogenesis and morphogenesis. FGFR3 is a negative regulator of chondrogenesis and multiple mutations with constitutive activity of FGFR3 result in achondroplasia, one of the most common dwarfisms in humans, but the molecular mechanism remains elusive. In this study, we found that chondrocyte-specific deletion of BMP type I receptor a (Bmpr1a) rescued the bone overgrowth phenotype observed in Fgfr3 deficient mice by reducing chondrocyte differentiation. Consistently, using in vitro chondrogenic differentiation assay system, we demonstrated that FGFR3 inhibited BMPR1a-mediated chondrogenic differentiation. Furthermore, we showed that FGFR3 hyper-activation resulted in impaired BMP signaling in chondrocytes of mouse growth plates. We also found that FGFR3 inhibited BMP-2- or constitutively activated BMPR1-induced phosphorylation of Smads through a mechanism independent of its tyrosine kinase activity. We found that FGFR3 facilitates BMPR1a to degradation through Smurf1-mediated ubiquitination pathway. We demonstrated that down-regulation of BMP signaling by BMPR1 inhibitor dorsomorphin led to the retardation of chondrogenic differentiation, which mimics the effect of FGF-2 on chondrocytes and BMP-2 treatment partially rescued the retarded growth of cultured bone rudiments from thanatophoric dysplasia type II mice. Our findings reveal that FGFR3 promotes the degradation of BMPR1a, which plays an important role in the pathogenesis of FGFR3-related skeletal dysplasia.
Keywords: FGFR3, BMPR1, Achondroplasia, Smurf1, Chondrocyte
1. Introduction
Fibroblast growth factors (FGFs) and their receptors (FGFRs) play essential roles in vertebrate organogenesis and morphogenesis by controlling cell proliferation, differentiation, migration, and apoptosis. Germline activation mutations of fibroblast growth factor receptor 3 (FGFR3) are responsible for achondroplasia (ACH) and several other types of chondrodysplasia including hypochondroplasia (HCH) and thanatophoric dysplasia (TD) [1–4]. Several groups have demonstrated that activation of FGFR3 inhibits chondrocyte proliferation and blocks its differentiation and matrix synthesis [5–14], which is thought to be responsible for the development of achondroplasia and chondrodysplasia. The skeletal development is precisely regulated by the integrated effects of diverse signaling pathways [15–17] and dysregulation of these signaling pathways can lead to a variety of skeletal dysplasia [18–20]. Currently, many signaling pathways including IGF, BMP, PTH, Wnt and Ihh have been also identified in regulation of endochondral ossification during the development of achondroplasia [20–24].
BMPs are members of the transforming growth factor-β (TGF-β) superfamily. BMPs trigger cellular responses through canonical (Smad) and noncanonical (non-Smad) pathways [25–28]. Smad ubiquitylation regulatory factor 1 (Smurf1) and Smurf2 are members of the Nedd4 (neural precursor cell-expressed developmentally downregulated gene 4) family and are key negative regulators of TGF-β/BMP signaling pathway [29]. It is well documented that Smurf1 binds to Smad1/Smad5 and BMPR1a, causing their ubiquitination and subsequent proteosomal degradation [30,31]. Accumulating evidence suggests that BMP/BMPR1 signaling is indispensable in regulation of chondrogenesis and endochondral ossification [21,32–37]. Conventional knockout of Bmpr1a leads to embryonic lethality [38] and cartilage-specific Bmpr1a null mice (Bmpr1a-cKO) shows a mild generalized chondrodysplasia with shortened long bones and delayed endochondral ossification, as well as smaller and flattened rib cages, which leads to distressed respiratory function and subsequent post-natal death [21].
Interestingly, both Bmpr1a-cKO and Fgfr3K644E germline activated mutant mice (mimicking human TDII) have chondrodysplasia and early post-natal death phenotype resulting from under-developed rib cages [21,39]. These observations support the notion that BMP and FGFR3 pathways are antagonistic in the regulation of chondrogenesis and endochondral development. It has been demonstrated that mice mimicking human ACH had decreased BMP-4 expression in post-natal growth plates [24], and the retarded development of the cultured embryonic limbs of ACH mice could be rescued by addition of BMP-2 [40]. These studies implied that BMP and FGFR3 signaling pathways might cross talk in regulation of chondrogenesis and development of achondroplasia.
A number of studies reported that FGF and BMP signaling interplay during the development of multiple cell lineages and tissues [41–43]. Activated MAP kinase cascades, especially ERK1/2 MAPK, as an important downstream pathway of FGFs/FGFRs signaling, can phosphorylate the linker region of Smad1/5, which not only causes Smurf1-dependent ubiquitination and degradation of the Smads, but also prevents Smad1/5 from translocation into the nucleus by inhibiting their interaction with the nuclear pore complex protein Nup214 [44]. However, the crosstalk between BMP and FGFR3 in chondrogenesis and achondroplasia remains unknown.
In this study, we report that FGFR3 antagonizes the BMP signaling during chondrogenesis and development of achondroplasia. We propose that FGFR3 inhibits chondrocyte differentiation by triggering Smurf1-mediated BMPR1 degradation, which may play an essential role in the pathogenesis of FGFR3-related skeletal dysplasia.
2. Materials and methods
2.1. Mice
Fgfr3G369C/+ mice (ACH) [7], Fgfr3 knockout mice (Fgfr3−/−) [6], Fgfr3+/K644Eneo mice [39], Bmpr1aflox/flox mice [45] and EIIa-Cre mice [46], Col2a1-Cre [47] were reported previously. Animals were handled in accordance with guidelines approved by the Institutional Animal Care and Use Committee of Daping Hospital, Third Military Medical University.
2.2. Skeletal preparation and histology
Skeletal preparations were performed as we previously described [48]. Briefly, mice were eviscerated and fixed in 95% EtOH at 4 °C overnight, followed by Alcian blue staining (30 mg/100 ml in 80% EtOH) at room temperature. Samples were stained in Alizarin red (5 mg/100 ml in 0.5% KOH) and cleared in a series of graded KOH in glycerol. For histology, samples were fixed in 4% paraformaldehyde, decalcified and embedded in paraffin. Paraffin sections (4–6 μm) were cut for histological analysis.
2.3. Primary chondrocyte culture
Primary chondrocytes were isolated from cartilage of knee joints of 5 day old mice. Briefly, dissected tissues with cartilage were first digested with 0.25% trypsinase/DMEM at 37 °C for 15 min to remove muscles, ligaments and bone tissues. Chondrocytes were isolated from the knee joints by further digesting with 0.1% collagenase II/DMEM for overnight in a CO2 incubator at 37 °C. Cells were plated in 12-well plates at a density of 1 × 105 cells/cm2, cultured in DMEM/F12 (1:1) medium supplemented with penicillin/streptomycin and 10% FBS until reaching sub-confluence.
2.4. Plasmids and cell lines
Full length mouse FGFR3 cDNA was cloned from SK-R3 and inserted into the pcDNA3.1/Myc vector (Clontech, USA) (Myc-FGFR3). Wild-type and Y373C, K650M and K508M-FGFR3 with flag-tag were kindly provided by Pavel Krejci (Masaryk University, Czech Republic) [49]. Myc-Smurf1 and Flag-Smurf1 (C699A) plasmids were provided by Lingqiang Zhang [50] (Beijing Institute of Radiation Medicine, China). HA-BMPR1a, HA-BMPR1b, caBMPR1b and GCCG-Luc plasmids were provided by Yeguang Chen (Tsinghua University, China). HA-caBMPR1a (Q227D) was a gift from Dr. Imamura (Cancer Institute-JFCR, Department of Biochemistry). pFR-Luc and pFA-ELK1 plasmids were purchased from Clontech.
HEK293T, HepG2, NIH3T3, HeLa and C3H10T1/2 cells were cultured in DMEM supplemented with 10% FBS. ATDC5 were cultured in DMEM/F12 (1:1) supplemented with 5% FBS. All the cells were kept at 37 °C in 5% CO2-containing atmosphere. The medium and serum were purchased from Hyclone.
2.5. Reagents and antibodies
FGF-2 and BMP-2 were purchased from R&D Systems, chloroquine and PD173074 were from Sigma. MG132 was obtained from Calbiochem. Antibodies were from various sources, including Anti-Actin (AC-15) and Anti-Flag M2 from Sigma, anti-Myc (9E10), anti-HA (F-7), anti-GFP (FL), anti-FGFR3, anti-BMPR1b, and anti-PCNA from Santa Cruz, anti-pSmad1/5, anti-Smad1, and anti-Smad5 from Cell Signal Corporation, anti-Smurf1 antibody was from Invitrogen and anti-BMPR1a antibody from R&D Systems. Anti-pSmad1S214 was kindly provided by Eddy De Robertis [51].
2.6. Western blot analysis
Cells were lysed in an ice-cold buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, and a cocktail of pro-tease inhibitors (Roche). Protein samples (10 μg) were resolved on a 10% or 12% SDS-PAGE gel and transferred onto a PVDF membrane (Millipore). Then indicated antibodies were used to probe the blots followed by ECL signal detection (Pierce).
2.7. Co-immunoprecipitation
HEK293T cells were plated in 60 mm dishes the day before transfection. Myc-Smurf1 and HA-BMPR1a (or other plasmids as indicated) were transfected individually or together. pcDNA3.1/Myc empty vector was added to balance the equal transfection efficiency. After transfection for 24–36 h, cells were harvested and lysed in 600 μl TNN buffer (50 mM TrisCl, 150 mM NaCl, 50 mM NaF, 0.5% NP-40, pH7.5) with protease inhibitors to prepare whole-cell lysates. Samples were centrifuged at 4 °C at top speed to isolate the supernatant, 500 μl lysate was mixed with indicated antibodies and incubated at 4 °C for 1–2 h, followed by addition of protein G or A-agarose beads to pellet the immune complex. The immunoprecipitants and 5% of lysates were analyzed by immunoblotting.
For endogenous coimmunoprecipitations, the whole lysates of HepG2 or siRNA-transfected HeLa cells were extracted. Aliquots were taken as lysate samples, and the remainders were subjected to immunoprecipitation with an antiserum against Smurf1 or with a preimmune serum (IgG) control. Purified immune complexes were probed for BMPR1a (or Smurf1, FGFR3) by immunoblotting.
2.8. Luciferase assay
NIH3T3 cells were plated in 24-well plates the day before transfection. 0.1 μg reporter plasmid (BRE-Luc, GCCG-Luc, pFR-Luc) together with 5 ng internal control plasmid (pRL-TK) was transfected each well. Constructs expressing HA-caBMPR1a, pcDNA3.1/Myc-FGFR3, Flag-FGFR3 or its mutants or empty vector were co-transfected as indicated, 1.0 μg each per well. 24–36 h after transfection, the reporter gene activity was assayed using the Dual Luciferase Assay System (E1910; Promega). For inhibitor assay, the transfected cells were treated by different inhibitors for 6 h before collecting the cells or as indicated.
2.9. Real-time RT-PCR
Total RNA was isolated by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. All reactions were performed in Mx3000P PCR machine (Stratagene) using the Two-Step QuantiTect SYBR Green RT-PCR Kit (Takara) and reaction conditions were optimized for each of the genes by changing the annealing temperature. Each run consisted of samples for genes of interest and cyclophilin A. The primers used were as follows: cyclophilin A (5′-CGAGCTCTGAGCACTGGAGA-3′ and 5′-TGGCGTGTAAAGTCACCACC-3′), Id-1 (5′-AGGTGAACGTCCTGCTCTAC-3′ and 5′-GTCCCGACTTCAGACTCCG-3′), Id-2 (5′-CGGTGAGGTCCGTTAGGAAAA-3′ and 5′-CATGTTGTAGAGCAGACTCATCG-3′), Id-3 (5′-CGACCGAGGAGCCTCTTAG-3′ and 5′-GCAGGATTTCCACCTGGCTA-3′), Ihh (5′-CAATCCCGACATCATCTTCA-3′and 5′-GCGGCCCTCATAGTGTAAAG-3′), Collagen II (5′-CTGGTGGAGCAGCAAGAGCAA-3′ and 5′-CAGTGGACAGTAGACGGAGGAAAG-3′), Sox9 (5′-TTCCTCCTCCCGGCATGAGTG-3′ and 5′-CAACTTTGCCAGCTTGCACG-3′), Col10 (5′-GCAGCATTACGACCCAAGAT-3′ and 5′-CATGATTGCACTCCCTGAAG-3′), Mmp13 (5′-CAGTTGACAGGCTCCGAGAA-3′ and 5′-CGTGTGCCAGAAGACCAGAA-3′). BMPR1a (5′-TCATGTTCAAGGGCAGAATCTAGA-3′ and 5′-GGCAAGGTATCCTCTGGTGCTA-3′), BMPR1b (5′-CCTCGGCCCAAGATCCTAC-3′ and 5′-CCTAGACATCCAGAGGTGACA-3′).
2.10. In vitro culture of metatarsal rudiment
Metatarsal rudiments were isolated from 18.5 pc pregnant female mice. Three to five left–right paired central bones were used in experiments. Rudiments were cultured in 24-well plates with 350 ml BGJb medium supplemented with 0.05 mg/ml ascorbic acid, 0.5 mg/ml L-glutamine, 10 mg/ml streptomycin, 10 units/ml penicillin, 1 mM β-glycerophosphate and 0.2% BSA (fraction V). Explants were grown at 37 °C in a humidified 5% CO2 incubator. FGF-2 (20 ng/ml) or dorsomorphin (10 μM) was added to cultures 16–20 h after dissection. Medium was changed on the second day of culture. The rudiments were observed and photographed under a dissecting microscope (Leica) at 0 and 4 or 7 d of treatment. The total, mineralization, hypertrophic and proliferation length of metatarsal rudiments were measured as described previously [52] with an eyepiece scale. Changes of bone length were expressed as percentage increase relative to the value before the treatment (percentage increase = length at day – length at day 0]/length at day 0). Data are expressed as mean ± SD, and the significance of differences was evaluated with Student’s t test.
2.11. Transfection of C3H10T1/2 and ATDC5 cells and chondrogenic differentiation assay
C3H10T1/2 cells were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. The transfected cells were trypsinized, inoculated as micromass, and induced for chon-drogenesis by applying recombinant BMP-2 (300 ng/ml). At the time point of 3 and 7 days, mRNA was collected for real-time RT-PCR analyses for chondrogenic differentiation marker genes.
ATDC5 cells were transfected with indicated plasmids. To induce chondrogenesis, cells were cultured in the medium supplemented with 10 μg/ml human insulin. The medium was replaced every 2–3 days. At the time point of 3 and 7 days, mRNA was collected for real-time RT-PCR analyses.
2.12. Immunohistochemistry
Skeletal preparations were immunostained using SP-9000 Histostain-Plus kits from Beijing Zhongshan Biotechnologies. Mice were sacrificed and the tibiae were fixed in 4% paraformaldehyde, decalcified in 15% EDTA (pH 7.4) and embedded in paraffin; paraffin sections (4–6 μm) were cut for histological analysis. Sections were pretreated with 0.1% trypsin for antigen demasking. Sections were blocked with 5% goat serum, incubated with indicated primary antibody overnight at 4 °C, and then incubated with the appropriate secondary antibody at room temperature. Color was developed with diaminobenzidine solution and sections were counterstained with Alcian blue.
2.13. RNAi protocols
The siRNA targeting Smurf1, FGFR3 and non-target control were ordered from Dharmacon (Thermo Fisher Scientific). HeLa cells or HepG2 was transfected with 50 nM siRNA using DharmoFect 4 (Thermo Fisher Scientific) according to the manufacturer’s instructions. 48 h later, the total protein was extracted and the protein level was assayed by IP or Western blotting.
2.14. Statistics
Data are presented as means ± standard deviation (SD). Data were evaluated using an independent sample Student’s t test or two-way analysis of variance (ANOVA) test. *p < 0.05, **p < 0.01 and ***p < 0.001 were considered to be statistically significant.
3. Results
3.1. Deletion of Bmp1a in chondrocytes rescues the phenotypes of Fgfr3−/− mice
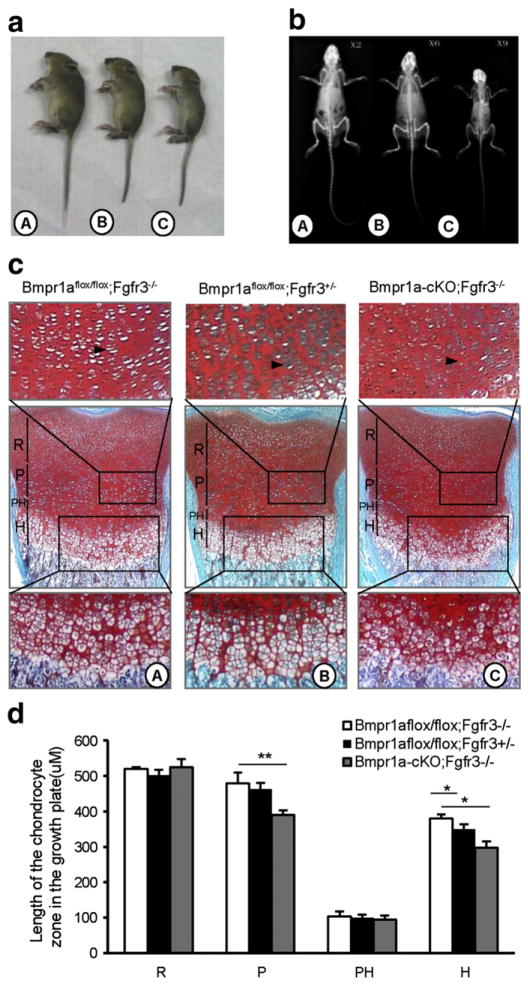
We and others have previously reported that Fgfr3 null mice had a bone overgrowth phenotype resulted from enhanced endochondral bone formation [5,6]. Mice with Bmpr1a deletion in chondrocytes exhibit defects in the growth plates and die shortly after birth as a result of respiratory failure caused by generalized chondrodysplasia [21,32]. We questioned whether the bone overgrowth phenotype of Fgfr3 deficient (Fgfr3−/−) mice is associated with BMPR1a-mediated BMP signaling. In this study, Fgfr3−/− [6], Bmpr1aflox/flox [45] and Col2a1-Cre [47] mice were used to mate each other based on Cre/LoxP conditional knockout strategy. To improve the birth rate of double knockout mice, BMPR1aflox/flox;Fgfr3−/− mice were mated with Bmpr1a+/flox;Col2a1-Cre;FGFR3+/− mice to generate BMPR1aflox/flox;Col2a1-Cre;FGFR3−/− mice (Bmpr1a-cKO;Fgfr3−/−). We found that when Bmpr1a was deleted in the chondrocytes of Fgfr3−/− mice, the mice were able to survive into adulthood, with a smaller skeleton and body size (Fig. 1a–b). The examination of the growth plate proliferative zones showed a marked reduction in proliferative zones in Bmpr1a-cKO;Fgfr3−/− mice compared to Bmpr1aflox/flox;Fgfr3−/− mice (Black arrow, Fig. 1c,d, C vs A). Long bone growth plates displayed a wider hypertrophic (h) zone in Bmpr1aflox/flox;Fgfr3−/− mice compared to the Bmpr1aflox/flox;Fgfr3+/− littermates (Fig. 1c,d, A vs B). The width of the hypertrophic zone of Bmpr1a-cKO;Fgfr3−/− mice was decreased compared to those of the Bmpr1aflox/flox;Fgfr3−/− mice (Fig. 1c, C vs A). Furthermore, the hypertrophic chondrocytes showed a smaller size and more-disorganized columns in Bmpr1a-cKO;Fgfr3−/− mice compared to Bmpr1aflox/flox; Fgfr3−/− mice (Fig. 1c, C vs A). Collectively, these data showed that depletion of Bmpr1a in chondrocytes rescued the bone overgrowth phenotype of Fgfr3 deficient mice by reducing chondrocyte differentiation.
Fig. 1.
Deletion of Bmpr1a in chondrocytes rescues the phenotypes of Fgfr3−/− mice by inhibiting chondrocyte differentiation. a,b. Deletion of Bmpr1a in chondrocytes rescued the bone overgrowth phenotype of Fgfr3 deletion mice. Side views (a) and the X-ray analysis (b) of the 30-day old mice. A: Bmpr1aflox/flox;Fgfr3−/−, B: Bmpr1aflox/flox;Fgfr3+/−, C: Bmpr1a-cKO;Fgfr3−/−. Note that Bmpr1a-cKO;FGFR3+/+ and Bmpr1a-cKO;FGFR3+/− mice were lethal after birth. c. Fgfr3 deletion results in increased width of hypertrophic zone but double deletion of Bmpr1a and Fgfr3 narrows the zone of hypertrophic chondrocytes. Alcian blue/alizarin red staining of proximal tibia growth plates of 1 day old indicated mice. (d) Quantitative analysis of length of the chondrocyte zones in growth plates (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001). R: resting zones, P: proliferation zones, PH: prehypertrophic zones; H: hypertrophic zones, black arrow indicates the proliferative columns.
3.2. FGFR3 inhibits BMPR1a-mediated chondrocyte differentiation
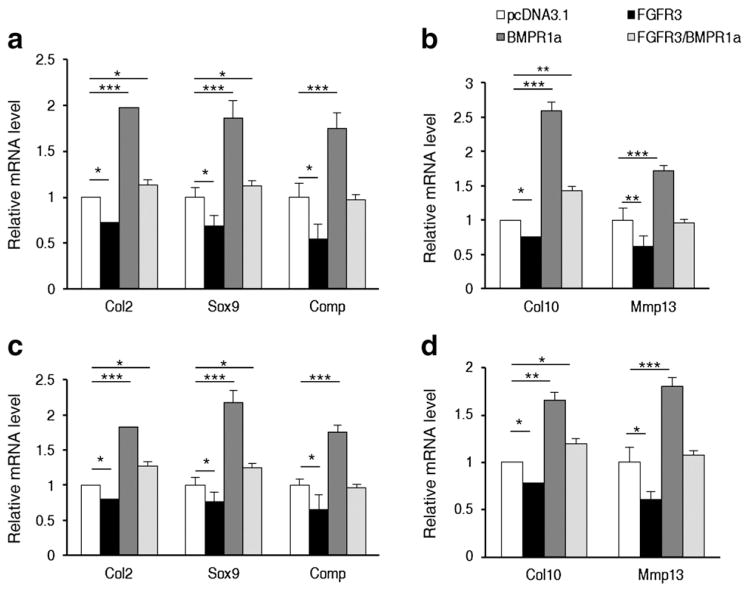
To further investigate the physiological role of FGFR3-mediated inhibition on BMPR1a signaling pathway in the regulation of chondrogenesis, we examined the effect of FGFR3 on BMPR1a-mediated chondrocyte differentiation. The chondrogenic differentiation of the mouse immature mesenchymal C3H10T1/2 and precursor chondrocyte ATDC5 cell line was used as model of chondrogenesis in the early and later stages of endochondral bone development [53]. The Real-time PCR results showed that transient transfection of FGFR3 resulted not only in the inhibition of the expressions of marker genes Col2, Sox9 and Comp involved in the early stage (3 days, Fig. 2a), but also in the inhibition of expressions of marker genes Col10 and Mmp13 involved in the later stage (7 days, Fig. 2b) of chondrocyte differentiation from immature mesenchymal C3H10T1/2 cells (Fig. 2a, b). Furthermore, FGFR3 also inhibited BMPR1a induced expression of genes involved in the early stage (3 days, Fig. 2c) or later stage (7 days, Fig. 2d) of chondrocyte differentiation in precursor chondrocyte ATDC5 (Fig. 2c,d). These data demonstrated that FGFR3 inhibited BMPR1a-mediated chondrocyte differentiation. Collectively, the in vivo and in vitro data suggest that FGFR3 inhibits BMPR1a-mediated signaling to regulate chondrocyte differentiation.
Fig. 2.
FGFR3 inhibits BMPR1a-mediated chondrocyte differentiation. a,b. FGFR3 inhibited BMPR1a induced expression of genes involved in the early stage (3 days, a) or later stage (7 days, b) of chondrocyte differentiation from mesenchymal cells (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001). c,d. FGFR3 inhibited BMPR1a induced expression of genes involved in the early stage (3 days, c) or later stage (7 days, d) of chondrocyte differentiation in precursor chondrocytes (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001).
3.3. FGFR3 inhibits BMP signaling in the chondrocytes of mouse growth plates
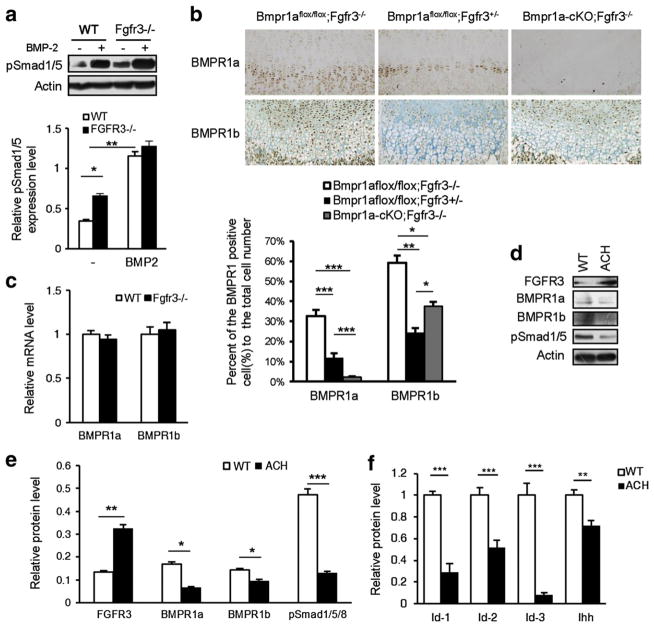
To examine the physiological function of FGFR3 in the inhibition of BMP signaling, we used Fgfr3 knockout (Fgfr3−/−) [6] and ACH mice (Fgfr3G369C/+; constitutive active FGFR3) [7] we generated previously. We found that the phosphorylation of Smad1/5/8 (p-Smad1/5/8) in primary chondrocytes from Fgfr3−/− mice was significantly increased compared to the wild-type controls with or without BMP-2 stimulation (Fig. 3a). The immunohistochemistry analysis also showed that the immunoreactivities of BMPR1a and 1b were increased in the primary chondrocytes of Bmpr1aflox/flox;Fgfr3−/− mice compared with that of Bmpr1aflox/flox;Fgfr3+/− mice (Fig. 3b), and the BMPR1a protein could not be detected in the growth plates of double knockout (Bmpr1a-cKO; Fgfr3−/−) mice (Fig. 3b). We further observed that depletion of FGFR3 did not affect the mRNA level of BMPR1a and 1b (Fig. 3c), which suggests that the influence of FGFR3 on BMPR1 is at the post-transcriptional level. Furthermore, an immunoblot showed that the protein levels of BMPR1a, BMPR1b and pSmad1/5 were decreased in the primary chondrocytes from ACH mice where the protein level of FGFR3 was increased and active (Fig. 3d,e). Consequently, BMP targeting genes including Id-1, Id-2, Id-3 and Ihh were down-regulated in the chondrocytes from ACH mice (Fig. 3f). All these data suggest that FGFR3 hyper-activation results in the decreased level of BMPR1 and impaired BMP signaling in chondrocytes of mouse growth plates.
Fig. 3.
FGFR3 inhibits BMP signaling in growth plate chondrocytes of mice. a. Western blot results of pSmad1/5 in the primary chondrocytes from Fgfr3−/− mice and their wild-type littermates stimulated with BMP-2 (25 ng/ml) for 45 min. The band signal intensity was quantified using software ImageJ (version 1.47). The error bars indicate the standard deviation (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01). b. Immunohistochemistry analysis results of BMPR1a and 1b in the growth plates of Bmpr1aflox/flox;Fgfr3−/−, Bmpr1aflox/flox;Fgfr3+/− and Bmpr1a-cKO;Fgfr3−/− mice. The percentage of the BMPR1 positive cells in the prehypertrophic and hypertrophic zones was calculated (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01). A: Bmpr1aflox/flox;Fgfr3−/−, B: Bmpr1aflox/flox;Fgfr3+/−, C: Bmpr1a-cKO;Fgfr3−/−. c. Real-time PCR results of BMPR1a and 1b from primary chondrocytes of 5-day-old Fgfr3−/− and wild-type mice. d. Western blot results of BMPR1a, 1b and pSmad1/5 in the primary chondrocytes from 5-day-old ACH mice and their wild-type littermates. e. Quantitative analysis of the band signal intensity in Fig. 3d (n = 3, Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001). f. Real-time PCR analysis of total mRNA from primary chondrocytes of 5-day-old ACH and wild-type mice for BMP signaling target genes Id-1, Id-2, Id-3 and Ihh (n = 3, Student’s t-test, **p < 0.01, ***p < 0.001).
3.4. FGFR3 inhibits BMP signaling independent of its tyrosine kinase activity
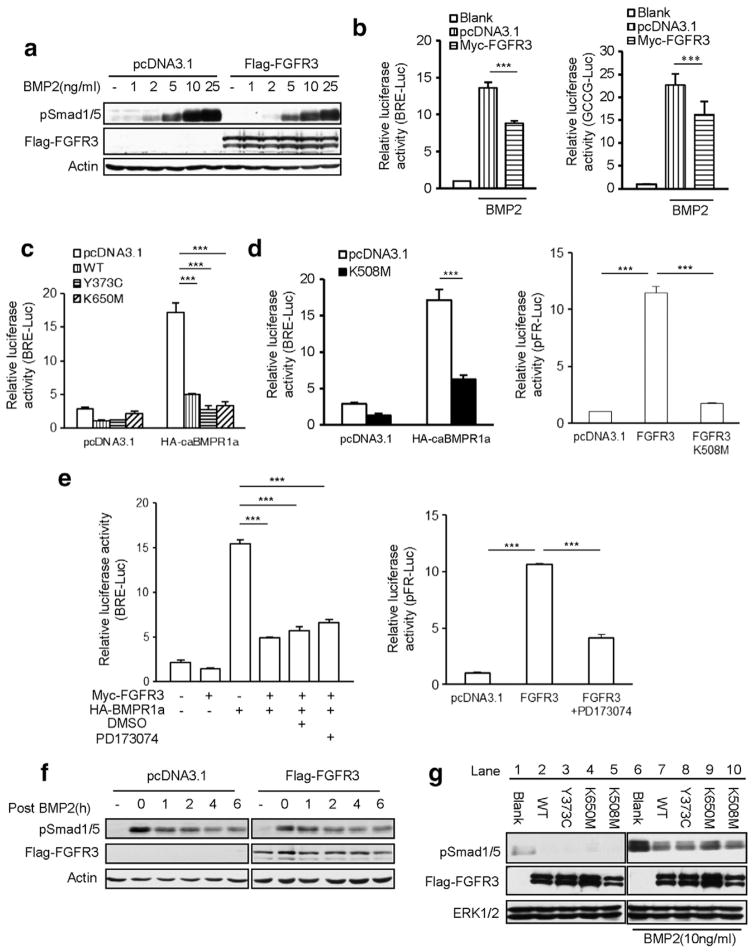
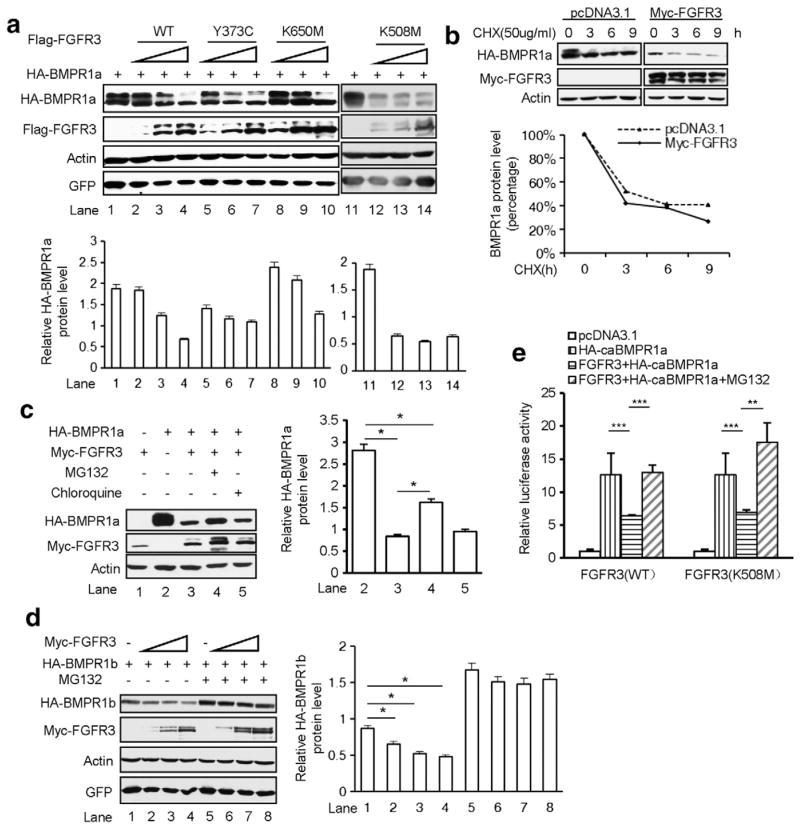
It is well documented that FGF/FGFR activated ERK-MAPK induces degradation of Smad1/5 by phosphorylating its linker region to inhibit BMP signaling [44]. Our above data showed that hyperactive FGFR3 decreased the protein levels of BMPR1s and its downstream signaling cascades in chondrocytes. To dissect the mechanism of FGFR3 inhibiting BMP signaling, we used another BMP-2-responsive cell lines to examine whether FGFR3 influences BMP signaling through its kinase activity. In this study, HepG2 and NIH3T3 cells were hired as the cell models. HepG2 is a human liver carcinoma cell line and its BMP-responsive characteristic is well established. NIH3T3 cell line is isolated from desegregated NIH Swiss mouse embryonic fibroblasts and has been proven to be very useful in DNA transfection studies. A Western blot analysis showed that HepG2 cells, transfected with FGFR3 required higher concentration of BMP-2 to elicit detectable Smad1/5 phosphorylation after 30 min of treatment (Fig. 4a). Luciferase assays showed that wild-type FGFR3 significantly inhibited BMP-2 induced BRE-Luc and GCCG-Luc reporter activity in NIH3T3 cells (Fig. 4b). Expression of two constitutively active FGFR3 (FGFR3-Y373C and FGFR3-K650M, causing TDI and TDII in humans, respectively) (Fig. 4c) and a kinase-inactivated FGFR3 (FGFR3-K508M) [49] (Fig. 4d) also impaired the BMP signaling induced by constitutively active BMPR1a (Q227D) (HA-caBMPR1a). Furthermore, we found that PD173074, a potent cell-permeable and ATP-competitive inhibitor of FGFR tyrosine kinase activity, inhibited the FGFR3-induced ERK luciferase activity effectively (Fig. 4e), but failed to rescue the inhibitory effect of FGFR3 on the caBMPR1a-induced Smad-transcriptional activity (Fig. 4e). These results suggest that the tyrosine kinase activity of FGFR3 is not required for the inhibition of FGFR3 on BMP signaling.
Fig. 4.
FGFR3 inhibits BMP signaling pathway in tyrosine kinase independent manner. a. Western blots of pSmad1/5 in different concentrations of BMP-2 treated HepG2 cells transfected with wild-type FGFR3 or pcDNA3.1-Flag. b. FGFR3 inhibited the BMP-2 stimulated BRE-Luc or GCCG-Luc reporter activity in NIH3T3 cells. Data are shown as means ± SD and are representative of three independent experiments (n = 3, two-way ANOVA, ***p < 0.001). c,d. Wild-type human FGFR3 (c) as well as Y373C (c), K650M (c) and K508M (d) mutants inhibited constitutive active receptor BMPR1a (Q227D) activated BRE-Luc reporter activity. FGFR3 K508M did not activated reporter gene activity for ERK MAPK (n = 3, two-way ANOVA, ***p < 0.001). e. PD173074 inhibited reporter gene activity for ERK MAPK, but could not rescue the inhibitory effect of FGFR3 to caBMPR1a activated BRE-Luc reporter activity (n = 3, two-way ANOVA, ***p < 0.001). f. Overexpression of FGFR3 had no significant effect on the turnover of phosphorylated Smad1/5. HepG2 cells transfected with wild-type FGFR3 or control (pcDNA3.1-myc) were stimulated with BMP-2 (25 ng/ml) for 1 h. Cells were then harvested at the indicated time after withdrawal of BMP-2 and total cell extracts were analyzed by immunoblot. g. Forced expression of FGFR3 significantly decreased the levels of c-terminal phosphorylated Smad1/5. HepG2 cells transfected with wild-type or mutated FGFR3 were stimulated with BMP-2 (10 ng/ml) for 30 min. Whole-cell lysates were extracted and subjected to Western blotting for c-terminal phosphorylated Smad1/5.
Due to the importance of both the peak intensity and the turnover of the phosphorylated proteins during signaling, we considered whether FGFR3 protein can affect the BMP-2-induced peak intensity or the turnover of the phosphorylated Smad1/5. HepG2 cells transfected with FGFR3 or control (pcDNA3.1-myc) were stimulated with BMP-2 (25 ng/ml) for 1 h. Cells were harvested at the indicated times after withdrawal of BMP-2 and total cell extracts were analyzed. Our results showed that overexpression of FGFR3 had no significant effect on the turnover of pSmad1/5 after BMP-2 stimulation (Fig. 4f), while FGFR3 significantly decreased the levels of phosphorylated Smad1/5 in HepG2 cells treated with BMP-2 (Fig. 4g, Lanes 7, 8, 9, and 10 vs Lane 6) or vehicle (Fig. 4g, Lanes 2, 3, 4, and 5 vs Lane 1). Overexpression of FGFR3 significantly decreased the BMP-2-induced peak intensity while had no effect on the turnover of pSmad1/5, suggesting that FGFR3 inhibits BMP-2-induced peak intensity of phosphorylated Smad1/5. Collectively, FGFR3 inhibits BMP-2- or constitutively activated BMPR1-induced phosphorylation of Smads through a mechanism independent of its tyrosine kinase activity.
3.5. FGFR3 enhances the ubiquitination and degradation of Type I BMP receptors
To explore the mechanism of FGFR3-induced impairment of BMP signaling, we further analyzed whether FGFR3 affects the protein level of BMPR1a in cells. Expression of Flag-tagged wild-type or mutant FGFR3 significantly reduced the HA-BMPR1a protein level in a dose-dependent manner in HEK293T cells (Fig. 5a), consistent with the results from primary chondrocytes of Fgfr3−/− or ACH mice (Fig. 3a–e). When HA-BMPR1a was co-expressed with Flag-FGFR3 in HEK293T cells in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX), BMPR1a turnover rates were markedly increased with expression of FGFR3 (Fig. 5b). Furthermore, MG132, an inhibitor of the proteasome, partially rescued the protein level of BMPR1a, while a lysosomal inhibitor chloroquine failed (Fig. 5c), suggesting that the decreased BMPR1a by FGFR3 is through the ubiquitination–degradation mechanism. In addition, we also observed that FGFR3 enhanced the degradation of BMPR1b through a ubiquitination–degradation mechanism (Fig. 5d). Consistently, MG132 also recovered the wild-type or kinase-inactivated FGFR3-reduced luciferase activity induced by HA-caBMPR1a (Fig. 5e). Our data indicate that FGFR3 boosts the degradation of type I BMP receptors in an ubiquitination-dependent manner.
Fig. 5.
FGFR3 enhances the degradation of type I BMP receptors through ubiquitination-dependent pathway. a. Increasing amounts of wild-type or mutant of FGFR3 affected the HA-BMPR1a in HEK293T cells. Quantitative analysis of HA-BMPR1a band signal intensity was performed. The error bars indicate the standard deviation from three separate experiments. b. FGFR3 shortened the half-life of BMPR1a. HEK293T cells transfected with pcDNA3.1 or Myc-FGFR3 were treated with CHX (50 μg/ml) for the indicated time. The levels of BMPR1a were assayed and quantified (lower panel). CHX, Cycloheximide. c,d. FGFR3 affected the protein level of HA-BMPR1a (c) and HA-BMPR1b (d) through a ubiquitination-dependent pathway. Quantitative analysis of the band signal intensity of HA-BMPR1a and HA-BMPR1b (n = 3, two-way ANOVA, *p < 0.05). e. MG132 (25 μM) recovered the wild-type or kinase-inactivated FGFR3 decreased luciferase activity induced by HA-caBMPR1a (n = 3, two-way ANOVA, **p < 0.01, ***p < 0.001).
3.6. FGFR3 promotes degradation of BMPR1a through Smurf1
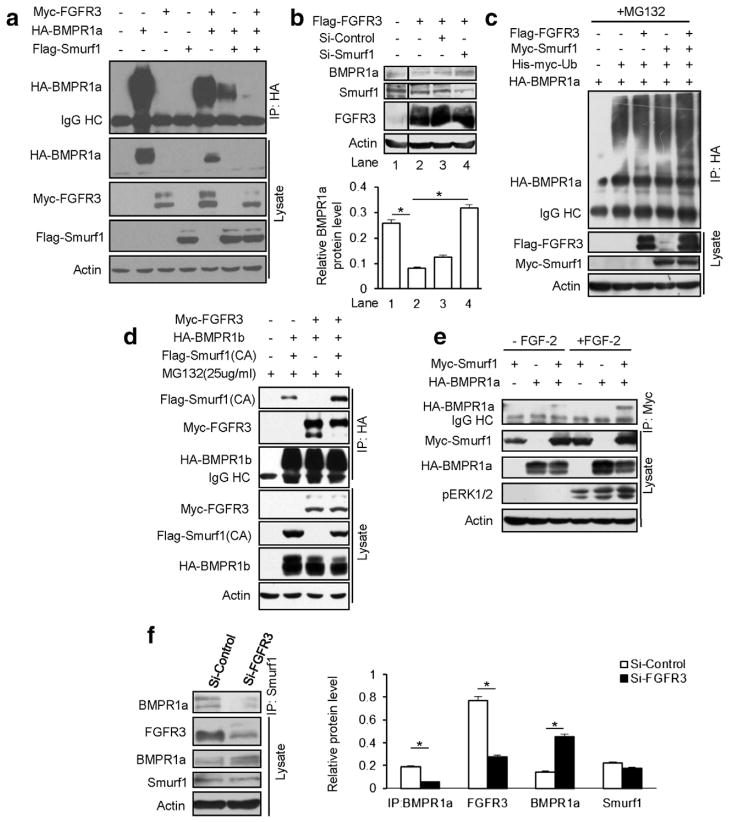
Smurf1, an E3 ligase, binds Smad1/Smad5 or BMPR1 [31], causing their ubiquitination and subsequent proteasomal degradation [29,30]. To address the mechanism of FGFR3 in the degradation of type I BMP receptors, we examined whether FGFR3 mediated degradation of BMPR1 through Smurf1. A Western blotting analysis showed that FGFR3 strongly promoted the degradation of BMPR1a in the presence of Smurf1 (Fig. 6a), whereas knock down of Smurf1 by an siRNA prevented FGFR3-mediated BMPR1a degradation in HepG2 cells (Fig. 6b). We further demonstrated that FGFR3 enhanced the ubiquitination of BMPR1a in HEK293T cells (Fig. 6c). We next examined whether FGFR3 enhances the interaction of BMPR1 and Smurf1. The IP results showed that the interaction between Smurf1 and BMPR1b was enhanced in the presence of FGFR3 (Fig. 6d). FGF-2 also enhanced the interaction of Smurf1 and BMPR1a (Fig. 6e). Conversely, we also found that in HeLa cells, where the protein levels of FGFR3, BMPR1a and Smurf1 are relatively higher, the interaction of Smurf1 and BMPR1a was impaired when FGFR3 was knocked down by an siRNA (Fig. 6f). All these data suggested that FGFR3 enhances the interaction of Smurf1 and BMPR1 and promotes the degradation of BMPR1.
Fig. 6.
FGFR3 prompts BMPR1a degradation through E3 ligase Smurf1. a. FGFR3 enhanced the ability of Smurf1 in the degradation of BMPR1a. b. Depletion of Smurf1 by siRNA prevented FGFR3-mediated BMPR1a degradation in HepG2 cells. The BMPR1a band signal intensity was quantitatively analyzed (n = 3, two-way ANOVA, *p < 0.05). c. FGFR3 enhanced the ubiquitination of BMPR1a. In vivo ubiquitination assay was performed in the HEK293T cells co-transfected with HA-BMPR1a, His-Myc–ubiquitin, Myc–Smurf1 and Flag-FGFR3. The ubiquitinated proteins were precipitated followed by immunoblotting with an anti-HA antibody. d. FGFR3 enhanced the interaction of BMPR1 and Smurf1. HEK293T cell was co-expressed with Smurf1 (CA), HA-BMPR1b and Myc-FGFR3 for 24 h. The cells were treated with MG132 (25 μM) for 6 h and coimmunoprecipitation experiments were performed. e. FGF-2 stimulated the association of HA-BMPR1a and Myc-Smurf1. HEK293T cells were co-transfected with HA-BMPR1a and Myc-Smurf1. After 24 h transfection, the cells were starved for 12–16 h and stimulated by FGF-2 (50 ng/ml) for 30 min. Whole-cell lysates were extracted and analyzed as described before. f. The interaction of Smurf1 and BMPR1a was impaired when FGFR3 was depleted in HeLa cells. The band signal intensity was quantified (n = 3, Student’s t-test, *p < 0.05).
3.7. BMP-2 rescues the retardation of the embryonic metatarsal growth of the TDII mice
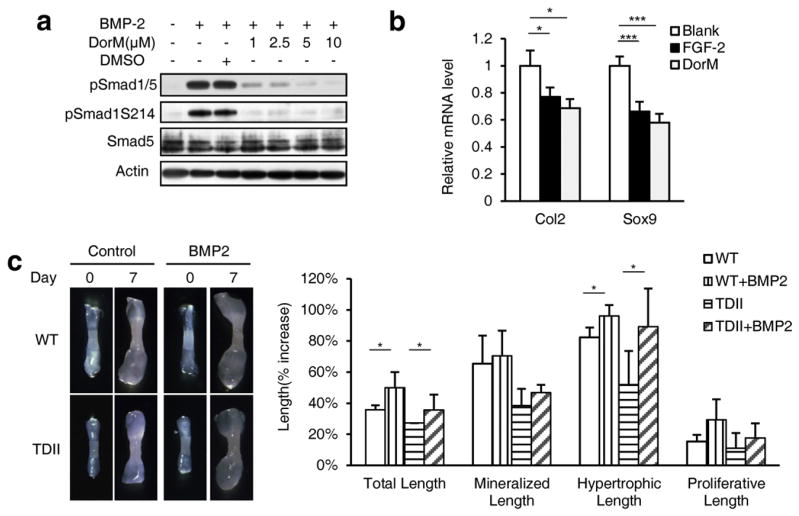
To investigate whether FGFR3 hyper-activation results in retardation of bone growth through inhibition of BMPR1-mediated BMP signaling, we analyzed the effect of dorsomorphin, a specific inhibitor for BMPR1a and BMPR1b [54], on chondrocyte differentiation. Firstly, we found that BMP-2 induced the c-terminal and linker region phosphorylation of Smad1/5 in a time-dependent manner and dorsomorphin decreased the phosphorylation of Smad1/5 in a dose-dependent manner (Fig. 7a) in ATDC5 cells. Furthermore, the Real-time PCR analysis results showed that dorsomorphin treatment inhibited the expression of the early chondrocyte differentiation marker genes Col2 and Sox9, similar to the effect of FGF-2 on ATDC5 cells (Fig. 7b). To further confirm the role of BMP signaling in FGFR3-mediated bone growth inhibition, we sought to rescue the retarded growth of the embryonic metatarsals of TDII mice with BMP-2. While the metatarsals from the TDII mice showed retarded bone growth (comparing with that from their wild-type littermates), addition of BMP-2 significantly promoted the growth of metatarsals (Fig. 7c). Intriguingly, BMP-2 significantly promoted the hypertrophic zone length of embryonic metatarsals, but not the mineralized and proliferation zone length (Fig. 7c). These results suggest that BMP-2 rescued the FGFR3-induced retardation of bone growth mainly by promoting chondrocyte differentiation. Taken together, FGFR3 inhibits the bone growth partially through inhibition of BMP signaling in chondrocytes.
Fig. 7.
BMP-2 rescues the retardation of the embryonic metatarsal growth of the TDII mice. a. BMPR1 inhibitor inhibits the phosphorylation of Smad1/5 in chondrocytes. DorM, dorsomorphin. b. ATDC5 cells were cultured for 3 days for chondrocyte differentiation in the presence of FGF-2 (25 ng/μl) or dorsomorphin (10 μM). The expression of Col2 and Sox9 was analyzed by Real-time PCR (n = 3, two-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001). c. BMP-2 ameliorated the cartilage growth retarded by hyper-activation of FGFR3. The metatarsals from wild-type and TDII mice were treated with BMP-2 (100 ng/ml) for 7 d. The percentage increase in the total, mineralization, hypertrophic and proliferation length of the cultured bones was measured and calculated (n = 5, two-way ANOVA, *p < 0.05).
4. Discussion
Achondroplasia is caused by gain-of-function mutations of FGFR3, which inhibits mainly chondrogenesis of endochondral ossification [4,7,24,55]. Although many signaling pathways have been reported in the regulation of chondrogenesis, it remains unclear how the development of chondrogenesis and achondroplasia is regulated coordinately by diverse signaling pathways. In this study, we revealed that FGFR3 inhibits chondrocyte differentiation through inhibition of BMP signaling. We found that FGFR3 negatively regulates BMP signaling in chondrocytes by facilitating BMPR1 for degradation.
The crosstalk between FGF and BMP pathways during chondrocyte development has been investigated in different studies [32,33,40,44, 56,57]. However, since multiple signaling components of FGF and BMP pathways are expressed in chondrocytes [10,58], it is unclear how the crosstalk between BMP and FGF signaling regulates chondrogenesis and development of ACH. Double Bmpr1a and Bmpr1b null mice have a phenotype of increased expression of FGFR1 in chondrocytes [32], suggesting the potential role of this crosstalk in chondrogenesis. However, the absence of gross phenotype in chondrogenesis with either gain-of-function mutation of FGFR1 or chondrocyte-specific deletion of FGFR1 indicates that FGFR1 is not the primary receptor for the crosstalk between FGF and BMP signaling during chondrogenesis [59,60]. FGFR3 is the major receptor in chondrocytes to regulate chondrogenesis [10]. More importantly, both FGFR3 and BMPR1a have relatively higher levels of expression at the prehypertrophic zone, an area critically controlling growth plate chondrocyte differentiation [32]. In this study, we found that chondrocyte-specific inactivation of Bmpr1a rescued the bone overgrowth phenotypes in FGFR3 deficient mice by alleviating their increased differentiation of chondrocytes, and FGFR3 hyper-activation results in impaired BMP signaling in chondrocytes and the specific inhibitor of BMPR1 mimics the effect of hyperactive FGFR3, leading to the retardation of bone growth. Furthermore, BMP-2 alleviated the retarded growth of cultured bones from TDII mice by increasing the chondrocyte differentiation. All these in vitro and in vivo results showed for the first time that direct down-regulation of BMPR1-mediated BMP signaling plays an important role in the regulation of chondrogenesis and achondroplasia by FGFR3. We believe that the counter balance between FGFR3 and BMP signaling pathways plays an important role in maintaining the normal chondrocyte development. Imbalance between these two signaling pathways, for example over-activation of FGFR3, results in developmental abnormalities such as achondroplasia.
It is well documented that FGF-2, by activating ERK1/2 MAPK pathway, promotes the phosphorylation of the linker region of Smad1/5/8, leading to their degradation in chondrocytes and other cells [33,44]. We found that FGFR3 resulted in a decrease of BMPR1 through a ubiquitin-mediated degradation mechanism. FGFR3 promoted the complex formation of BMPR1/Smurf1, mediating its degradation, which led to impair the BMP signaling as measured by the phosphorylation of c-terminal region of Smad1/5. Although FGFR3 induces ERK1/2 phosphorylation and further phosphorylates Smad1/5 linker region for the protein degradation, FGFR3-mediated degradation of membrane BMPR1 affects the upper stream of BMP signaling, which is a novel mechanism and likely more important in negative regulation of the BMP signaling pathway during chondrogenesis.
Modulation of BMP signaling occurs at different levels, including degradation of BMPR1 [61–63] through an internalization process (endosome) mediated by caveolin-1 [64]. Smurf1 and Smurf2 were reported to antagonize the TGF-β/BMP pathway by targeting Smads, TGF-β/BMP receptors or TGF-β-responsive transcription factors for ubiquitination-mediated degradation [30,31,65,66]. We showed that FGFR3 promotes the association between BMPR1 with Smurf1. FGFR3-mediated degradation of BMPR1 through Smurf1 ubiquitination process is different from degradation of BMPRs by a caveolin-1-mediated internalization process [64]. However, the mechanisms by which FGFR3 promotes the association between BMPR1 with Smurf1 need more investigations in the future study.
5. Conclusion
In these studies, we propose a novel model that FGFR3 signaling inhibits chondrocyte differentiation through down-regulation of BMP signaling by Smurf1-mediated ubiquitination and degradation of BMPR1. Our study provides novel insights for understanding the mechanisms of the chondrogenesis and FGFR3-related chondrodysplasia, as well as the crosstalk between FGF and BMP pathways, two essential signaling pathways in the development of a variety of organs and diseases.
Supplementary Material
Acknowledgments
We thank Drs. Pavel Krejci (Masaryk University, Czech Republic), Yeguang Chen (Tsinghua University, China) and Lingqiang Zhang (Beijing Institute of Radiation Medicine, China) for providing constructs. The work was supported by the Tsinghua Internal Research Foundation (20091081322), the National Natural Science Foundation of China (No. 81030036, No. 30800652), the Special Funds for Major State Basic Research Program of China (973 program) (No. 2012CB518100), and the Foundation of the State Key Laboratory of Trauma, Burns and Combined Injury (No. SKLZZ200902).
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamcr.2014.03.011.
References
- 1.Horton WA, Hall JG, Hecht JT. Achondroplasia. Lancet. 2007;370:162–172. doi: 10.1016/S0140-6736(07)61090-3. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 3.Bellus GA, Hefferon TW, Ortiz de Luna RI, Hecht JT, Horton WA, Machado M, Kaitila I, McIntosh I, Francomano CA. Achondroplasia is defined by recurrent G380R mutations of FGFR3. Am J Hum Genet. 1995;56:368–373. [PMC free article] [PubMed] [Google Scholar]
- 4.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 5.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 6.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Adar R, Yang X, Monsonego EO, Li C, Hauschka PV, Yayon A, Deng CX. Gly369Cys mutation in mouse FGFR3 causes achondroplasia by affecting both chondrogenesis and osteogenesis. J Clin Invest. 1999;104:1517–1525. doi: 10.1172/JCI6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwata T, Li CL, Deng CX, Francomano CA. Highly activated Fgfr3 with the K644M mutation causes prolonged survival in severe dwarf mice. Hum Mol Genet. 2001;10:1255–1264. doi: 10.1093/hmg/10.12.1255. [DOI] [PubMed] [Google Scholar]
- 9.L’Hote CGM, Knowles MA. Cell responses to FGFR3 signalling: growth, differentiation and apoptosis. Exp Cell Res. 2005;304:417–431. doi: 10.1016/j.yexcr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Ornitz DM. FGF signaling in the developing endochondral skeleton. Cytokine Growth Factor Rev. 2005;16:205–213. doi: 10.1016/j.cytogfr.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahni M, Ambrosetti DC, Mansukhani A, Gertner R, Levy D, Basilico C. FGF signaling inhibits chondrocyte proliferation and regulates bone development through the STAT-1 pathway. Genes Dev. 1999;13:1361–1366. doi: 10.1101/gad.13.11.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su N, Sun Q, Li C, Lu X, Qi H, Chen S, Yang J, Du X, Zhao L, He Q, Jin M, Shen Y, Chen D, Chen L. Gain-of-function mutation in FGFR3 in mice leads to decreased bone mass by affecting both osteoblastogenesis and osteoclastogenesis. Hum Mol Genet. 2010;19:1199–1210. doi: 10.1093/hmg/ddp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He L, Horton W, Hristova K. Physical basis behind achondroplasia, the most common form of human dwarfism. J Biol Chem. 2010;285:30103–30114. doi: 10.1074/jbc.M109.094086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olsen BR, Reginato AM, Wang W. Bone development. Annu Rev Cell Dev Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- 16.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genomics Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 17.Karaplis AC, Luz A, Glowacki J, Bronson RT, Tybulewicz VL, Kronenberg HM, Mulligan RC. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8:277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- 18.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 19.Lanske B, Amling M, Neff L, Guiducci J, Baron R, Kronenberg HM. Ablation of the PTHrP gene or the PTH/PTHrP receptor gene leads to distinct abnormalities in bone development. J Clin Invest. 1999;104:399–407. doi: 10.1172/JCI6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kronenberg HM. PTHrP and skeletal development. Ann N Y Acad Sci. 2006;1068:1–13. doi: 10.1196/annals.1346.002. [DOI] [PubMed] [Google Scholar]
- 21.Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:5062–5067. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike M, Yamanaka Y, Inoue M, Tanaka H, Nishimura R, Seino Y. Insulin-like growth factor-1 rescues the mutated FGF receptor 3 (G380R) expressing ATDC5 cells from apoptosis through phosphatidylinositol 3-kinase and MAPK. J Bone Miner Res. 2003;18:2043–2051. doi: 10.1359/jbmr.2003.18.11.2043. [DOI] [PubMed] [Google Scholar]
- 24.Naski MC, Colvin JS, Coffin JD, Ornitz DM. Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development. 1998;125:4977–4988. doi: 10.1242/dev.125.24.4977. [DOI] [PubMed] [Google Scholar]
- 25.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Massague J. Mechanisms of TGF-[beta] signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–924. doi: 10.1016/j.molcel.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenblatt MB, Shim JH, Zou W, Sitara D, Schweitzer M, Hu D, Lotinun S, Sano Y, Baron R, Park JM, Arthur S, Xie M, Schneider MD, Zhai B, Gygi S, Davis R, Glimcher LH. The p38 MAPK pathway is essential for skeletogenesis and bone homeostasis in mice. J Clin Invest. 2010;120:2457–2473. doi: 10.1172/JCI42285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
- 31.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- 32.Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133:4667–4678. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 33.Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136:1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsumaki N, Nakase T, Miyaji T, Kakiuchi M, Kimura T, Ochi T, Yoshikawa H. Bone morphogenetic protein signals are required for cartilage formation and differently regulate joint development during skeletogenesis. J Bone Miner Res. 2002;17:898–906. doi: 10.1359/jbmr.2002.17.5.898. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi T, Lyons KM, McMahon AP, Kronenberg HM. BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc Natl Acad Sci U S A. 2005;102:18023–18027. doi: 10.1073/pnas.0503617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, Gygi S, Glimcher LH. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28:2028–2041. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi SE, Daluiski A, Pederson R, Rosen V, Lyons KM. The type I BMP receptor BMPRIB is required for chondrogenesis in the mouse limb. Development. 2000;127:621–630. doi: 10.1242/dev.127.3.621. [DOI] [PubMed] [Google Scholar]
- 38.Mishina Y, Suzuki A, Ueno N, Behringer RR. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995;9:3027–3037. doi: 10.1101/gad.9.24.3027. [DOI] [PubMed] [Google Scholar]
- 39.Iwata T, Chen L, Li C-l, Ovchinnikov DA, Behringer RR, Francomano CA, Deng C-X. A neonatal lethal mutation in FGFR3 uncouples proliferation and differentiation of growth plate chondrocytes in embryos. Hum Mol Genet. 2000;9:1603–1613. doi: 10.1093/hmg/9.11.1603. [DOI] [PubMed] [Google Scholar]
- 40.Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev Cell. 2002;3:439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, Chang JY, Huang Y, Lin X, Luo Y, Schwartz RJ, Martin JF, Wang F. The FGF-BMP signaling axis regulates outflow tract valve primordium formation by promoting cushion neural crest cell differentiation. Circ Res. 2010;107:1209–1219. doi: 10.1161/CIRCRESAHA.110.225318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003;422:625–629. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 43.Maier E, von Hofsten J, Nord H, Fernandes M, Paek H, Hebert JM, Gunhaga L. Opposing Fgf and Bmp activities regulate the specification of olfactory sensory and respiratory epithelial cell fates. Development. 2010;137:1601–1611. doi: 10.1242/dev.051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Mishina Yuji, Hanks MC, Miura S, Tallquist MD, Behringer RR. Generation of Bmpr/Alk3 conditional knockout mice. Genesis. 2002;32:69–72. doi: 10.1002/gene.10038. [DOI] [PubMed] [Google Scholar]
- 46.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hao ZM, Yang X, Cheng X, Zhou J, Huang CF. Generation and characterization of chondrocyte specific Cre transgenic mice. Yi Chuan Xue Bao. 2002;29:424–429. [PubMed] [Google Scholar]
- 48.Yin L, Du X, Li C, Xu X, Chen Z, Su N, Zhao L, Qi H, Li F, Xue J, Yang J, Jin M, Deng C, Chen L. A Pro253Arg mutation in fibroblast growth factor receptor 2 (Fgfr2) causes skeleton malformation mimicking human Apert syndrome by affecting both chondrogenesis and osteogenesis. Bone. 2008;42:631–643. doi: 10.1016/j.bone.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 49.Krejci P, Salazar L, Kashiwada TA, Chlebova K, Salasova A, Thompson LM, Bryja V, Kozubik A, Wilcox WR. Analysis of STAT1 activation by Six FGFR3 mutants associated with skeletal dysplasia undermines dominant role of STAT1 in FGFR3 signaling in cartilage. PLoS One. 2008;3:e3961. doi: 10.1371/journal.pone.0003961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu K, Yin X, Weng T, Xi S, Li L, Xing G, Cheng X, Yang X, Zhang L, He F. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat Cell Biol. 2008;10:994–1002. doi: 10.1038/ncb1760. [DOI] [PubMed] [Google Scholar]
- 51.Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Li C, Qiao W, Xu X, Deng C. A Ser(365) -> Cys mutation of fibroblast growth factor receptor 3 in mouse downregulates Ihh/PTHrP signals and causes severe achondroplasia. Hum Mol Genet. 2001;10:457–465. doi: 10.1093/hmg/10.5.457. [DOI] [PubMed] [Google Scholar]
- 53.Atkinson BL, Fantle KS, Benedict JJ, Huffer WE, Gutierrez-Hartmann A. Combination of osteoinductive bone proteins differentiates mesenchymal C3H/10T1/2 cells specifically to the cartilage lineage. J Cell Biochem. 1997;65:325–339. [PubMed] [Google Scholar]
- 54.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C, Chen L, Iwata T, Kitagawa M, Fu XY, Deng CX. A Lys644Glu substitution in fibroblast growth factor receptor 3 (FGFR3) causes dwarfism in mice by activation of STATs and ink4 cell cycle inhibitors. Hum Mol Genet. 1999;8:35–44. doi: 10.1093/hmg/8.1.35. [DOI] [PubMed] [Google Scholar]
- 56.Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsushita T, Wilcox WR, Chan YY, Kawanami A, Bukulmez H, Balmes G, Krejci P, Mekikian PB, Otani K, Yamaura I, Warman ML, Givol D, Murakami S. FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Hum Mol Genet. 2009;18:227–240. doi: 10.1093/hmg/ddn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Cao XU. BMP signaling and skeletogenesis. Ann N Y Acad Sci. 2006;1068:26–40. doi: 10.1196/annals.1346.006. [DOI] [PubMed] [Google Scholar]
- 59.Zhou YX, Xu X, Chen L, Li C, Brodie SG, Deng CX. A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum Mol Genet. 2000;9:2001–2008. doi: 10.1093/hmg/9.13.2001. [DOI] [PubMed] [Google Scholar]
- 60.Jacob AL, Smith C, Partanen J, Ornitz DM. Fibroblast growth factor receptor 1 signaling in the osteo-chondrogenic cell lineage regulates sequential steps of osteo-blast maturation. Dev Biol. 2006;296:315–328. doi: 10.1016/j.ydbio.2006.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurozumi K, Nishita M, Yamaguchi K, Fujita T, Ueno N, Shibuya H. BRAM1, a BMP receptor-associated molecule involved in BMP signalling. Genes Cells. 1998;3:257–264. doi: 10.1046/j.1365-2443.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- 62.Nishanian TG, Waldman T. Interaction of the BMPR-IA tumor suppressor with a developmentally relevant splicing factor. Biochem Biophys Res Commun. 2004;323:91–97. doi: 10.1016/j.bbrc.2004.08.060. [DOI] [PubMed] [Google Scholar]
- 63.Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-[beta] signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 64.Nohe A, Keating E, Underhill TM, Knaus P, Petersen NO. Dynamics and interaction of caveolin-1 isoforms with BMP-receptors. J Cell Sci. 2005;118:643–650. doi: 10.1242/jcs.01402. [DOI] [PubMed] [Google Scholar]
- 65.Kavsak P, Rasmussen R, Causing C, Bonni S, Zhu H, Thomsen G, Wrana J. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFβ receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 66.Thien CBF, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.