Abstract
Methyltransferase expression and DNA methylation are linked to aging and age-related disease. We utilized 3-, 12-, and 24-month-old Ames dwarf and their wild-type siblings to examine the genotype and age-related differences in the expression of methyltransferase enzymes related to DNA methylation in the liver, glycine-N-methyltransferase and DNA methyltransferase (DNMT). We found that DNMT proteins and transcripts are differentially expressed in dwarf mice compared with wild-type siblings that can be attributed to age and/or genotype. However, DNMT1 protein expression is drastically reduced compared with wild-type controls at every age. DNMT3a protein levels coincide with differences observed in DNMT activity. Growth hormone appears to modulate expression of DNMT1 and 3a in dwarf liver tissue and primary hepatocytes. Therefore, growth hormone may contribute to age-related processes, DNA methylation, and, ultimately, longevity.
Key Words: Ames dwarf mice, DNA methylation, DNA methyltransferase, Glycine-N-methyltransferase, Aging, Liver.
Epigenetic markers are important for the regulation of gene expression, cell differentiation, development, chromatin structure, aging, and carcinogenesis (1–6). One theory of aging suggests that the loss of epigenetic stability through alterations in epigenetic factors, such as DNA methylation, contributes to altered gene expression over time (7). Changes in DNA methylation and DNA methyltransferase (DNMT) expression are well documented in age-related diseases including cancer, autoimmune disease, atherosclerosis, rheumatoid arthritis, and Alzheimer’s disease (8). DNMTs contribute to epigenetic stability and consist of three known catalytically active isoforms that either maintain DNA methylation (DNMT1) or methylate DNA de novo (DNMT3a and DNMT3b; 9,10). The majority of studies have investigated DNA methylation alterations in embryonic development, stem cells, and cancer-derived cell lines. Though expression of DNMTs is postulated to contribute to epigenetic instability during aging, and aging murine liver tissue exhibits altered DNA methylation (11), these factors have not been studied in mice that exhibit a delayed aging phenotype and life-span extension.
DNMTs utilize S-adenosylmethionine (SAM) that serves as the primary methyl group donor provided by the methionine pathway. Glycine-N-methyltransferase (GNMT) directly regulates SAM availability and is altered in cancer cell lines and hepatocellular carcinoma (12–14). Activation of GNMT with retinoic acid alters DNA methylation in rat liver (15). DNMT is the only methyltransferase known to directly methylate DNA and is an important regulator of cellular responses at the epigenetic level. Thus, the role of GNMT on DNA methylation is indirect but critical in terms of maintaining appropriate epigenetic patterns and normal tissue function.
Components of the methionine pathway are altered in rats and mice fed methyl-deficient diets, and in turn, DNA methylation and DNMT protein and activity are affected (16–18). Methionine supplementation also increases DNA methylation and increases or decreases DNMT activity in HEPG2 cells depending on whether GNMT expression is induced (19). In addition, it is also known that methionine-restricted diets extend life span in rodents (20). Over time, alterations in DNA methylation via methionine deficiency could become irreversible potentially causing a permanent alteration in the plasticity of cellular responses to the environment and epigenetic stability (21). Taken together, this evidence suggests the possibility that altered GNMT expression and activity help to direct DNA methylation through its relationship to DNMT activity and expression. DNMTs may also play a presently undiscovered role in the longevity attributed to alterations in the methionine pathway and aging.
The Ames dwarf is a product of a single nucleotide mutation in the prop1 gene resulting in an underdeveloped pituitary gland and deficiencies of circulating growth hormone (GH), prolactin, and thyroid-stimulating hormone (22,23). Importantly, these diminutive mice live 49%–68% longer than their wild-type siblings, males and females, respectively (24). In addition, dwarf mice have a significantly decreased tumor incidence and burden, concurrent with their extended life spans compared with those of wild-type mice (24,25).
We have shown that dwarf mice exhibit atypical methionine metabolism, lower basal levels of SAM, higher basal levels of SAH, and a reduced methyl donor availability ratio without signs of liver pathology unlike other animal models (26,27). GH-deficient Ames mice exhibit significant enzyme expression and activity differences in the methionine pathway including GNMT compared with normal, wild-type mice (26,28,29). Importantly, GH is known to modulate the activity of GNMT (30), and we have found that both GH-treated and untreated mice exhibit a reduction in GNMT activity between 3 and 12 months of age (31). Our previous studies have focused on the methionine pathway in relation to oxidative metabolism and stress resistance. Our current interest in understanding mechanisms of aging and longevity has expanded to include this novel investigation of the role of DNMTs, GNMT, and the effects of GH on methylation in long-living dwarf mice.
Methods
Animals and Tissue Preparation
Ames dwarf and age-matched wild-type mice were obtained from a closed colony at the University of North Dakota. The Ames dwarf mice (df/df) were produced by crossing heterozygous (df/+) or homozygous (df/df) male mice with carrier females (df/+). The mice were kept under standard laboratory conditions with a 12-hour light/dark cycle and fed ad libitum (8640 Teklad 22/5) with free access to water. Liver tissue was collected from mice at 3, 12, and 24 months of age and divided into portions, rapidly frozen, and stored at −80°C. These tissues were utilized to evaluate mRNA expression, protein abundance, and enzyme activity levels. All procedures involving animals were reviewed and approved by the UND Institutional Animal Care and Use Committee in accordance with the NIH guidelines for the care and use of laboratory animals.
Immunoblot Analysis
Liver tissue and hepatocyte samples were analyzed using standard immunoblotting procedures previously published (32,36). Protein abundance was determined using the Bradford assay (33). Specific proteins were detected with antibodies to DNMT 1 (Abcam, Cambridge, MA, ab13537), DNMT3a (ab14291), DNMT3b (ab13604), and GNMT (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, sc-166834) using chemiluminescence. Ponceau S staining was used to assess equivalent loading. Mean optical density was measured and used for comparative analysis. To maximize the genotype comparisons in our immunoblotting system, blots were assayed primarily across genotype.
DNMT Activity Assay
Liver nuclear extracts were prepared from frozen liver tissue using an Epiquick Nuclear Extraction Kit (OP-0002, Epigentek, Farmingdale, NY). Protein abundance was measured using the Bradford assay (33). Activity of DNMT was measured using Epigentek DNA Methyltransferase Activity/Inhibition Assay (P-3009, Epigentek) following manufacturer’s directions. Samples were run in triplicate and analyzed using dual-wave spectrophotometric analysis (630 and 450nm).
Global Methylation Assay
Genomic DNA was isolated from frozen liver tissue (Gentra Puregene Tissue Kit; Qiagen, Germantown, MD) and quantified by spectrophotometry (Epoch Micro-Volume Spectrophotometer System, BioTek Instruments, Winooski, VT). Global methylation was measured using the Methylamp Global DNA Methylation Quantification Ultra Kit (Epigentek) according to the manufacturer’s instructions. Samples were run in triplicate and analyzed using dual-wave spectrophotometric analysis (630 and 450 nm).
mRNA Expression via Reverse Transcriptase–Polymerase Chain Reaction
Total RNA was isolated and used to measure gene expression of the methyltransferases using real-time quantitative reverse transcriptase–polymerase chain reaction) employing methods published previously (28). Primers were obtained from previous literature (26,34,35), tested using NCBI primer blast, and optimized (Table 1). Melt curve analysis was used for quality control of the polymerase chain reaction and detection. The amount of tested gene complementary DNA relative to a control gene (β2-microglobulin) was determined using the ΔΔC t (comparative C t method; 28).
Table 1.
Primer Pairs Utilized for Gene-specific Real-time Reverse Transcriptase–Polymerase Chain Reaction
| Gene | GeneBank Accession No. | Primer 5′-3′ | Annealing Temperature |
|---|---|---|---|
| β2M | NM_00975 | Forward—AAGTATACTCACGCCACCCA | 60°C |
| Reverse—AAGACCAGTCCTTG | |||
| GNMT | NM_010321 | Forward—GCTGGACGTAGCCTGTGG | 60°C |
| Reverse—CACGCTCATCACGCTGAA | |||
| DNMT1 | NM_010066 | Forward—AAAGTGTGATCCCGAAGATCAAC | 60°C |
| Reverse—TGGTACTTCAGGTTAGGGTCGTCTA | |||
| DNMT3a | NM_007872 | Forward—TGCTACATGTGCGGGCATAA | 58°C |
| Reverse—GGAGTCGAGAAGGCCAGTCTT | |||
| DNMT3b | NM_001003961 | Forward—CCCAAGTTGTACCCAGCAATTC | 59°C |
| Reverse—TGCAATTCCATCAAACAGAGACA |
In Vivo GH Treatment of 6-Month-Old Ames Dwarf Mice
Porcine GH (NIDDK, National Hormone and Peptide Program, Torrance, CA) was administered to 6-month-old Ames dwarf mice to determine whether GH affects DNMT expression in the liver (36). Porcine GH in alkaline saline mixed with 50% polyvinylpyrrolidone (pH 9, 1:1) was injected subcutaneously twice daily (50 µL/injection) into one group of dwarf mice (n = 12). The vehicle, saline–polyvinylpyrrolidone, was injected subcutaneously (50 µL/injection) into another group of dwarf mice (n = 11) and age-matched wild-type mice (n = 11). All mice were injected twice daily (8:00 am and just before lights out at 8:00 pm) for 7 days (total of 13 injections). On the morning of the seventh day, 1 hour following the last injection of saline–polyvinylpyrrolidone or GH, liver tissue was collected from each group of mice, frozen, and stored at −80°C until analysis. The body weight of each mouse was recorded prior to starting the experiment, during, and following the last GH injection to validate weight gain in GH-treated animals. Liver weights of all the mice were also recorded.
Hepatocyte Isolation and GH Treatment In Vitro
Primary hepatocyte cultures were prepared with liver tissue isolated from 5- to 6-month-old Ames dwarf and wild-type mice using previously published preperfusion, collection, and culturing methods (36,37). Briefly, each animal was anesthetized with 2.5% tribromoethanol (i.p. 100 μL/10g body weight), the inferior vena cava was cannulated, and the liver was perfused with 37°C calcium-free HEPES preperfusion buffer (pH 7.65) for 15 minutes followed by perfusion with a 37°C collagenase solution (0.03% collagenase [Worthington, Lake Park, NJ]). Following perfusion, livers were removed, and the cells were transferred, washed, and cultured in serum-free HepatoStim Media (Becton Dickinson, Mountain View, CA). Trypan blue dye exclusion was used to assess cell viability and integrity. Cell collections with ≥95% viability were used. Cells for each genotype were pooled (n = 2–5 per genotype), resuspended in attachment medium (Life Technologies, Grand Island, NY), and seeded (2×106) into Matrigel-coated dishes. Following cell attachment (2 hours at 37°C and 5% CO2), the medium was exchanged for serum-free HepatoStim media (Life Technologies) containing 1% pen/strep and 2mM glutamine. Following overnight recovery, fresh serum-free media were added and the cells allowed to equilibrate (2 hours) before GH treatment (0.1, 1.0, 10, and 20 μg porcine GH per milliliter media). Twenty-four hours following treatment, cells were collected by washing with cold (4°C) HEPES and applying MatriSperse (BD Biosciences, Franklin Lakes, NJ) for 1 hour on ice. Dishes were scraped and cells pelleted and washed via centrifugation before homogenization for sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotting.
Statistical Analysis
Real-time quantitative polymerase chain reaction and global methylation data were analyzed with one-way or two-way analysis of variance (factors: genotype and age; GraphPad Prism 5.0) with Bonferroni’s or Dunnett’s post hoc tests as appropriate. Global methylation data in the in vivo GH experiment did not pass Levene’s test for equal variances and, thus, were analyzed using a Kruskal–Wallis test and Dunn’s multiple comparison posttest. Immunoblotting and DNMT activity data were analyzed using Student’s t test (GraphPad Prism 5.0). All data are shown as mean ± standard error of the mean. Sample values (n) are listed in the figure legends.
Results
Given that GNMT is linked to DNA methylation and possible DNMT activity via the methionine pathway, we investigated GNMT expression in dwarf and wild-type mice at three age groups. Similar to previous findings, GNMT liver mRNA is more abundantly expressed across all three age groups in the Ames dwarf compared with wild-type mice with significance at 3 and 12 months of age (Figure 1). The GNMT mRNA expression is influenced by genotype (p = .0001) and age (p = .0176). Protein levels of GNMT are also higher in dwarf mice at 12 months of age compared with age-matched wild-type controls (p = .0408).
Figure 1.
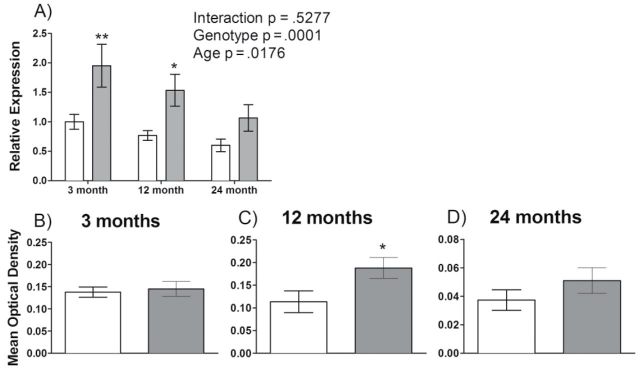
GNMT mRNA and protein expression are higher in Ames dwarf mice compared with their wild-type siblings. (A) Liver GNMT mRNA expression is shown for dwarf (gray bars) and wild-type mice (open bars) at 3, 12, and 24 months of age (n = 7–8). (B) GNMT protein levels at 3 (n = 10–11, p = .7248), (C) 12 (n = 8–10, p = .0408) and (D) 24 months (n = 6–8, p = .6618) of age. Mean optical densities are shown ± SEM. *p < .05 and **p < .01.
We examined mRNA expression of DNMT1, DNMT3a, and DNMT3b using reverse transcriptase–polymerase chain reaction analysis of isolated liver RNA. Surprisingly, we found that dwarf mice show a much higher mRNA expression of both DNMT1 (105%) and DNMT3a (238%) at 3 months of age (p < .0001; Figure 2). Gene expression of DNMT3b mRNA was similar between genotypes at all age groups studied. Age appeared to be an important factor in the expression of all three DNMTs. A significant effect of genotype was detected for DNMT1 and DNMT3a; however, the genotype by age interaction was significant confounding the main effects. The DNMT expression overall shows an interesting U-shaped pattern with the lowest levels at 12 months of age in both genotypes.
Figure 2.
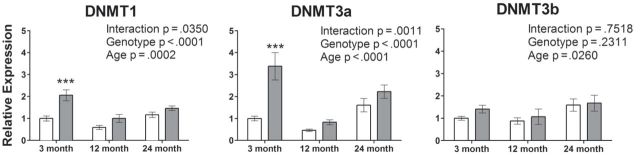
DNMT1 and DNMT3a mRNA expressions are higher in Ames dwarf mice at 3 months of age. Liver DNMT mRNA expression is shown for wild-type (open bars) and dwarf mice (gray bars) at 3, 12, and 24 months of age (n = 7–8). Mean relative changes in expression are shown ± SEM. ***p < .001.
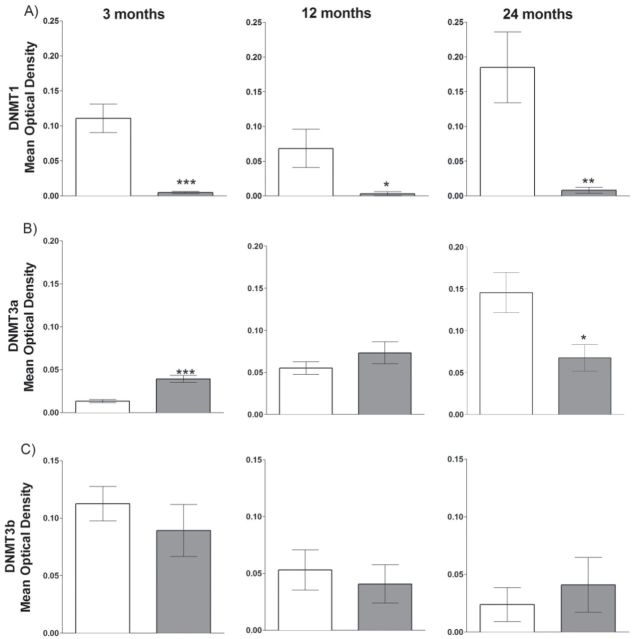
Protein levels of DNMT1, DNMT3a, and DNMT3b were compared between dwarf and wild-type mice at 3, 12, and 24 months of age (Figure 3). To maximize the genotypic comparisons, each immunoblotting assay included only one age group. Very low levels of DNMT1 protein were observed in the liver tissue of dwarf mice compared with wild-type mice at each age group examined (Figure 3A). The variability of protein levels between animals was more pronounced in wild-type compared with dwarf mice. DNMT3a protein levels were higher (193%) in the dwarf mice at 3 months of age (p < .001), similar at 12 months of age (p = .2464), and significantly lower (54%) at 24 months of age (p = .0172). Compared with wild-type controls, DNMT3b levels were not different between genotypes at any age, similar to mRNA findings at each age group.
Figure 3.
DNMT protein levels in Ames dwarf and wild-type mice liver tissue at 3, 12, and 24 months of age. Wild-type mice represented by open bars and dwarf mice by gray bars. (A) DNMT1 protein differences at 3 months (n = 10–11, p < .0001), 12 months (n = 9, p = .0427), and 24 months (n = 7–9, p = .0015). (B) DNMT3a protein differences at 3 months (n = 10–11, p < .0001), 12 months (n = 12, p = .2464), and 24 months (n = 8, p = .0172). (C) DNMT3b protein differences at 3 months (n = 10, p < .4000), 12 months (n = 11–12, p = .6244), and 24 months (n = 9–10, p = .5621). Mean optical densities are shown ± SEM. *p < .05, **p < .01, and ***p < .001.
Overall, DNMT activity was measured with an ELISA-based colorimetric assay using nuclear lysates extracted from liver tissue, as DNMT isoform-specific activity assays are not available (Figure 4). Independent assays comparing dwarf versus wild type for each age group were performed. Remarkably, dwarf mice displayed higher liver DNMT activity in young mice and similar DNMT activity in middle-aged mice (3 months, p = .0214; 12 months, p = .1704) but lower activities in old Ames dwarf mice (24 months, p = .0491) compared with that of wild-type mice (Figure 4). The DNMT activity in the liver more closely resembles DNMT3a protein expression than DNMT1 or DNMT3b.
Figure 4.
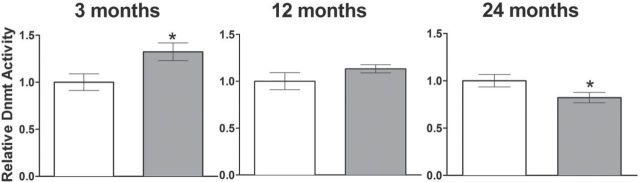
DNMT1 activity is higher in Ames dwarf mice at 3 months and lower at 24 months than age-matched wild-type siblings. ELISA-based colorimetric activity assay of nuclear liver lysate showing differences in total DNMT activity in 3-month-old (n = 11, p = .0214), 12-month-old (n = 8–11, p = .1704), and 24-month-old (n = 9–12, p = .0491) wild-type (open bars) and Ames dwarf mice (gray bars). Mean activities expressed as optical density per milligram per hour are shown ± SEM. *p < .05.
Porcine GH was administered to one group of 6-month-old Ames dwarf mice, whereas saline vehicle was administered to wild type and an additional group of dwarf mice as controls. In this study, GH-treated dwarf mice gained body weight over the course of 1 week (+4.19±0.06g; p < .0001) compared with saline-treated dwarf mice. Wild-type mouse body weights were not different pre- and postsaline injection. Liver weight was recorded as an additional indicator of GH action, and the mean liver weight of GH-injected dwarf mice was approximately 60% higher than saline-injected dwarf mice (0.70±0.13g and 0.44±0.05 g, respectively; p < .0001).
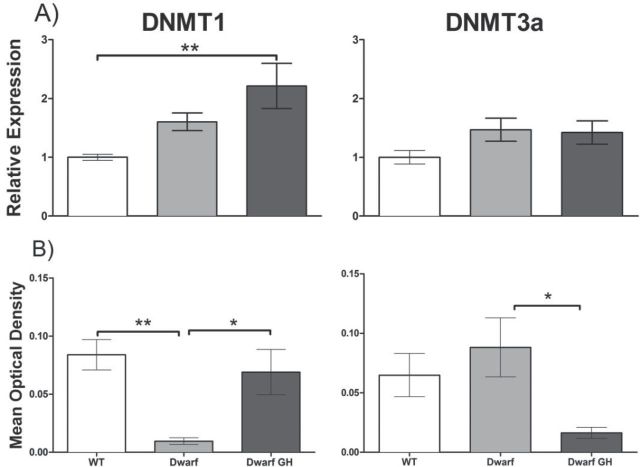
The expression of DNMT1 mRNA and protein were compared between GH- and saline-treated Ames dwarf and wild-type controls. Liver DNMT1 mRNA expression was higher in saline-treated dwarf mice than in saline-treated wild-type mice and increased further after GH administration (Figure 5A). Protein levels of DNMT1 were lower in saline-treated dwarf than in the saline-treated wild-type mice. However with GH treatment, protein expression of DNMT1 increased in dwarf mice (Figure 5B; p < .05). Expression of DNMT3a mRNA levels was higher in saline-treated dwarf mice compared with saline-treated wild-type mice, and no change was observed after GH treatment. However, DNMT3a protein levels were decreased following GH treatment in Ames dwarf mice (p < .05).
Figure 5.
DNMT mRNA and protein expression are altered by growth hormone in Ames dwarf liver. (A) Liver DNMT mRNA expression is shown for wild-type mice injected with saline (n = 10–11, open bars), dwarf mice injected with saline (n = 10–11, gray bars), and dwarf mice injected with growth hormone (n = 10, black bars) at 5–6 months of age. One-way analysis of variance for DNMT1 p = .0054 (left graph) and for DNMT3a p = .1202 (right graph). (B) Western blot analysis of whole liver lysate showing differences in overall DNMT protein expression (n = 5–8). One-way analysis of variance for DNMT1 protein p = .0021 and for DNMT3a protein p = .0367. *p < .05 and **p < .01 using Bonferroni’s Multiple Comparison test. Bars represent means ± SEM.
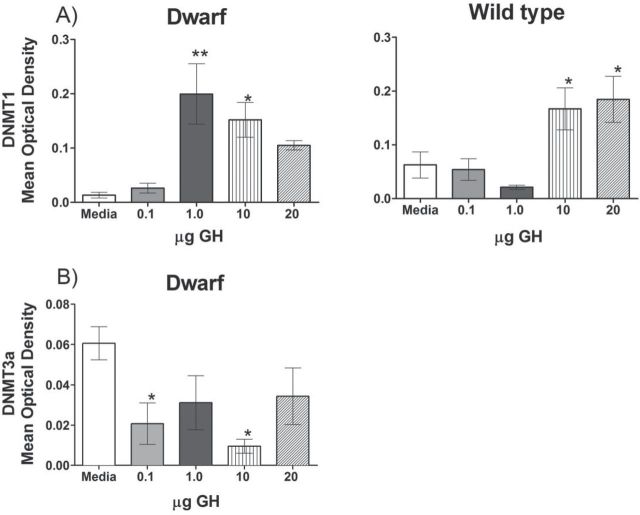
Hepatocytes were isolated from Ames dwarf and age-matched wild-type mice and treated with porcine GH. Significant differences were detected in the means for DNMT1 due to increasing GH concentrations in dwarf (p = .0026) and wild-type (p = .0014) mouse hepatocytes (Figure 6). Ames dwarf mice showed a significant increase in DNMT1 protein expression following 1.0 and 10 μg of GH per plate compared with a media-only control. Though the higher concentration of GH (20 μg) also showed visibly greater protein expression, only two samples were present for quantification and statistically difficult to compare. Protein expression of DNMT1 in hepatocytes from wild-type mice did not show a significant increase compared with a media-only control until a dose 10 times higher, at 10 μg GH per plate. The levels of DNMT3a protein were decreased in primary dwarf hepatocytes following GH treatment (one-way analysis of variance, p = .0464) independent of GH concentration. Wild-type hepatocytes showed very little protein expression of DNMT3a and no difference following GH treatment that could be quantified by our methods (data not shown).
Figure 6.
Ames dwarf hepatocytes are more sensitive to growth hormone treatment, which alters DNMT expression. (A) DNMT1 expression in wild-type (one-way analysis of variance, p = .0014) and dwarf (one-way analysis of variance, p = .0026) hepatocytes with increasing concentrations of growth hormone (0, 0.1,1.0, 10, and 20 µg/ml). (B) DNMT3a expression in dwarf (one-way analysis of variance, p = .0464) hepatocytes with increasing concentrations of growth hormone (0, 0.1,1.0, 10, and 20 µg/ml). *p < .05 and **p < .01 using Dunnett’s Multiple Comparison Test to a media-only control. Error bars represent means ± SEM.
Global DNA methylation in liver tissue was measured using an ELISA-based method with antibodies for methylated cytosines (Figure 7). Comparison of dwarf versus wild-type mice at all three age groups showed independent factors of genotype (p = .0635) and age (p = .0534) approaching significance. At every age, dwarf mice retained methylation at 100% or above compared with 3-month-old wild-type mice and becoming equivalent at 24 months of age. Dwarf mice treated with GH for 1 week lost approximately 15% of global DNA methylation. Figure 7C shows a simple schematic of the DNMT protein and global methylation changes demonstrated in the presence and absence of GH in Ames dwarf mice and potential pathways that remain to be studied. DNMT3b was not included as no changes in this protein were observed between genotypes.
Figure 7.
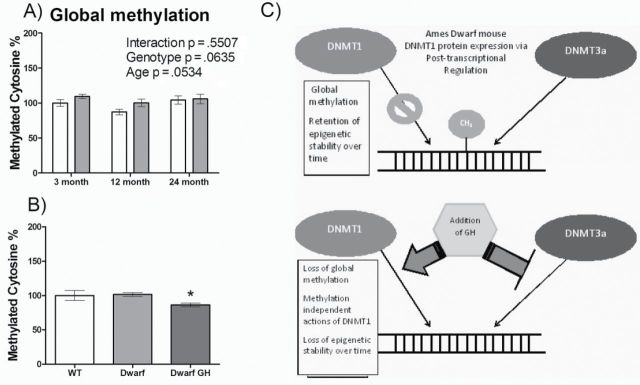
Global methylation in dwarf mice and in response to in vivo growth hormone treatment. (A) ELISA-based assay of methylated cytosine normalized to 3-month-old wild-type mice is shown for dwarf (gray bars) and wild-type mice (open bars) at 3, 12, and 24 months of age (n = 7–8). (B) Percent methylated cytosine shown for wild-type mice injected with saline (n = 10–11, open bars), dwarf mice injected with saline (n = 10–11, gray bars), and dwarf mice injected with growth hormone (n = 10, black bars) at 5–6 months of age (Kruskal–Wallis test, p = .0392). *p < .05 using Dunn’s Multiple Comparison Test. (C) A simple schematic representing the changes in DNMT protein expression and DNA methylation in Ames dwarf mice liver without (top) and with (bottom) growth hormone treatment.
Discussion
Several lines of long-living mice display deficiencies in GH or GH signaling. GH influences the methionine pathway by decreasing the expression and activity of several enzymes, including methionine adenosyltransferase and GNMT, which promote the production of SAM and S-adenosylhomocysteine, respectively. GNMT facilitates the conversion of SAM to SAH; thus, it is a major regulator of SAM in the liver. SAM is the major methyl donor for most methyltransferase reactions in cells. Although many methyltransferases, including DNMTs, are negatively regulated by rising SAH levels, GNMT regulates DNMT by a mechanism that is yet unclear (19). Previously we showed that GNMT activity decreased between 3 and 12 months of age in Ames dwarf mice (31). In this study, we investigated cross-sectional GNMT expression in both genotypes and expanded our findings to include protein levels. We found age-related decreases in GNMT mRNA expression in both genotypes and confirmed increased GNMT gene expression in dwarf mice at all three age groups, most significant at 3 and 12 months of age, in comparison to wild-type mice. Protein levels of GNMT were elevated in dwarf mice (12 months); thus, our GNMT expression relates well with current literature and known GNMT activity level differences between dwarf and wild-type mice (28,31).
To determine whether alterations in DNMTs were associated with GNMT, we examined DNMTs in liver tissue of GH-deficient dwarf and normal, wild-type mice. Interestingly, all three known catalytically active DNMTs (1, 3a, and 3b) showed a significant difference in transcriptional expression due to age that did not follow the linear decrease exhibited in GNMT mRNA expression, though in dwarf mice, DNMT1 and 3a expression was decreased after 3 months of age. In both genotypes, the lowest levels of DNMT mRNA were observed at 12 months of age, whereas in dwarf mice, the highest levels were expressed in the youngest mice. Thus, the expression of methyltransferases may be related to aging in both long-living and normal mice.
Very few studies have reported age-related differences in murine DNMT expression. Similar to our findings, one study using C57Bl/6- and DNMT1-deficient mouse T cells showed a decrease in DNMT1 expression with age (38). Folate- and methyl-deficient diets alter methionine metabolism in rats that, in turn, also decrease DNMT1 and 3a gene expression with age (16). This evidence substantiates a link between DNMT transcription, the methionine pathway, and age-related expression of DNMTs. The elevated DNMT transcription in young dwarf mice is a novel finding. At 6 months of age, the basal transcription of FOS and c-Jun in Ames dwarf mice is significantly higher than in wild-type mice, and both genes are known to positively affect DNMT1 transcription (39). The transcription of FOS is positively affected by DNMT1 transcription, and the transformation of cells in cancer by c-fos overexpression requires DNMT1 (40). The levels of FOS and Jun transcription are also higher in mice or rats fed methyl-deficient diets, altering components of the methionine pathway and methyl donor availability (41,42). In this study, protein abundance assays were performed to detect primarily genotype differences; thus, age-related differences were not directly ascertained. Liver protein levels of DNMT1 in the dwarf mice were much lower than in their wild-type counterparts at every age examined, even though transcriptional expression was significantly higher in the dwarf mice. Comparison between long-living (C57Bl/6) and short-living (DBA2J) mouse strains indicate that liver DNMT1 decreases in both lines when fed a lipogenic methyl-deficient diet, but the C57Bl/6 mice showed higher liver DNMT3a protein levels. High levels of DNMT3a are thought to be protective against the more pronounced demethylation of repetitive elements observed in the DBA2J, thereby protecting epigenetic stability (18). Ablation of DNMT3a in the nervous system of mice causes shortened life spans even though at birth they appear healthy (43). Ablation of DNMT3a also shortens the life span of conditional K-ras knockout mice (44). Taken together, our data and that of others indicate that DNMT3a levels in young mice correlate with longevity in that the high levels observed in relatively young liver tissue may indeed be protective. DNMT1 is required for the activity of de novo DNMT methylation of repetitive elements, which make up a large part of murine and human DNA (45). Further studies are under way to investigate whether the genotype differences discovered in our study contribute to epigenetic stability as postulated.
In general, DNMT1 is considered the main DNMT expressed in adulthood and is responsible for maintaining DNA methylation in adult dividing cells; therefore, it is considered the largest contributor of DNMT activity. Overall, liver DNMT activity was comparatively higher in the young and lower in old dwarf mice compared with age-matched controls. The pattern of activity in dwarf mice may be significantly affected by the contribution of DNMT3a because DNMT1 protein is present at markedly lower levels in dwarf mice. Increased DNMT activity has been widely associated with cancer, loss of GNMT expression, and methyl-deficient diets (19). However, cancer initiation and progression are influenced by the magnitude and timing of DNMT protein expression changes that can alter the disease phenotype considerably (44,46,47). A comparably diminished level of DNMT activity concurrent with high GNMT expression in dwarf mice might delay cancer and aging in dwarf mice in comparison to wild-type mice. It is also possible that DNMT activity is under greater inhibition at 24 months in the liver of dwarf mice due to higher SAH levels. Nonetheless, in young animals, increased DNMT3a may override the comparatively higher inhibitory SAH levels and GNMT competition for SAM in the liver of dwarf mice allowing greater DNMT activity. Unfortunately, current DNMT activity assays are not able to distinguish between DNMT1, 3a, or 3b activity in animal tissues.
Global methylation was also examined in Ames dwarf mice (Figure 7). In an earlier study using a different method, we found no differences in DNA methylation between 18-month-old dwarf and wild-type mice but now have data indicating alterations in global DNA methylation between genotypes (24). DNA methylation in Ames mice was similar or higher than that observed in wild-type mice at all three age groups. Wild-type mice exhibited a U-shaped pattern in methylation comparable to their DNMT transcription. In agreement with the earlier study using 18-month-old mice, the older 24-month-old groups did not show any difference between genotype. We did not see a linear progression in hypomethylation in this strain of mice. In a widely cited landmark study, C57Bl6 mouse liver was shown to lose DNA methylation with age using a linear model (48). However, in that same study older mice showed significant variation in methylation after 20 months of age, and some data points exhibited higher methylation. In addition, another report found a U-shaped pattern of global methylation in the same strain of mice, indicating that methylation does not necessarily decrease in a linear fashion with age (49). It may appear counterintuitive; however, it has been shown that increased liver DNMT1 protein and activity results in lower global methylation (50).
The examination of GH’s role on liver DNMT expression is novel. GH regulates GNMT and appears to have a relationship with DNMT expression and activity in Ames dwarf mouse liver. In this study, in vivo and in vitro GH administration to GH-deficient dwarf mice markedly increased DNMT1 and decreased DNMT3a levels. A lower concentration of GH was required by dwarf hepatocytes to stimulate DNMT expression compared with wild-type cells, suggesting increased sensitivity in the GH-deficient state. This supports earlier findings that dwarf liver is more sensitive to GH stimulation than wild-type tissue in pathways related to stress response and cellular proliferation (36,51). As noted, wild-type cells did not produce enough DNMT3a protein for us to detect with our methods and may be a consequence of culture conditions. GH-treated dwarf mice show significantly lower global DNA methylation compared with saline-injected wild-type and dwarf mice (Figure 7). Determining the genomic location of these DNA methylation changes will provide insight on the magnitude of gene expression alterations, the level of epigenetic stability due to the methylation of repetitive elements, and possible heterochromatic architectural changes between dwarf and wild-type mice. These investigations have been initiated by our laboratory.
Interestingly, our DNMT hepatocyte data does not correspond to those found in rat primary hepatocytes (52). Both species and culture condition differences likely account for this discrepancy. For instance, DNMT1 is regulated by glucocorticoid receptor stimulation by dexamethasone and corticosterone (53). Rat hepatocyte culture protocols require these compounds, whereas mouse primary hepatocyte protocols do not.
Several other factors have also been shown to modulate DNMT expression and activity and include PCNA, PKA, PKB (Akt), PKC, and GSK3β. Independently, diquat and GH treatment of dwarf mice increased liver pAkt protein levels while GH treatment also raised pGSK3β protein levels compared with wild-type mice or untreated dwarf mice (39,51,54,55). Phosphorylation of p21cip/waf by pAkt releases PCNA, resulting in increased DNMT1 translocation to the nucleus. GSK3β regulates pAkt activity and can stabilize DNMT1 affecting its expression and activity (56). PI3K and pAkt are known to be involved in cell proliferation and aging. Because liver DNMT activity is higher in the dwarf at 3 months and lower at 24 months, there may be additional regulation by PKA, PKB (Akt), PKC, and other pathways where Ames dwarf mice exhibit increased responses to diquat and GH treatment in the liver. Another related pathway that contributes to posttranscriptional regulation is PI3K/Akt/mTOR. This pathway controls protein translation and has been shown to be downregulated in Ames dwarf mice and activated by GH administration (55,57).
Recently, a DEAD box protein (DDX20) was reported to regulate DNMT1 levels posttranscriptionally via the regulation of microRNA-140. This pathway stimulates degradation of DNMT1 mRNA transcripts and affects protein levels (58). We are currently investigating the expression of DDX20 and MiRNA-140 in Ames dwarf liver. Finally, GH regulates STAT3 expression and activation in dwarf mice (55). STAT3 is also known to be involved in DNMT1 expression; however, there is no indication of a posttranscriptional role at this time (59).
Together, these pathways could modulate DNMT expression and represent potential avenues of investigation for posttranscriptional regulation of DNMT in the liver of Ames dwarf mice and in aging studies, in general. GH deficiency in the Ames dwarf mouse may contribute to epigenetic stability by decreasing DNMT1 protein through a variety of aging-associated pathways and increasing DNMT3a protein at young ages (Figure 7). Our current data suggest that there are differences between genotypes in global DNA methylation. We also have preliminary results showing differences in specific gene regions (Brown-Borg, unpublished data). The very low levels of DNMT1 protein may play a role in dwarf mouse longevity. However, DNMT1-deficient mice while exhibiting protective effects from cancer in certain organs have other organs that exhibit higher incidence of cancer (38). In addition, DNMT1-deficient mice do not exhibit an increase in longevity so it is unlikely that a systemic deficiency of DNMT1 is beneficial to any organism. Whether Ames dwarf mice exhibit low DNMT1 in other tissues remains to be studied, but we suspect that DNMT1 and DNMT3a differences will be tissue specific. Furthermore, GH signaling may also be important to differences in tissues other than liver such as muscle and fat. The results of this study do not exclude potential IGF-1 effects on DNMT expression. DNMT1 also has DNA methylation–independent actions that may be different in dwarf mice and contribute to cell cycle regulation (60).
Epigenetic instability may contribute to the aging phenotype. DNA methylation plays a significant role in development and likely in responses to early life events, either of which may avert or exacerbate the aging phenotype. However, due to the lack of GH signaling, Ames dwarf mice remain able to resist and delay many pathologies of aging such as the hepatocellular carcinoma found in their wild-type siblings (25). Our study demonstrates that Ames dwarf mice exhibit significant differences in DNMT expression and activity that change with age. We also demonstrate that GH administration can have a significant impact on the methylation mechanisms and status of the Ames dwarf. The differential expression of both de novo and maintenance DNMTs between wild-type and Ames dwarf mice likely affects the methylation of repetitive elements and specific genes and therefore contribute to epigenetic stability. These novel findings contribute to our understanding of the mechanisms underlying the relationships between GH, longevity, and epigenetic stability associated with aging and aging pathologies.
Funding
This work was supported by the National Institutes of Health R01 AG034206 and KO2 AG038509 (H.M.B.B.), the Ellison Medical Foundation AG-SS-2376-09 (H.M.B.B.), and North Dakota Experimental Program to Stimulate Competitive Research (NDEPSCoR) NSF grant # EPS-0184442.
Conflict of Interest
The authors have no conflicts of interest to declare.
References
- 1. Golbus J, Palella TD, Richardson BC. Quantitative changes in T cell DNA methylation occur during differentiation and ageing. Eur J Immunol. 1990;20:1869–1872 [DOI] [PubMed] [Google Scholar]
- 2. Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ehrlich M. Expression of various genes is controlled by DNA methylation during mammalian development. J Cell Biochem. 2003;88:899–910 [DOI] [PubMed] [Google Scholar]
- 4. Holliday R. Epigenetics: an overview. Dev Genet. 1994;15:453–457 [DOI] [PubMed] [Google Scholar]
- 5. Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H. The role of DNA methylation in setting up chromatin structure during development. Nat Genet. 2003;34:187–192 [DOI] [PubMed] [Google Scholar]
- 6. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(suppl):245–254 [DOI] [PubMed] [Google Scholar]
- 7. Berdasco M, Esteller M. Hot topics in epigenetic mechanisms of aging: 2011. Aging Cell. 2012;11:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pogribny IP, Vanyushin BF. Age-related genomic hypomethylation. In: Tollefsbol TO, ed. Epigenetics of Aging. New York, NY: Springer; 2010:11–27 [Google Scholar]
- 9. Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–220 [DOI] [PubMed] [Google Scholar]
- 11. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418 [DOI] [PubMed] [Google Scholar]
- 12. Martínez-Chantar ML, Vázquez-Chantada M, Ariz U, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen YM, Shiu JY, Tzeng SJ, et al. Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int J Cancer. 1998;75:787–793 [DOI] [PubMed] [Google Scholar]
- 14. Yeo EJ, Wagner C. Tissue distribution of glycine N-methyltransferase, a major folate-binding protein of liver. Proc Natl Acad Sci U S A. 1994;91:210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rowling MJ, McMullen MH, Schalinske KL. Vitamin A and its derivatives induce hepatic glycine N-methyltransferase and hypomethylation of DNA in rats. J Nutr. 2002;132:365–369 [DOI] [PubMed] [Google Scholar]
- 16. Ghoshal K, Li X, Datta J, et al. Nutrition and disease A folate- and methyl-deficient diet alters the expression of DNA methyltransferases and methyl CpG binding proteins involved in. J Nutr. 2006; 136:1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopatina NG, Vanyushin BF, Cronin GM, Poirier LA. Elevated expression and altered pattern of activity of DNA methyltransferase in liver tumors of rats fed methyl-deficient diets. Carcinogenesis. 1998;19:1777–1781 [DOI] [PubMed] [Google Scholar]
- 18. Pogribny IP, Tryndyak VP, Bagnyukova TV, et al. Hepatic epigenetic phenotype predetermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Tang F, Chen S, Chen Y, Chiang EI. Glycine-Nmethyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation. J Nutr. 2011;141:777–782 [DOI] [PubMed] [Google Scholar]
- 20. Cavuoto P, Fenech MF. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 2012;38:726–736 [DOI] [PubMed] [Google Scholar]
- 21. Pogribny IP, Ross SA, Wise C, et al. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–87 [DOI] [PubMed] [Google Scholar]
- 22. Bartke A. Histology of the anterior hypophysis, thyroid and gonads of two types of dwarf mice. Anat Rec. 1964;149:225–235 [DOI] [PubMed] [Google Scholar]
- 23. Sornson MW, Wu W, Dasen JS, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333 [DOI] [PubMed] [Google Scholar]
- 24. Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. [DOI] [PubMed] [Google Scholar]
- 25. Ikeno Y, Bronson RT, Hubbard GB, Lee S, Bartke A. Delayed occurrence of fatal neoplastic diseases in ames dwarf mice: correlation to extended longevity. J Gerontol A Biol Sci Med Sci. 2003;58:291–296 [DOI] [PubMed] [Google Scholar]
- 26. Uthus EO, Brown-Borg HM. Methionine flux to transsulfuration is enhanced in the long living Ames dwarf mouse. Mech Ageing Dev. 2006;127:444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Uthus EO, Brown-Borg HM. Altered methionine metabolism in long living Ames dwarf mice. Exp Gerontol. 2003;38:491–498 [DOI] [PubMed] [Google Scholar]
- 28. Brown-Borg HM, Rakoczy SG, Sharma S, Bartke A. Long-living growth hormone receptor knockout mice: potential mechanisms of altered stress resistance. Exp Gerontol. 2009;44:10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Amador-Noguez D, Dean A, Huang W, Setchell K, Moore D, Darlington G. Alterations in xenobiotic metabolism in the long-lived Little mice. Aging Cell. 2007;6:453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aida K, Tawata M, Negishi M, Onaya T. Mouse glycine N-methyltransferase is sexually dimorphic and regulated by growth hormone. Horm Metab Res. 1997;29:646–649 [DOI] [PubMed] [Google Scholar]
- 31. Brown-Borg HM, Rakoczy SG, Uthus EO. Growth hormone alters methionine and glutathione metabolism in Ames dwarf mice. Mech Ageing Dev. 2005;126:389–398 [DOI] [PubMed] [Google Scholar]
- 32. Brown-Borg HM, Rakoczy SG. Catalase expression in delayed and premature aging mouse models. Exp Gerontol. 2000;35:199–212 [DOI] [PubMed] [Google Scholar]
- 33. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254 [DOI] [PubMed] [Google Scholar]
- 34. Chen HW, Chen JJ, Yu SL, et al. Transcriptome analysis in blastocyst hatching by cDNA microarray. Hum Reprod. 2005;20:2492–2501 [DOI] [PubMed] [Google Scholar]
- 35. Sharma S, Rakoczy S, Dahlheimer K, Brown-Borg H. The hippocampus of Ames dwarf mice exhibits enhanced antioxidative defenses following kainic acid-induced oxidative stress. Exp Gerontol. 2010;45:936–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown-Borg HM, Rakoczy SG. Growth hormone administration to long-living dwarf mice alters multiple components of the antioxidative defense system. Mech Ageing Dev. 2003;124:1013–1024 [DOI] [PubMed] [Google Scholar]
- 37. Kreamer BL, Staecker JL, Sawada N, Sattler GL, Hsia MT, Pitot HC. Use of a low-speed, iso-density percoll centrifugation method to increase the viability of isolated rat hepatocyte preparations. In Vitro Cell Dev Biol. 1986;22:201–211 [DOI] [PubMed] [Google Scholar]
- 38. Ray D, Wu A, Wilkinson JE, et al. Aging in heterozygous Dnmt1-deficient mice: effects on survival, the DNA methylation genes, and the development of amyloidosis. J Gerontol A Biol Sci Med Sci. 2006;61:115–124 [DOI] [PubMed] [Google Scholar]
- 39. Sun LY, Bokov AF, Richardson A, Miller RA. Hepatic response to oxidative injury in long-lived Ames dwarf mice. FASEB J. 2011;25:398–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–390 [DOI] [PubMed] [Google Scholar]
- 41. Wainfan E, Poirier LA. Methyl groups in carcinogenesis : Effects on DNA methylation and gene expression. Cancer Res. 1992;53(7 suppl):2071s–2077s [PubMed] [Google Scholar]
- 42. Zhou W, Alonso S, Takai D, et al. Requirement of RIZ1 for cancer prevention by methyl-balanced diet. PLoS One. 2008;3:e3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen S, Meletis K, Fu D, Jhaveri S, Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–1676 [DOI] [PubMed] [Google Scholar]
- 44. Gao Q, Steine EJ, Barrasa MI, et al. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108:18061–18066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liang G, Chan MF, Tomigahara Y, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kinney SR, Moser MT, Pascual M, Greally JM, Foster BA, Karpf AR. Opposing roles of Dnmt1 in early- and late-stage murine prostate cancer. Mol Cell Biol. 2010;30:4159–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson VL, Smith RA, Mag S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951 [PubMed] [Google Scholar]
- 49. Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210 [DOI] [PubMed] [Google Scholar]
- 50. Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutat Res. 2004;548:53–59 [DOI] [PubMed] [Google Scholar]
- 51. Miquet JG, Muñoz MC, Giani JF, et al. Ames dwarf (Prop1(df)/Prop1(df)) mice display increased sensitivity of the major GH-signaling pathways in liver and skeletal muscle. Growth Horm IGF Res. 2010;20:118–126 [DOI] [PubMed] [Google Scholar]
- 52. Vinken M, Snykers S, Fraczek J, et al. DNA methyltransferase 3a expression decreases during apoptosis in primary cultures of hepatocytes. Toxicol In Vitro. 2010;24:445–451 [DOI] [PubMed] [Google Scholar]
- 53. Yang X, Ewald ER, Huo Y, et al. Glucocorticoid-induced loss of DNA methylation in non-neuronal cells and potential involvement of DNMT1 in epigenetic regulation of Fkbp5. Biochem Biophys Res Commun. 2012;420:570–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Panici JA, Harper JM, Miller RA, Bartke A, Spong A, Masternak MM. Early life growth hormone treatment shortens longevity and decreases cellular stress resistance in long-lived mutant mice. FASEB J. 2010;24:5073–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Masternak MM, Panici JA, Wang F, Wang Z, Spong A. The effects of growth hormone (GH) treatment on GH and insulin/IGF-1 signaling in long-lived Ames dwarf mice. J Gerontol A Biol Sci Med Sci. 2010;65:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun L, Zhao H, Xu Z, et al. Phosphatidylinositol 3-kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell Signal. 2007;19:2255–2263 [DOI] [PubMed] [Google Scholar]
- 57. Sharp ZD, Bartke A. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol A Biol Sci Med Sci. 2005;60:293–300 [DOI] [PubMed] [Google Scholar]
- 58. Takata A, Otsuka M, Yoshikawa T, et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology. 2013;57:162–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927 [DOI] [PubMed] [Google Scholar]