Abstract
Background and Objectives.
Reduced lean mass and physical function is a characteristic of frailty. However, it is currently unknown if proteolysis through the E3 ubiquitin ligases and the autophagic lysosomal pathway is dysregulated in inactive frail older women. The purpose of this study was to determine the expression of key markers of ubiquitin-mediated and autophagic lysosomal proteolysis in inactive (N = 7) compared with active (N = 7) older women.
Methods.
Strength, mobility, leg lean mass, and physical activity assessment were used to characterize activity levels and frailty in older women. Vastus lateralis biopsies were collected after an overnight fast and were assessed for gene and protein targets related to E3 ubiquitin ligases and the autophagic lysosomal system.
Results.
We found that AMP-activated protein kinase alpha (Thr172) was increased (p = .045), and forkhead box O3A (FOXO3A) gene expression (p = .047) was lower in inactive frail older women. Foxo3a (Ser253), Beclin1 (Ser93/96), and class III phosphatidylinositol-3-kinase (VPS34) protein expression were not different between the groups (p > .05). Neural precursor cell-expressed developmentally downregulated protein 4, muscle ring finger 1, muscle atrophy F-box, and the autophagy/mitophagy gene expression markers, Beclin1, autophagy-related-7, BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3), dynamin-related protein 1, and Parkinson protein 2 (PARKIN) were lower in inactive frail older women (p < .05). Autophagy/mitophagy markers were positively correlated with the 6-minute walk and leg lean mass (p < .05).
Conclusions.
We conclude that physical inactivity in frail older women is associated with a downregulation of ubiquitin-mediated and autophagic lysosomal skeletal muscle gene expression, perhaps related to low muscle mass and poor physical function.
Key Words: Aging, Autophagy, Mitochondrial dysfunction, Atrophy, Mobility.
Sarcopenia—the decline in skeletal muscle mass and function—is a common phenotypic trait of frail older adults. Sarcopenia can underlie and contribute to an increased risk of falls (1) and death (2), whereas physical inactivity can accelerate loss in lean tissue and ensuing disability (3). However, the molecular basis underpinning sarcopenia in older adults and the role of physical inactivity in this process are complex and require further investigation.
The autophagic lysosomal system is a major catabolic cellular pathway activated during human muscle atrophy. Yet, far less information exists as to the role of autophagy in skeletal muscle of older adults (4,5). Autophagy involves the bulk degradation of cytoplasm and organelles through the lysosomal system (6) and can be organelle selective, as demonstrated with the mitochondria (mitophagy). Interestingly, mitochondrial dysfunction is linked to aging (7), low muscle mass (8), and deteriorating levels of physical performance (4), suggesting a potential dysregulation of the autophagy system (4). It is unclear if regulators of skeletal muscle autophagy and mitophagy are impaired in frail older adults with low physical activity levels.
Muscle atrophy can also occur through ubiquitin-mediated proteolysis. Activation of the muscle-specific E3 ubiquitin ligases, muscle ring finger 1 (MuRF1), and muscle atrophy F-box (MAFbx) are observed in rodent (9) and human models of muscle wasting (10–12). Recently, neural precursor cell-expressed developmentally downregulated protein 4 (NEDD4-1) has gained attention as another E3 ubiquitin ligase that is important in activating muscle atrophy (13,14), but it is unknown if NEDD4-1 expression is altered in the skeletal muscle of inactive frail older women.
Forkhead box 3a (Foxo3a) plays a unique role in the coordinated upregulation of ubiquitin-mediated and lysosomal proteolysis under conditions of muscle loss (15). Not only is Foxo3a capable of upregulating MuRF1 and MAFbx, but Foxo3a can transcriptionally regulate several autophagy/mitophagy markers including microtubule-associated protein 1B light chain 3 (LC3), class III phosphatidylinositol-3-kinase (VPS34), autophagy-related-7 (ATG7), BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3), PTEN-induced putative kinase 1 (PINK1) (16), and two recently identified upstream regulators of autophagy/mitophagy: mitochondrial E3 ubiquitin ligase 1 (MUL1) (17) and growth arrest and DNA damage-inducible 45a (GADD45A) (18). The relationship of Foxo3a and mediators of skeletal muscle ubiquitin proteolysis and autophagy may provide novel insight on potential mechanisms leading to sarcopenia and physical dysfunction in inactive frail older adults.
To date, no study has evaluated the relationship of physical activity and frailty and skeletal muscle ubiquitin-mediated autophagic lysosomal proteolysis in older women. The purpose of this study was to determine if key molecular markers related to ubiquitin-mediated and autophagy/mitophagy proteolysis are differentially regulated in the locomotor skeletal muscle of physically inactive frail older women. Our secondary goal was to determine if the AMP-activated protein kinase alpha (AMPKα)/Beclin1/VPS34 signaling pathway could clarify the downstream gene expression in muscle autophagy in inactive frail older women because this cellular axis is known to induce autophagy and regulate atrophy (19). We hypothesized that Foxo3a and AMPKα phosphorylation and downstream E3 ubiquitin ligase and autophagy/mitophagy gene expression would be dysregulated in muscle of inactive frail older women.
Methods
Participants
Fourteen older female volunteers (>65 y and <30 body mass index [BMI]) were recruited based upon the following inclusion/exclusion criteria: free of known diseases including diabetes, chronic obstructive pulmonary disease, chronic kidney disease, human immunodeficiency virus, and heart disease and free of any tobacco use and medications such as corticosteroids, nonsteroidal antiinflammatories, hormone replacements, and anticoagulants. Two groups of individuals were enrolled, active (N = 7) and inactive (N = 7) older women, and characterized by the results of both performance-based and self-report assessments. The active older women contained individuals who had modified physical performance test scores > 32 (not frail) and reported at least 90 minutes of moderate activity a week over the last year (20). The inactive older women contained individuals who had modified physical performance test scores of <25 (moderately frail or frail) and reported little to no planned physical activity over the last year (20). Enrolled participant characteristics are found in Table 1. All participants read and signed the informed consent approved by the University of Utah Institutional Review Board.
Table 1.
Participant Characteristics
| Active Women | Inactive Women | |
|---|---|---|
| N | 7 | 7 |
| Age (y) | 77.3 (1.7) | 83.0 (1.8)* |
| BMI (kg/m2) | 22.6 (0.6) | 25.0 (1.3) |
| PASE | 221.0 (23.7) | 58.6 (11.5)* |
| MPPT | 35.7 (0.2) | 17.6 (2.8)* |
| Leg Lean CSA (%) | 88.2 (0.6) | 82.0 (2.0)* |
| 6MW (m) | 518.6 (28.6) | 308.7 (49.1)* |
| GS (m/s) | 1.12 (0.1) | 0.72 (0.1)* |
| Stair Ascent (s) | 4.7 (0.2) | 24.1 (8.9)* |
| MVIC (N) | 267.3 (27.3) | 167.4 (19.2)* |
| MQ (N/cm2) | 2.4 (0.34) | 3.3 (0.25)* |
| Serum insulin (pmol/L) | 41.7 (8.84) | 77.4 (13.84)* |
| Serum glucose (mg/dL) | 5.1 (0.24) | 5.2 (0.10) |
Notes: Data are presented as mean (SE). BMI = body mass index; PASE = physical activity scale for the elderly; MPPT = modified physical performance test score; Leg Lean CSA = percentage of thigh lean tissue average cross-sectional area; 6MW = 6-min walk test; GS = self-selected gait speed; MVIC = maximal voluntary isometric contraction of knee extensor muscles; MQ = muscle quality determined by MVIC and lean mass cross-sectional area. Serum insulin and glucose values were sampled following an overnight fast. *Significant difference between groups (p ≤ .05).
Physical Activity, Strength, and Mobility Measurements
To quantify physical activity levels, participants were asked to complete the physical activity scale for the elderly. The physical activity scale for the elderly is a self-report measure that has been shown to be reliable and valid method for estimating levels of physical activity in older adults and not only includes planned physical activity but also considers the contribution of leisure time, volunteer, and housework activities (21). Additionally, participants underwent knee extension strength testing via a maximum voluntary isometric contraction on a KinCom dynamometer (Chattanooga Inc. Hixon, TN). Mobility was determined using three tests: (a) a 6-minute walk (6MW), (b) stair ascent, and (c) self-selected gait speed. These performance tests have been shown to be both valid and reliable in this population (22).
Lean Muscle Mass Analysis
Magnetic resonance imaging was used to determine the quadriceps’ lean tissue cross-sectional area in cm2 from the magnetic resonance imaging scans using the 3-point Dixon method, after excluding subcutaneous adipose tissue and bone (23). Muscle quality was determined by the ratio of maximum voluntary isometric contraction and lean mass cross-sectional area reported as N/cm2.
Insulin and Glucose
Plasma glucose concentrations were measured on fasting blood samples using an automated glucose analyzer (YSI, Yellow Springs, OH). Plasma insulin was measured by Associated Regional and University Pathologists laboratories using a quantitative chemiluminescent immunoassay.
Muscle Biopsy
All individuals arrived at the University of Utah Center for Clinical and Translational Sciences in the morning after a 12-hour fast for a muscle biopsy. All participants were asked to refrain from strenuous activity for 48 hours prior to the biopsy and from taking statins for at least 7 days prior to the muscle biopsy. Statin medication usage, as recorded during initial screening were N = 2 for active healthy older women and N = 3 or inactive frail older women. Muscle biopsies were taken from the vastus lateralis using aseptic technique, local anesthesia (1% lidocaine), and a 5-mm Bergström biopsy needle. All muscle tissue was immediately blotted, flash-frozen in liquid nitrogen, and stored at −80°C for further analysis.
Western Blotting
Western blotting procedures have been described elsewhere (24). All western blot data were normalized to the internal control, and replicate samples were averaged. Alpha tubulin was used as a loading control.
Primary antibodies against protein targets, purchased from Cell Signaling, Danvers, MA, were the following (catalog #): phospho-Foxo3a Ser253 (#9466), Foxo3a (#2497), phospho-AMPKα Thr172 (#2531), AMPKα (#2532), phospho-Beclin-1 Ser93/96 (#12476), Beclin1 (#3495), and VPS34 (#3358). Alpha-tubulin (#F2168) was purchased from Sigma–Aldrich (St. Louis, MO). Donkey antirabbit immunoglobin G horseradish peroxidase-conjugated secondary antibody (#sc-2313) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Gene Expression
Detailed methods can be found elsewhere (25). Cycle threshold values of target genes were normalized to beta 2-microglobulin (β2M), then fold change values were calculated (2−∆∆Ct). We carried out validation of β2M to determine if this reference gene was unchanged between the two groups (26). The validation of β2M produced a fold change of 1.07 ± 0.16 and a slope of 0.069, indicating that β2M is a suitable reference gene.
Predesigned primers for GADD45A, microtubule-associated protein 1B light chain 3 (LC3), and BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3) were purchased from Life Technologies (CA). Unpublished custom primers were designed with Beacon Designer (Premier Biosoft International, Palo Alto, CA), purchased from Life Technologies, and carefully optimized (see Supplemental Table 1).
Statistical Analysis
Analysis of the subject’s characteristics and gene and protein expressions between active and inactive groups were conducted using a paired t test. A linear regression was used to determine relationships between functional measurements (6MW) and molecular regulators of autophagy/mitophagy (SigmaPlot v12.5, Sysstat software inc., San Jose, CA). Multiple stepwise regression was performed with SAS PROC REG utilizing SAS software package (Carrey, NC). Significance was set at p ≤ .05. All values are presented as mean ± SE.
Results
Subject Characteristics, Lean Area, Strength, and Mobility (Table 1)
By design, inactive older women (n = 7) scored lower on the physical activity scale for the elderly and modified physical performance tests designating them as moderately frail (p < .05). Further, inactive older women had lower lean leg muscle area, isometric knee extensor strength and muscle quality, covered less distance on the 6MW, and had slower gait speed and stair accent time than the active group (p < .05). Active and inactive women had similar BMI (p > 0.05); however, the inactive women were on average 6 years older than the active older women (p = .039) (age range: active: 71–83 y; Inactive: 76–91 y). Finally, inactive frail older women had higher fasting insulin levels than that of the active healthy older women (p < .05), but fasting glucose levels were not different between the groups (p > .05).
Foxo3a Ser253 phosphorylation tended to be increased by approximately 70% in inactive frail older women but was not statistically different between the groups (p = .072). We were unable to detect Foxo3a total protein levels within the current samples. However, we found that skeletal muscle AMPKα Thr172 phosphorylation was increased by approximately 50% in inactive frail compared with active healthy older women (p = .045). There were no differences in Beclin1 Ser93/96 phosphorylation (p = .146) or total VPS34 protein levels (p = .774) between the two groups. Total protein levels were unchanged between active healthy and inactive frail older women for AMPKα (p = .306) and Beclin1 (p = .553). FOXO3A gene expression was decreased (p = .047) in skeletal muscle of inactive frail women. As for E3 ubiquitin ligase expression, we found that NEDD4-1 (p = .014), MURF1 (p = .018), and MAFBX (p = .011) muscle gene expression was drastically lower (~40%) in inactive frail compared with active healthy older women. However, MUL1 (p = .105) gene expression was not different between the groups. Data for protein and select gene expression can be found in Table 2.
Table 2.
Protein and Select Gene Expression in Active Healthy and Inactive Frail Older Women
| Active | Inactive | |
|---|---|---|
| Protein expression | ||
| Foxo3a (Ser253) | 1.50±0.39 | 2.61±0.41 |
| AMPKα (Thr172) | 1.55±0.25 | 2.37±0.27* |
| AMPKα | 2.57±0.40 | 3.22±0.47 |
| Beclin1 (Ser93/96) | 3.07±0.48 | 4.03±0.40 |
| Beclin1 | 4.32±0.63 | 3.84±0.48 |
| VPS34 | 1.96±0.18 | 1.88±0.23 |
| Gene expression | ||
| FOXO3A | 1.05±0.15 | 0.50±0.10* |
| MUL1 | 1.01±0.07 | 0.83±0.08 |
| MAFBX | 1.07±0.15 | 0.58±0.07* |
| MURF1 | 1.08±0.16 | 0.59±0.08* |
| NEDD4-1 | 1.03±0.10 | 0.66±0.09* |
Notes: AMPKα = AMP-activated protein kinase alpha. Data are mean ± SE. *Indicates significantly different from active healthy older women (p < .05).
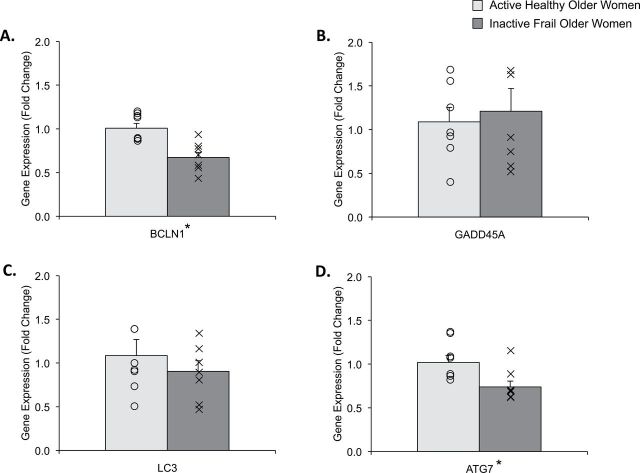
We found that Beclin1 (BCLN1; Figure 1A; p = .001) and autophagy-related 7 (ATG7; Figure 1D; p = .021) gene expression was lower (~40%) in inactive frail compared with active healthy older women. GADD45A (Figure 1B; p = .698) and LC3 (Figure 1C; p = .435) gene expression remained unchanged between the groups.
Figure 1.
Figure represents skeletal muscle gene expression data for (A) BCLN1, (B) GADD45A, (C) LC3, and (D) ATG7 in active healthy (light gray) and inactive frail (dark gray) older women. Data (mean ± SE) are reported as fold change from active healthy older women. The “O” and “X” represent individual data points for the active healthy and inactive frail older women, respectively. *Indicates significantly different from active healthy older women (p < .05).
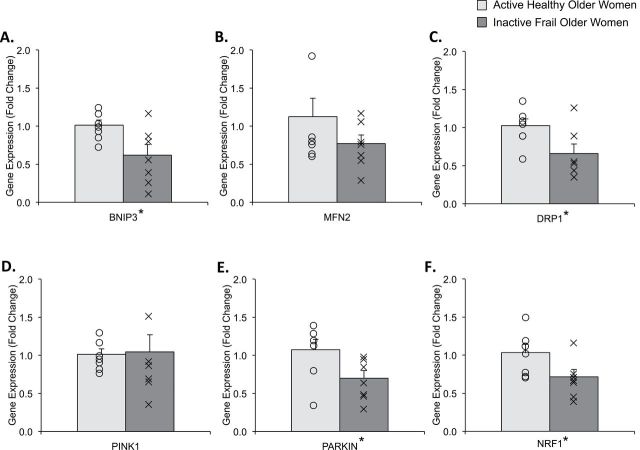
We found that BNIP3 (Figure 2A; p = .026), dynamin-1 like (DRP1; Figure 2C; p = .034), Parkinson protein 2 (PARKIN), E3 ubiquitin protein ligase (Figure 2E; p = .044), and nuclear respiratory factor-1 (NRF1; Figure 4F; p = .047) gene expression was lower (~30–40%) in inactive frail compared with active healthy older women. However, there were no differences in the gene expression for mitofusin 2 (MFN2; Figure 2B; p = .210) and PINK1 (Figure 2D; p = .897) between the two groups.
Figure 2.
Figure represents skeletal muscle gene expression data for (A) BNIP3, (B) MFN2, (C) DRP1, (D) PINK1, (E) PARKIN and (F) NRF1 in active healthy (light gray) and inactive frail (dark gray) older women. Data (mean ± SE) are reported as fold change from active healthy older women. The “O” and “X” represent individual data points for the active healthy and inactive frail older women, respectively. *Indicates significantly different from active healthy older women (p < .05).
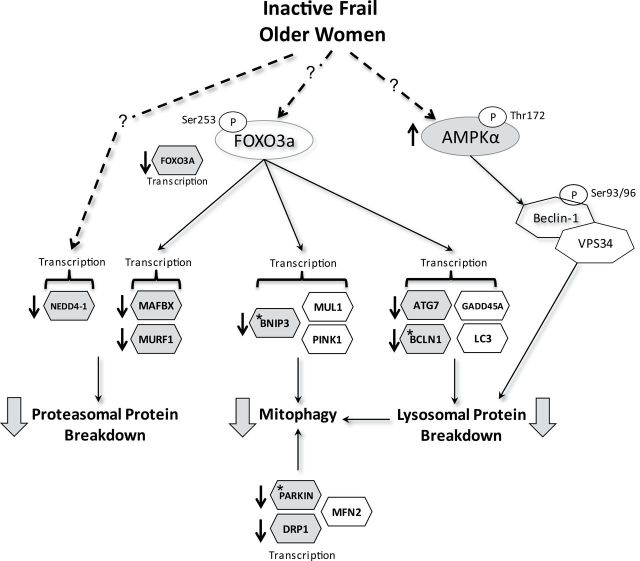
Figure 4.
Physical inactivity and frailty in older women is associated with downregulation of key regulators of the proteosomal and autophagy/mitophagy pathways (large shaded arrows). Schematic represents regulation of skeletal muscle ubiquitin-mediated and autophagic lysosomal proteolysis by Foxo3a and AMPKα activity. Molecules identified in schematic were examined in the following experiment. Dashed lines with question marks represent unknown upstream regulators of Foxo3a, AMPKα, and NEDD4-1. Shaded molecules with arrows indicate differences observed in skeletal muscle of inactive frail compared with active healthy older women (p < .05). Stars within shaded molecules represent a positive correlation with the 6-min walk test (ie, lower expression associated with lower 6MW distance [p < .05]). Leg lean mass was positively correlated with ATG7 and BCLN1 expression (not indicated on figure). We believe that these molecular events may be related to mitochondrial dysfunction, low muscle volume, and physical function deficits in inactive, frail older women.
To assess the potential effect of age and BMI on these measured outcomes, we performed multiple stepwise regression with age, BMI, and physical inactivity + frailty as independent variables. In this analysis, AGE entered as a significant covariate for only FOXO3A (p = .03) and NEDD4.1 (p = .001) gene expression. BMI was not a significant covariate for any variable.
Finally, we found a positive correlation with the 6MW and BCLN1 (Figure 3A; R = 0.597, p = .024), BNIP3 (Figure 3C; R = 0.570, p = .033), and PARKIN (Figure 3E; R = 0.618, p = .019) gene expression. ATG7 (Figure 3B; R = 0.428, p = .127) and DRP1 (Figure 3D; R = 0.411, p = .145) gene expression was not correlated with 6MW. Additionally, a positive correlation with leg lean muscle mass (%) was detected for ATG7 (R = 0.528; p = .052) and PARKIN (R = 0.538; p = .047) gene expression. DRP1 (R = 0.266; p = .358), BCLN1 (R = 0.460; p = .098), and BNIP3 (R = 0.447; p = .109) gene expression was not statistically correlated with leg lean muscle mass (data not shown). Outcomes in this study are summarized in Figure 4.
Figure 3.
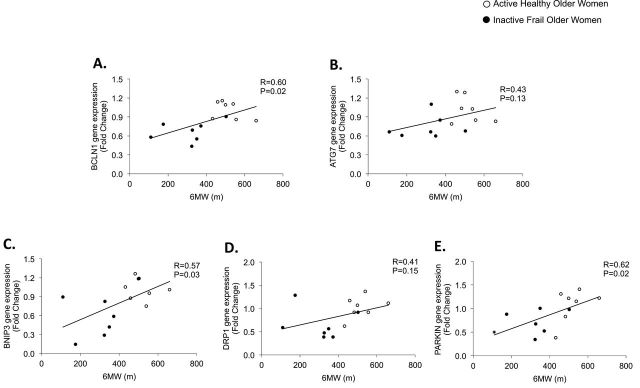
Data represent correlations and p values for the 6-min walk in relation to the following genes in skeletal muscle of active healthy and inactive frail older women: (A) BCLN1, (B) ATG7, (C) BNIP3, (D) DRP1, and (E) PARKIN. Gene expression data are reported as fold change and 6-min walk are reported in meters.
Discussion
The novel finding from this study was that several genes linked to skeletal muscle autophagy and mitophagy were correlated to physical function and leg lean mass in inactive frail older women. Specifically, decreased gene expression of BCLN1, BNIP3, and PARKIN was positivity correlated with lower distance covered on the 6MW, and lower PARKIN and ATG7 gene expression was correlated to low leg lean mass. These data suggest that skeletal muscle of inactive frail older women is characterized by a decrease activity in two major muscle catabolic pathways (Figure 4), suggesting that the decreased autophagy and proteolysis are associated with muscle loss and low physical function.
Several genes related to autophagy and mitophagy were robustly (~40%) decreased in skeletal muscle of inactive frail older women. Although we found that FOXO3A gene expression was downregulated in muscle of inactive frail older women, Foxo3a phosphorylation in whole muscle homogenates was not significantly different between the groups. We did observe an elevated AMPKα phosphorylation in muscle of inactive frail women but was not associated with phosphorylation of Beclin1. It is possible that an increased AMPKα activity in inactive frail older women may reflect decreased skeletal muscle protein synthesis by inhibition of the mTORC1 signaling pathway (27) or perhaps AMPKα could be upregulating FOXO3a-mediated autophagy via Ser413/588 phosphorylation (28), but this information was not examined in this experiment. Together, our data indicate for the first time that the skeletal muscle autophagy/mitophagy gene expression is downregulated in inactive frail older women and does not appear to be related to the AMPKα/beclin1/VPS34 signaling axis. The role of skeletal muscle Foxo3a in response to physical inactivity and frailty in older women remains to be determined.
Several reports have provided evidence that muscle autophagy is reduced with aging (29–31), and we add that this downregulated response may be also triggered by physical inactivity and frailty. In contrast to our findings, short-term muscle disuse (bed rest) in healthy younger adults increased skeletal muscle autophagy (ie, Beclin1) (32). However, it is likely that chronic physical inactivity in frail older adults (our study) affected skeletal muscle autophagic lysosomal system differently, compared with short-term physical inactivity in young participants (study by Brocca and colleagues.) (32). Interestingly, exercise training in old rodents normalized the age-related decrease in skeletal muscle autophagy (ie, Beclin1 and ATG7) to levels found in young rodents (33), suggesting that downregulated autophagy may be caused by inactivity and may be reversible; however, this has not yet been investigated in frail older adults.
We suggest that a downregulation of key regulators of skeletal muscle mitophagy (BNIP3, DRP1, PARKIN) may indicate a degree of mitochondrial dysfunction in the muscle of inactive older women as also reported with aging (7). In support, we found a downregulation of muscle NRF1 gene expression (a positive transcriptional regulator of mitochondrial proteins) (34) and higher fasting blood insulin levels in inactive frail older women (as observed in type 2 diabetic patients with low mitochondrial volume) (35) providing indirect evidence that mitochondrial abundance may be reduced in this population. Further, we also found an imbalance in mitochondrial fission (ie, decreased DRP1) and fusion (ie, unchanged MFN2) regulators, perhaps inferring an accumulation of nonfunctional mitochondria. However, with such limited fusion/fission data, further investigation in necessary to support this hypothesis. Importantly, we note that several downregulated mitophagy and autophagy genes were positively correlated with poor physical function (ie, 6MW) and reduced lean tissue percentage in inactive frail older women. Others have observed a decline in mitochondrial function in older adults with low mobility (4). Mitochondrial dysfunction is also related to muscle loss (8) as represented by increased muscle mitochondrial apoptotic events in community-dwelling older adults with low muscle volume (36). Together, decreased expression of mitophagy key regulators may be related to mitochondrial dysfunction and the associated low lean muscle mass and physical function deficits found in inactive frail older women.
We also noted a approximately 40% decrease in MURF1 and MAFBX gene expression in inactive frail older women. Contrary to our findings, Raue and colleagues (37) identified an increase in skeletal muscle MURF1 and MAFBX gene expression in healthy older women compared with younger women. Likely, the discrepancy between Raue and colleagues (37) and our study is that the older women in our study were frail and likely less physically active than the healthy older women cited in Raue and colleagues (37). An interesting observation in our study was that some participants were on statins. Chronic statin medication usage may induce muscle proteolysis through an increased MAFBX expression as observed in statin-treated patients (38). However, we do not believe this can explain our findings since approximately an equal number of participants in the active (N = 2) and inactive (N = 3) older women were taking statins, and the inactive frail older women had lower expression of MURF1 and MAFBX and less lean mass. In agreement with our interpretation of the autophagic lysosomal molecular markers, it is also possible that low expression of E3 ubiquitin ligases may represent an impaired proteolytic system that has slowed in its ability to turnover nonfunctional/damaged proteins.
Another unique finding in this study was that NEDD4-1 gene expression was also decreased in inactive frail older women. Some targets of NEDD4-1-mediated ubiquination are regulators of muscle tissue regeneration such as the Smad transcription factors (39) and Notch-1 (13,14). Perhaps, physical inactivity and frailty in older women alters normal healthy muscle regeneration (eg, proliferation, differentiation) in response to tissue injury.
A limitation of this study was the relatively small sample size for the two groups. Additionally, we recognize the difference in age between the two groups although age (nor BMI) was not a significant covariate for any of the molecular markers except FOXO3A and NEDD4.1, using multiple stepwise regression. Finally, it is not possible for us to tease apart the individual effects of physical inactivity and frailty on muscle E3 ubiquitin ligase and autophagy/mitophagy markers measured in this study, but it is quite common to see physical inactivity to coexist in frail older adults.
In summary, we report that skeletal muscle of inactive frail older women has a transcriptional profile characterized by a decreased E3 ubiquitin ligase and autophagy/mitophagy system that may be related to mitochondrial dysfunction, low muscle mass, and poor physical function. Further examination is necessary to determine if these molecular regulators are valuable therapeutic targets in frail older women who are classified as sarcopenic.
Supplementary Material
Supplementary material can be found at: http://biomedgerontology.oxfordjournals.org/
Funding
This work was supported by grants from the National Institute on Aging (K01AG038556 to M.J.D.), National Center for Advancing Translational Sciences (1ULTR001067 to D.A.M.), and the University of Utah Research Foundation (10020526 to R.L.M.).
Acknowledgments
We thank the volunteers for participating in this study and the Center for Clinical and Translational Sciences nursing and medical staff for assisting with the screening and biopsy procedure.
References
- 1. Landi F, Liperoti R, Russo A, et al. Sarcopenia as a risk factor for falls in elderly individuals: results from the ilSIRENTE study. Clin Nutr. 2012;31:652–658. 10.1016/j.clnu.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 2. Rockwood K, Mitnitski A, Song X, Steen B, Skoog I. Long-term risks of death and institutionalization of elderly people in relation to deficit accumulation at age 70. J Am Geriatr Soc. 2006;54:975–979. 10.1111/j.1532-5415.2006.00738.x [DOI] [PubMed] [Google Scholar]
- 3. English KL, Paddon-Jones D. Protecting muscle mass and function in older adults during bed rest. Curr Opin Clin Nutr Metab Care. 2010;13:34–39. 10.1097/MCO.0b013e328333aa66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joseph AM, Adhihetty PJ, Buford TW, et al. The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell. 2012;11:801–809. 10.1111/j.1474-9726.2012.00844.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fry CS, Drummond MJ, Glynn EL, et al. Skeletal muscle autophagy and protein breakdown following resistance exercise are similar in younger and older adults. J Gerontol A Biol Sci Med Sci. 2013;68:599–607. 10.1093/gerona/gls209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. 10.1016/j.febslet.2010.01.056 [DOI] [PubMed] [Google Scholar]
- 7. Hiona A, Leeuwenburgh C. The role of mitochondrial DNA mutations in aging and sarcopenia: implications for the mitochondrial vicious cycle theory of aging. Exp Gerontol. 2008;43:24–33. 10.1016/j.exger.2007.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marzetti E, Hwang JC, Lees HA, et al. Mitochondrial death effectors: relevance to sarcopenia and disuse muscle atrophy. Biochim Biophys Acta. 2010;1800:235–244. 10.1016/j.bbagen.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bodine SC, Stitt TN, Gonzalez M, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. 10.1038/ncb1101-1014 [DOI] [PubMed] [Google Scholar]
- 10. Constantin D, McCullough J, Mahajan RP, Greenhaff PL. Novel events in the molecular regulation of muscle mass in critically ill patients. J Physiol. 2011;589(Pt 15):3883–3895. 10.1113/jphysiol.2011.206193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suetta C, Frandsen U, Jensen L, et al. Aging affects the transcriptional regulation of human skeletal muscle disuse atrophy. PLoS One. 2012;7:e51238. 10.1371/journal.pone.0051238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones SW, Hill RJ, Krasney PA, O’Conner B, Peirce N, Greenhaff PL. Disuse atrophy and exercise rehabilitation in humans profoundly affects the expression of genes associated with the regulation of skeletal muscle mass. FASEB J. 2004;18:1025–1027. 10.1096/fj.03-1228fje [DOI] [PubMed] [Google Scholar]
- 13. Koncarevic A, Jackman RW, Kandarian SC. The ubiquitin-protein ligase Nedd4 targets Notch1 in skeletal muscle and distinguishes the subset of atrophies caused by reduced muscle tension. FASEB J. 2007;21:427–437. 10.1096/fj.06-6665com [DOI] [PubMed] [Google Scholar]
- 14. Nagpal P, Plant PJ, Correa J, et al. The ubiquitin ligase Nedd4-1 participates in denervation-induced skeletal muscle atrophy in mice. PLoS One. 2012;7:e46427. 10.1371/journal.pone.0046427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. 10.1016/j.cmet.2007.11.004 [DOI] [PubMed] [Google Scholar]
- 16. Sandri M. Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int J Biochem Cell Biol. 2013;45:2121–2129. 10.1016/j.biocel.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lokireddy S, Wijesoma IW, Teng S, et al. The ubiquitin ligase Mul1 induces mitophagy in skeletal muscle in response to muscle-wasting stimuli. Cell Metab. 2012;16:613–624. 10.1016/j.cmet.2012.10.005 [DOI] [PubMed] [Google Scholar]
- 18. Ebert SM, Dyle MC, Kunkel SD, et al. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J Biol Chem. 2012;287:27290–27301. 10.1074/jbc.M112.374777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown M, Sinacore DR, Binder EF, Kohrt WM. Physical and performance measures for the identification of mild to moderate frailty. J Gerontol A Biol Sci Med Sci. 2000;55:M350–M355. 10.1093/gerona/55.6.M350 [DOI] [PubMed] [Google Scholar]
- 21. Washburn RA, McAuley E, Katula J, Mihalko SL, Boileau RA. The physical activity scale for the elderly (PASE): evidence for validity. J Clin Epidemiol. 1999;52:643–651. 10.1016/S0895-4356(99)00049-9 [DOI] [PubMed] [Google Scholar]
- 22. Enright PL, McBurnie MA, Bittner V, et al. ; Cardiovascular Health Study. The 6-min walk test: a quick measure of functional status in elderly adults. Chest. 2003;123:387–398. 10.1378/chest.123.2.387 [DOI] [PubMed] [Google Scholar]
- 23. Kovanlikaya A, Guclu C, Desai C, Becerra R, Gilsanz V. Fat quantification using three-point dixon technique: in vitro validation. Acad Radiol. 2005;12:636–639. 10.1016/j.acra.2005.01.019 [DOI] [PubMed] [Google Scholar]
- 24. Drummond MJ, Timmerman KL, Markofski MM, et al. Short-term bed rest increases TLR4 and IL-6 expression in skeletal muscle of older adults. Am J Physiol Regul Integr Comp Physiol. 2013;305:R216–R223. 10.1152/ajpregu.00072.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drummond MJ, Glynn EL, Fry CS, Timmerman KL, Volpi E, Rasmussen BB. An increase in essential amino acid availability upregulates amino acid transporter expression in human skeletal muscle. Am J Physiol Endocrinol Metab. 2010;298:E1011–E1018. 10.1152/ajpendo.00690.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 27. Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. AMP-activated protein kinase suppresses protein synthesis in rat skeletal muscle through down-regulated mammalian target of rapamycin (mTOR) signaling. J Biol Chem. 2002;277:23977–23980. 10.1074/jbc.C200171200 [DOI] [PubMed] [Google Scholar]
- 28. Greer EL, Oskoui PR, Banko MR, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. 10.1074/jbc.M705325200 [DOI] [PubMed] [Google Scholar]
- 29. Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. 10.4161/auto.1.3.2017 [DOI] [PubMed] [Google Scholar]
- 30. Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- 31. Wohlgemuth SE, Seo AY, Marzetti E, Lees HA, Leeuwenburgh C. Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp Gerontol. 2010;45:138–148. 10.1016/j.exger.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brocca L, Cannavino J, Coletto L, et al. The time course of the adaptations of human muscle proteome to bed rest and the underlying mechanisms. J Physiol. 2012;590(Pt 20):5211–5230. 10.1113/jphysiol.2012.240267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim YA, Kim YS, Oh SL, Kim HJ, Song W. Autophagic response to exercise training in skeletal muscle with age. J Physiol Biochem. 2013;69:697–705. 10.1007/s13105-013-0246-7 [DOI] [PubMed] [Google Scholar]
- 34. Viña J, Gomez-Cabrera MC, Borras C, et al. Mitochondrial biogenesis in exercise and in ageing. Adv Drug Deliv Rev. 2009;61:1369–1374. 10.1016/j.addr.2009.06.006 [DOI] [PubMed] [Google Scholar]
- 35. Giebelstein J, Poschmann G, Højlund K, et al. The proteomic signature of insulin-resistant human skeletal muscle reveals increased glycolytic and decreased mitochondrial enzymes. Diabetologia. 2012;55:1114–1127. 10.1007/s00125-012-2456-x [DOI] [PubMed] [Google Scholar]
- 36. Marzetti E, Lees HA, Manini TM, et al. Skeletal muscle apoptotic signaling predicts thigh muscle volume and gait speed in community-dwelling older persons: an exploratory study. PLoS One. 2012;7:e32829. 10.1371/journal.pone.0032829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Proteolytic gene expression differs at rest and after resistance exercise between young and old women. J Gerontol A Biol Sci Med Sci. 2007;62:1407–1412 [DOI] [PubMed] [Google Scholar]
- 38. Hanai J, Cao P, Tanksale P, et al. The muscle-specific ubiquitin ligase atrogin-1/MAFbx mediates statin-induced muscle toxicity. J Clin Invest. 2007;117:3940–3951. 10.1172/JCI32741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao S, Alarcón C, Sapkota G, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–468. 10.1016/j.molcel.2009.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]