Abstract
Background and Aims
The taxonomic complexity of Crataegus (hawthorn; Rosaceae, Maleae), especially in North America, has been attributed by some to hybridization in combination with gametophytic apomixis and polyploidization, whereas others have considered the roles of hybridization and apomixis to be minimal. Study of the chemical composition and therapeutic value of hawthorn extracts requires reproducible differentiation of entities that may be difficult to distinguish by morphology alone. This study sought to address this by using the nuclear ribosomal spacer region ITS2 as a supplementary DNA barcode; however, a lack of success prompted an investigation to discover why this locus gave unsatisfactory results.
Methods
ITS2 was extensively cloned so as to document inter- and intraindividual variation in this locus, using hawthorns of western North America where the genus Crataegus is represented by only two widely divergent groups, the red-fruited section Coccineae and the black-fruited section Douglasia. Additional sequence data from selected loci on the plastid genome were obtained to enhance further the interpretation of the ITS2 results.
Key Results
In the ITS2 gene tree, ribotypes from western North American hawthorns are found in two clades. Ribotypes from diploid members of section Douglasia occur in one clade (with representatives of the east-Asian section Sanguineae). The other clade comprises those from diploid and polyploid members of section Coccineae. Both clades contribute ribotypes to polyploid Douglasia. Data from four plastid-derived intergenic spacers demonstrate the maternal parentage of these allopolyploids.
Conclusions
Repeated hybridization between species of section Douglasia and western North American members of section Coccineae involving the fertilization of unreduced female gametes explains the observed distribution of ribotypes and accounts for the phenetic intermediacy of many members of section Douglasia.
Keywords: Reticulate evolution, nrITS, cpDNA, Crataegus, Douglasia, Coccineae, Rosaceae, hawthorn, gametophytic apomixis, allopolyploid, autopolyploid, concerted evolution, hybrids, taxonomy
INTRODUCTION
North American hawthorns (Crataegus L., Rosaceae) are well known for being taxonomically difficult. ‘The Crataegus problem’ consists of having large numbers of species, often with very limited geographical ranges, that are distinguishable only by relatively minor morphological differences (Palmer, 1932; Camp, 1942). Palmer and Camp were able to point to limited evidence implicating apomixis, hybridization and polyploidy in the origin of these difficulties, but it is only in the past 35 years that all three of these processes have been causally linked to each other (Dickinson et al., 2007; Whitton et al., 2008), and polyploidy and gametophytic apomixis are now well documented in this genus (Muniyamma and Phipps, 1979a, b; Talent and Dickinson, 2005, 2007a, b, c). Also, molecular data have provided evidence for hybridization and auto- and allopolyploidization in Crataegus (Lo et al., 2009b, 2010). Taxonomic revision of the genus has employed numerical phenetics in order to place morphological taxonomy on a sound footing (e.g. Dickinson et al., 2008), and many species names have been placed in synonymy on this basis (e.g. Phipps, 1998).
Because of the potential therapeutic value of hawthorn-derived natural health products (NHPs; Guo et al., 2008; Edwards et al., 2012), we have collaborated with natural product chemists, physiologists and growers in comparing Crataegus spp. native to the Pacific Northwest with some of the Eurasian taxa used in the manufacture of most commercial NHPs. Such a study requires reproducible differentiation of entities that may be difficult to distinguish by morphology alone. Moreover, certification of hawthorn NHPs will require taxon-specific markers with which to authenticate the species composition of the NHPs. As described elsewhere (M. Zarrei et al., unpubl. res.), we investigated the utility of plant DNA barcode loci as possible markers. The loci tested included the second nuclear internal transcribed spacer (nrITS2), and our lack of success with this locus came as a surprise, given the results obtained earlier with ITS by Lo et al. (2007, 2009a) and others in phylogenetic studies.
Here we describe the results of extensively cloning ITS2 so as to document inter- and intraindividual variation in this locus. Additional sequence data from selected loci on the plastid genome further enhance our interpretation of the ITS2 results. Specifically, we ask (1) whether ITS2 sequences are homogeneous within individuals; (2) whether sequence variation in the plastid loci parallels that seen in ITS2; and (3) whether ITS2 homogeneity varies with taxon, ploidy and breeding system. Answers to these questions are evaluated in the light of data from recent hybrids of known parentage and data on ITS2 secondary structure. In turn, we use our results to evaluate the role of reticulate evolution in a group of mainly western North American hawthorns. In this way, our results will help us to evaluate the way in which taxonomists have minimized the role of hybridization in Crataegus (e.g. Phipps, 2005) in describing new taxa for the genus from the Pacific Northwest.
MATERIALS AND METHODS
Plant materials
Our exploration of inter- and intraindividual ITS2 variation focuses on individuals (our operational taxonomic units; OTUs) of the diploid and polyploid black-fruited hawthorns of western North America, Crataegus section Douglasia Loud. (Table 1; Supplementary Data Table S1). Because this is a geographically relatively isolated group, it has been intensively studied, and is now probably the best known group of hawthorns, especially with respect to its cytotypic, molecular and morphological variation (Dickinson et al., 1996, 2008; Talent and Dickinson, 2005, 2007a, b; Lo et al., 2007, 2009b, 2010). Section Douglasia is also a group in which recent botanical exploration in western North America has led to the description of several new species over the past 20 years (Table 1; Phipps, 1998, 1999; Phipps and O'Kennon, 1998, 2002, 2007). Several of these newly described species exemplify the concerns referred to above in having limited geographical ranges (Table 1), and being distinguishable only by relatively minor morphological differences.
Table 1.
Ingroup species included in the current study for ITS2 and plastid locus sequencing (Table 2)
| Section | Series | Species | Ploidy (x = 17) | Distribution | Accessions |
|---|---|---|---|---|---|
| Brevispinae Beadle ex C.K.Schneid. | Brevispinae (Beadle) Rehder | C. brachyacantha Sarg. & Engelm. | 2x | Regional (LA, TX) | 3 |
| Coccineae Loudon | Crus-galli (Loud.) Rehder | C. crus-galli L. | 2x | E North America | 1 |
| Chrysocarpeae J.B. Phipps | C. chrysocarpa Ashe | 4x | N America | 2 | |
| Macracanthae (Loud.) Rehder | C. macracantha Lodd. ex Loud. | 4x | N America | 3 | |
| Punctatae (Loud.) Rehder | C. punctata Jacq. | 2x | E North America | 3 | |
| Triflorae (Beadle) Rehder | C. triflora Chapm. | 4x | SE USA | 1 | |
| Crataegus | Crataegus | C. monogyna Jacq. | 2x | W Eurasia | 4 |
| Pentagynae (C.K. Schneid.) Russanov | C. pentagyna Waldst. & Kit. | 2x | W Eurasia | 1 | |
| Pinnatafidae (Zabel ex C.K. Schneider) Rehder | C. pinnatifida Bunge | 2x | E Asia (China, Korea) | 3 | |
| Douglasia Loud. | Cerrones J.B.Phipps | C. erythropoda Ashe | 4x | Regional (CO, NM; SK?) | 2 |
| C. rivularis Nutt. | 4x | Regional (CO, ID, UT, WY) | 5 | ||
| C. saligna Greene | 2x | Regional (CO, UT) | 3 | ||
| Douglasianae (Loud.) Poletiko | C. castlegarensis J.B. Phipps & O'Kennon | 4x | Regional (Pacific Northwest) | 4 | |
| C. douglasii Lindl. | 4x | Pacific Northwest, Great Lakes basin | 13 | ||
| 5x | Local (BC) | 1 | |||
| C. gaylussacia A. Heller | 3x | Marin and Sonoma Counties, CA | 5 | ||
| C. okennonii J.B. Phipps | 4x | Regional (BC, ID, MT, WA) | 2 | ||
| C. shuswapensis J.B. Phipps & O'Kennon | 4x | Local (BC, MT) | 1 | ||
| C. suksdorfii (Sarg.) Kruschke | 2x | W of Cascades (CA, OR, WA) | 14 | ||
| 3x | Regional (Pacific Northwest) | 5 | |||
| 4x | Local (ID, MT) | 1 | |||
| 5x | Local (BC) | 2 | |||
| Montaninsulae J.B. Phipps & O'Kennon | C. rivuloadamensis J.B. Phipps & O'Kennon | 4x | Local (Cypress Hills) | 1 | |
| C. rivulopugnensis J.B. Phipps & O'Kennon | 4x | Local (Cypress Hills) | 1 | ||
| Purpureofructi J.B. Phipps & O'Kennon | C. aquacervensis J.B. Phipps & O'Kennon | 4x | Local (Cypress Hills) | 1 | |
| C. atrovirens J.B. Phipps & O'Kennon | 4x | Local (BC) | 1 | ||
| C. cupressocollina J.B. Phipps & O'Kennon | 4x | Local (Cypress Hills) | 1 | ||
| C. enderbyensis J.B. Phipps & O'Kennon | 4x | Local (BC) | 2 | ||
| C. okanaganensis J.B. Phipps & O'Kennon | 4x | Regional (BC, ID, WA) | 1 | ||
| C. orbicularis J.B. Phipps & O'Kennon | 4x | Local (BC) | 1 | ||
| C. phippsi O'Kennon | 4x | Regional (BC, MT, WA) | 1 | ||
| Mespilus (L.) T.A. Dickinson & E.Y.Y. Lo | C. germanica (L.) Koch | 2x | W Eurasia | 2 | |
| Sanguineae Zebel ex Schneid. | Nigrae (Loudon) Russanov | C. kansuensis E.H.Wilson | 4x | E Asia (China) | 1 |
| C. maximowiczii C.K.Schneid. | 2x | E Asia (China, Japan, Korea, Mongolia, Russia) | 2 | ||
| C. nigra Waldst. & Kit. | 2x | Central Europe | 1 | ||
| Sanguineae (Zebel ex Rehder) C.K. Schneid. | C. wilsonii Sarg. | 2x | E Asia (China) | 1 | |
| Hybrid | C. × cogswellii K.I.Chr. & T.A.Dickinson | 2x | Local (OR) | 2 | |
| Hybrid | C. × ninae-celottiae K.I.Chr. & T.A.Dickinson | 2x | Local (ON) | 2 | |
The infrageneric classification follows Phipps et al. (1990), Phipps and O'Kennon (1998, 2007) and Lo et al. (2007); also shown are ploidy level (Talent and Dickinson, 2005; Lo et al., 2007, 2010; J. Coughlan and N. Talent unpubl. data), geographical distribution [Christensen, 1992; Gu and Spongberg, 2003 (online version); Phipps et al., 1990; and specimens in TRT, including the J. B. Phipps Hawthorn Research Collection], and the number of individuals studied per species (see Supplementary Data Table S1 for voucher information and outgroup taxa).
Crataegus kansuensis and two C. douglasii accessions sampled from Montana (JC281 and JC296; Table S1) were not amplified for the plastid markers. Crataegus triflora was not amplified for ITS2.
Our use of two of the older, well-established names in the section warrants comment. Crataegus gaylussacia is understood here in the restricted sense of the original description by Heller (1903), rather than in the wide sense employed in the standard list for Canada (Brouillet et al., 2010 +). We argue elsewhere (T. A. Dickinson et al., unpubl. res.) that this narrower circumscription is warranted because individuals corresponding to the entity seen by Heller in California (without determining the number of stamens per flower) are ecologically and cytologically distinct from other 20-stamen, black-fruited hawthorns of western North America. For this reason, Crataegus suksdorfii is understood here in the wide sense of Dickinson et al. (2008) as encompassing 20-stamen, black-fruited diploid, triploid, tetraploid and pentaploid cytotypes found from northern California to central British Columbia, and as far east as the continental divide. Elsewhere (T. A. Dickinson et al., unpubl. res.) we justify this wide circumscription by noting that it includes the type material of the first 20-stamen black-fruited entity to be described (C. douglasii var. suksdorfii Sarg.).
Based on previous phylogenetic results (Campbell et al., 1997; Lo et al., 2007), we used Amelanchier, Pyrus, Malus, Cotoneaster and Sorbus as outgroups, obtaining sequences from individuals in earlier studies deposited in GenBank (Benson et al., 2011; Supplementary Data Tables S2 and S3). We also included black- and red-fruited members of Crataegus sections Brevispinae, Coccineae, Crataegus, Mespilus (previously considered as a distinct genus) and Sanguineae to provide context for our Douglasia results. Only a few of the red-fruited species that are numerous and abundant in eastern North America have ranges that extend into the Rocky Mountains and farther west (Phipps, 1998), and we included these in our sample in order to be able to evaluate hypotheses of hybridization (Table 1). We also included naturalized diploid C. monogyna (section Crataegus; Table 1) and its two documented hybrids (Christensen et al., 2014) with native diploids in sections Coccineae (C. punctata; Wells and Phipps, 1989) and Douglasia (C. suksdorfii; Love and Feigen, 1978). By including sequences from known hybrid accessions and from the parental species, we are able to observe the effects of recent hybridization on intraindividual variation in recoverable ITS2 sequences, and the extent of their concerted evolution. Inclusion of additional species from section Coccineae enables us to test whether one or more of the entities in section Douglasia could have also arisen as intersectional hybrids, as suggested by Brunsfeld and Johnson (1990) and Talent (2009). Our current sampling thus encompasses representatives of all major Crataegus clades (Table 1; Lo et al., 2007, 2009a) and almost all Crataegus taxa found in western North America.
To infer the maternal phylogeny of species studied for ITS2, four plastid intergenic spacers were analysed here: trnL-F, trnG-trnS, rpl2-trnH and rpl20-rps12. For these markers, only one accession per species per ploidy level was studied (as indicated by an asterisk in Supplementary Data Table S1).
Molecular methods
Total genomic DNAs were extracted from young silica-gel dried leaves (Chase and Hills, 1991) and occasionally from herbarium specimens using a method modified from that of Tsumura et al. (1995). All genomic DNAs were further purified using a Bio Basic EZ-10 Spin Column PCR purification Kit (Bio Basic Inc., Markham, Canada) to remove polysaccharides and polyphenols that prevent amplification, particularly if the tissues were collected later in the growing season (Demeke and Adams, 1992).
Amplification of ITS2 and portions of downstream and upstream regions of ITS2 used the previously published primers, ITS-S2F (Chen et al., 2010) and ITS4 (White et al., 1990). Plant systematic studies at lower phylogenetic levels often use the entire nrITS region. However, only ITS2 was adopted as one of the supplementary universal markers for plant DNA barcoding purposes (Yao et al., 2010). Our interest in this particular region was aroused by the difficulties we encountered in using it for barcoding (Dickinson et al., 2011). In addition, there is an extensive database available for ITS2 that provides information on secondary structure (Koetschan et al., 2012). Amplification of four plastid markers (trnL-F, trnG-trnS, rpl2-trnH and rpl20-rps12) used the previously published primers as follows: c and f of Taberlet et al. (1991) for trnL-F; trnG (UCC) and trnS (GCU) of Hamilton (1999) for trnG-trnS; rpl2 and trnH of Vaillancourt and Jackson (2000) for rpl2-trnH; and rpl20 and 5′-rps12 of Hamilton (1999) for rpl20-rps12 (see Heinze, 2007 for PCR primer sequences). Polymerase chain reaction (PCR) volumes of 25 μL were prepared following the protocol detailed in Stefanović et al. (2007). DMSO (dimethylsulphoxide; 4 % final concentration) was added to ITS2 PCR to reduce problems common in this region due to secondary structure (Winship, 1989; Baldwin et al., 1995; Zarrei et al., 2009). The amplicons were purified using a Bio Basic EZ-10 Spin Column PCR purification Kit (Bio Basic Inc.).
By direct sequencing of ITS2 amplicons, variation in both the sequence length and additive polymorphic sites (APS) was detected. Because of complete conservation of the ITS2 flanking regions and presence of no clear variation patterns in the ITS2, it was not possible to design specific PCR primers to target different copies selectively. To ensure that all distinct copies of ITS2 were amplified, different sets of PCR amplifications including changes in annealing temperature, elongation time, and DMSO and MgCl2 concentrations were tested. Highly denaturing PCR conditions with DMSO, for example, have been shown to amplify both low- and high-stability paralogues of ribosomal DNA well (Buckler et al., 1997). Amplicons were carefully checked on agarose gels, but no sign of significant length variation (40 + bp) was found. All purified amplicons were cloned using the AccepTor™ Vector cloning kit (Novagen, EMD Chemicals Inc., CA, USA). ITS2 was amplified from the successfully transformed colonies using the vector primers. To maximize the probability of capturing the allelic variation in the genome, we sequenced a minimum of eight, 12, 16 and 20 clones from diploid, triploid, tetraploid and pentaploid individuals, respectively. The PCR products were then cleaned using the 96-well silica plate system (Wizard® SV 96 PCR Clean-Up System; Promega, Madison, USA). Given the relatively short length of ITS2, generally only one strand was sequenced. However, a number of amplicons were sequenced for both strands to ensure accuracy. Plastid amplicons were directly sequenced using both the forward and reverse PCR primers. Cycle sequencing reactions were performed using the BigDye® Terminator v3.1 kit (Applied Biosystems Inc., Foster City, CA, USA). Cleaned cycle sequencing products were sequenced on an ABI 3730 (Applied Biosystems) DNA Analyzer at the Royal Ontario Museum (Toronto, Canada). Sequences were proofed and edited using either Sequencher 4·1 (Gene Codes, Corp.) or Geneious Pro. v.5.6 (Drummond et al., 2012), and assembled using BioEdit v.7.0.5.3 (Hall, 1999) or Geneious Pro. v.5.6 (Drummond et al., 2012).
In ten cases out of the 1247 ITS2 clones, DNA sequences showed extremely high divergence compared with ingroup and outgroup taxa. We conducted BLAST searches for those sequences against GenBank. In most of these cases, the matches came back from only distantly related families, with a low percentage identity (78–88 %) and coverage (80–89 %); in some cases, no similarity was found to the database. We suspect that those sequences might result from PCR artefacts such as: PCR-generated chimeras, high number of PCR cycles and template concentration (see the Discussion in Qiu et al., 2001; Acinas et al., 2005) or be derived from highly divergent pseudogenes. These sequences would not fold into the secondary structure known for angiosperms (Mai and Coleman, 1997). All of them were removed from subsequent data analyses.
Sequence analyses
A total of 1237 ITS2 and 175 plastid DNA sequences were analysed. The sequences were submitted to, and accession numbers were obtained from, GenBank (Supplementary Data Tables S2 and S3). Sequences were aligned with CLUSTAL_X (Thompson et al., 1997) or the Geneious alignment option in Geneious Pro v.5.6 (Drummond et al., 2012). This initial alignment was adjusted manually in BioEdit v.7.0.5.3 or Geneious Pro v.5.6 to minimize steps in the most-parsimonious trees (e.g. Koch et al., 2010).
To reduce uninformative repetition in the ITS2 matrix, all sequences identical to each other for any given accession were collapsed into a single ribotype using DnaSP v.5 (Librado and Rozas, 2009). The matrix reduced in this fashion consists of 874 ITS2 ribotypes. The ribotype diversity per individual (Rd; Nei, 1987), nucleotide diversity per site (Nd; Jukes and Cantor; Nei, 1987), sequence length variation and guanine plus cytosine (GC) content were calculated for each accession using DnaSP v.5. A recombination test was performed using RDP4 Beta 4·14 (Martin et al., 2010). No signs of recombination were detected in the data set. In addition, the Phi test (Bruen et al., 2006) was performed using SplitsTree v.4.12.3 (Huson and Bryant, 2006). This test also failed to find statistically significant evidence for recombination (P = 0·797).
Initially, separate parsimony analyses were conducted for each plastid region to investigate possible incongruence between data sets (Bull et al., 1993). Because no substantial incongruence was detected, and because all four plastid markers used in this study are linked together on a single chromosome, the concatenated sequence matrix was analysed.
Indels were coded as separate binary characters for both nuclear and plastid data sets using SeqState version 1.4.1 (Müller, 2005) with a modified complex coding option (Simmons and Ochoterena, 2000). Lastly, the ITS2 sequences were annotated following Keller et al. (2009). The secondary structure of ITS2 was then predicted using the hidden Markov model-based method (Keller et al., 2009) and the web interface ITS2 prediction tool (Koetschan et al., 2012). The presence of the five structural motifs (see the Results) was investigated for all ribotypes. The secondary structures were displayed with PseudoViewer v.3 (Han et al., 2002).
Phylogenetic analyses
Three different phylogenetic analyses were run for the ITS2 and plastid data sets: (1) maximum parsimony (MP) analyses using PAUP* v.4b10 (Swofford, 2002); (2) Bayesian analyses (BI; Yang and Rannala, 1997) using MrBayes v.3.2.0 (Ronquist and Huelsenbeck, 2003; Ronquist et al., 2010), and (3) Neighbor-Net (NN; Bryant and Moulton, 2004) network reconstruction using SplitsTree v.4.12.6 (Huson and Bryant, 2006).
The original ribotype matrix contained more than twice as many ribotypes compared with the number of characters (874 ribotypes; 369 characters). This prevented the production of meaningful bootstrap support and reaching convergence in the Bayesian analysis even after 35 million generations. A safe deletion rule, originally introduced as the safe taxonomic reduction (STR) strategy by Wilkinson (1995) and modified by Zarrei et al. (2012), was used to reduce the size of the original data set. In this fashion, the only sequences retained for a given individual were those that were found in different clades in one of the most-parsimonious trees. The reduced matrix, containing 299 ribotypes, including those from the seven outgroups, was used for further phylogenetic analyses. The concatenated plastid matrix included 47 OTUs, including four outgroups.
For the Fitch parsimony analyses (Fitch, 1971), the character state changes were equally weighted and character changes were interpreted under ACCTRAN optimization (Agnarsson and Miller, 2008). A two-stage search strategy was undertaken following Stefanović et al. (2007). First, analyses involved 1000 replicates of random taxon addition sequences (RAS), tree bisection and reconnection (TBR) branch swapping and MULTREES option off to search for multiple islands of the most-parsimonious trees (Maddison, 1991). To reduce search time, ten trees per replicate were saved. The second round of analyses was performed on all trees saved in memory with the same settings, except the MULTREES option in effect. Both stages of analyses were conducted either to completion or until a minimum of 100 000 trees were found. The bootstrap support (BS) was estimated using 1000 bootstrap pseudoreplicates (Efron, 1979; Felsenstein, 1985; Holmes, 2003) with simple taxon addition and TBR swapping but permitting only ten trees per replicate to be held. The consistency index (CI), rescaled consistency index (RC), and Farris' (1989) retention index (RI) were calculated to measure the amount of homoplasy in the data set.
The general time reversible (GTR; Yang, 1994) model of DNA evolution, with rate variation among sites following gamma distribution and proportion of invariable sites (GRT + G + I), was selected as the best fit model for the ITS2 nucleotide sequence partition by Akaike information criterion (AIC; Akaike, 1974), as implemented in MrModeltest v.2.3 (Nylander, 2004). The best fit model for the concatenated plastid data set selected by AIC was GTR + I. The Markov k-state (Mk) model, a generalized JC69 model, allowing any particular changes to be equally probable (Lewis, 2001), was used for the indel partition for both plastid and nuclear data sets.
For the Bayesian analyses, two simultaneous runs with four chains each were run for 36 million generations for ITS2 and for 2 million generations for the plastid data sets. All shared parameters were unlinked between DNA sequence and coded indel partitions. In each run, every 2000th and 200th tree was sampled, resulting in 36 000 and 20 000 trees for the ITS2 and plastid matrices, respectively. The completion of the Bayesian analysis was assumed when the average standard deviation of split frequencies was ≤0·05 (Ronquist and Huelsenbeck, 2003) for the two runs combined, and the convergence between the Bayesian MCMC (Markov chain Monte Carlo) runs was reached. Convergence of an independent search was further explored by plotting likelihood scores vs. generations using Tracer v.1.5 (Rambaut and Drummond, 2007). The effective sample size (ESS) values ≥ 200 were considered the absolute convergence between runs. The burn-in phase for each run (the first 25 % of sampled trees far after −lnL stationary distribution has been achieved) was discarded, and the 50 % majority rule consensus tree of the remaining trees (27 000 trees for ITS2 and 15 000 trees for plastid data) was computed using PAUP*. Support for Bayesian topologies was estimated using node posterior probabilities (PP) from the posterior distribution of topologies.
To investigate the relationships further, the network reconstruction using the NN algorithm with uncorrected P-distances was used for ITS2. Due to the maternal inheritance of the plastid genome, no network reconstruction was performed for these markers in this study. The network analyses were performed on both the original ribotype matrix (874 ribotypes) and the reduced matrix of 299 ribotypes.
RESULTS
Sequence divergence and ribotype diversity
All hawthorn taxa investigated here, including diploids, possess more than one ribotype. The minimum number of distinct ribotypes found was three, in C. suksdorfii (2n = 3x) collected from Idaho (EL-172), and the maximum number was 20, in C. douglasii (2n = 5x) collected from British Columbia (2010-31), (Supplementary Data Table S1). No sign of APS or electropherogram displacements (indicating the presence of indels in the amplicon) was detected in the plastid sequences. Moreover, the cloned sequences for trnL-F in pentaploid C. douglasii sampled from British Columbia (2010-38; Supplementary Data Table S1) showed no sign of the potential biparental plastid inheritance proposed by Zhang and Sodmergen (2010). Only three private purine transitional mutations were found in five clones sequenced. Considering 4750 bp sequences (five clones × 950 bp for the trnL-F length) and that the frequency of error introduced by Taq polymerase was probably 1 in 1500 bp (Eyre-Walker et al., 1998), these mutations were probably not genuine mutations. Allopolyploids exhibited greater nucleotide diversity than diploids and autotriploids (Fig. 1A). Ribotype diversity exhibited a similar, but less marked, contrast (Fig. 1B). A total of 586 unique ribotypes (47 % of the total sequences) were found in the entire data set when the identity of accessions was not considered. This number was 874 (71 % of the total sequences) when the identities of accessions were considered separately. The GC content of triploids and pentaploids was lower than in diploids and tetraploids (Fig. 1C; Table 2). Because differences in GC content do not appear to be significant (Fig. 1C), they will not be discussed further.
Fig. 1.
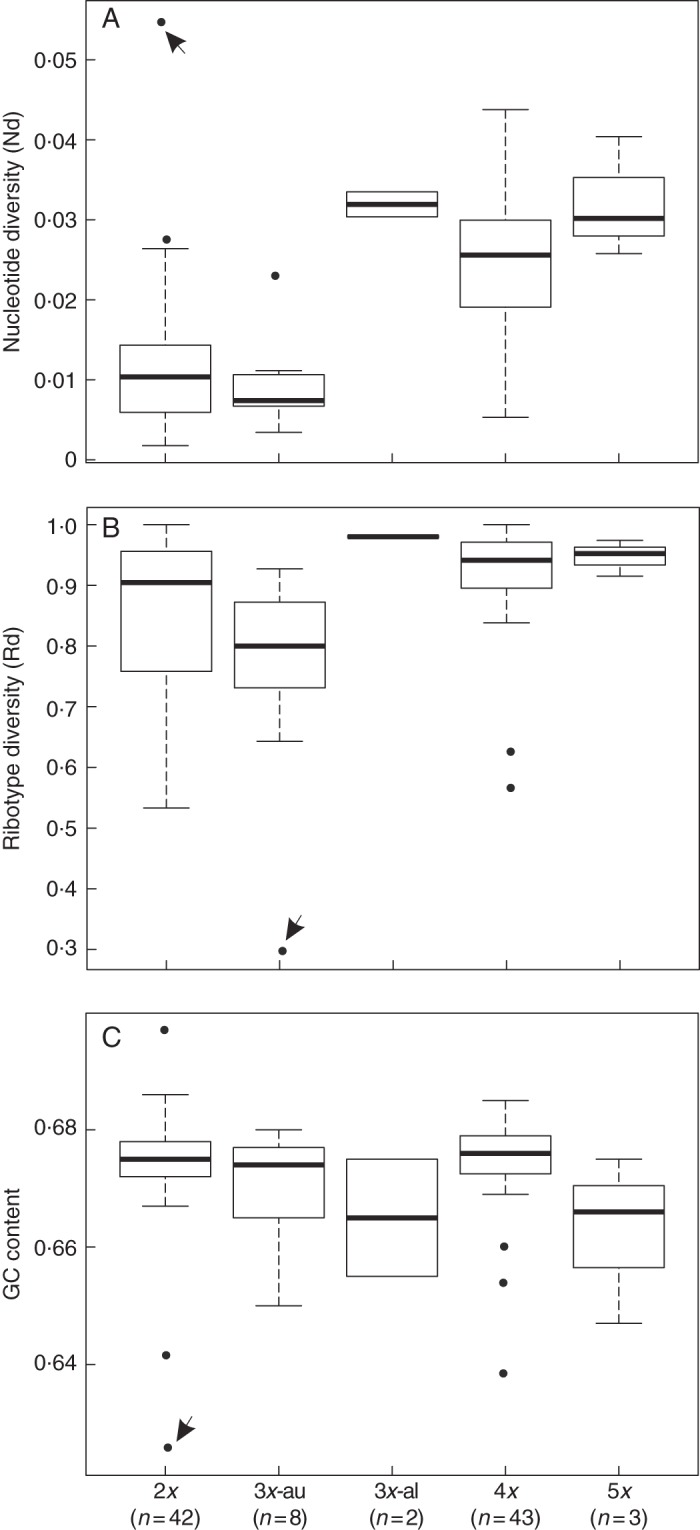
The distribution of various genetic values (nucleotide diversity, ribotype diversity and GC content) as a function of ploidy. Autotriploid (indicated by 3x-au) and allotriploid (indicated by 3x-al) C. suksdorfii were analysed separately. (A) Box plot of ploidy vs. nucleotide diversity, (B) box plot of ploidy vs. ribotype diversity, (C) box plot of ploidy vs. GC content. Arrowheads indicate (A) C. brachyacantha sampled from Louisiana (2011-02), (B) C. suksdorfii sampled from Idaho (EL-172) and (C) C. brachyacantha sampled from Louisiana (2011-02) and C. suksdorfii sampled from Oregon (JC117).
Table 2.
Comparison of sequence variation among different markers utilized for the current study (outgroups excluded)
| Matrix |
No. of sequences | Ungapped length of sequences* | Gapped length of sequences | % pairwise identity | % identical sites | % GC content | Nucleotide frequencies |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | G | T | ||||||||
| ITS2 | Original matrix | 1237 | 265 (342·2 ± 6·3) 352 | 386 | 93·6 | 3·5 | 60 | 0·155 | 0·361 | 0·316 | 0·169 |
| Haplotype matrix | 874 | 272 (342·3 ± 5·7) 352 | 357 | 95·5 | 9·2 | 64·9 | 0·155 | 0·361 | 0·316 | 0·169 | |
| Reduced matrix | 291 | 295 (342·4 ± 5 .8) 345 | 346 | 95·9 | 44·5 | 67·1 | 0·155 | 0·36 | 0·316 | 0·168 | |
| Plastid | trnL-F | 43 | 936 (938·6 ± 2·8) 942 | 950 | 99·3 | 96·7 | 30·3 | 0·364 | 0·156 | 0·151 | 0·329 |
| trnG-trnS | 43 | 638 (654·3 ± 16) 688 | 703 | 97·1 | 86·7 | 30·4 | 0·397 | 0·129 | 0·169 | 0·304 | |
| rpl2-trnH | 43 | 264 (323·8 ± 77·9) 533 | 623 | 77·3 | 40·1 | 13·1 | 0·411 | 0·13 | 0·121 | 0·338 | |
| rpl20-rps12 | 42 | 740 (744·2 ± 3·3) 760 | 760 | 99·2 | 95·7 | 32·6 | 0·29 | 0·182 | 0·151 | 0·376 | |
| Concatenated | 43 | 2579 (2598 ± 94·4) 2707 | 2707 | 97·5 | 66·1 | 30 | 0·355 | 0·156 | 0·155 | 0·328 | |
The ungapped sequence lengths are shown as minimum (mean ± s.d.) maximum.
ITS2 secondary structure
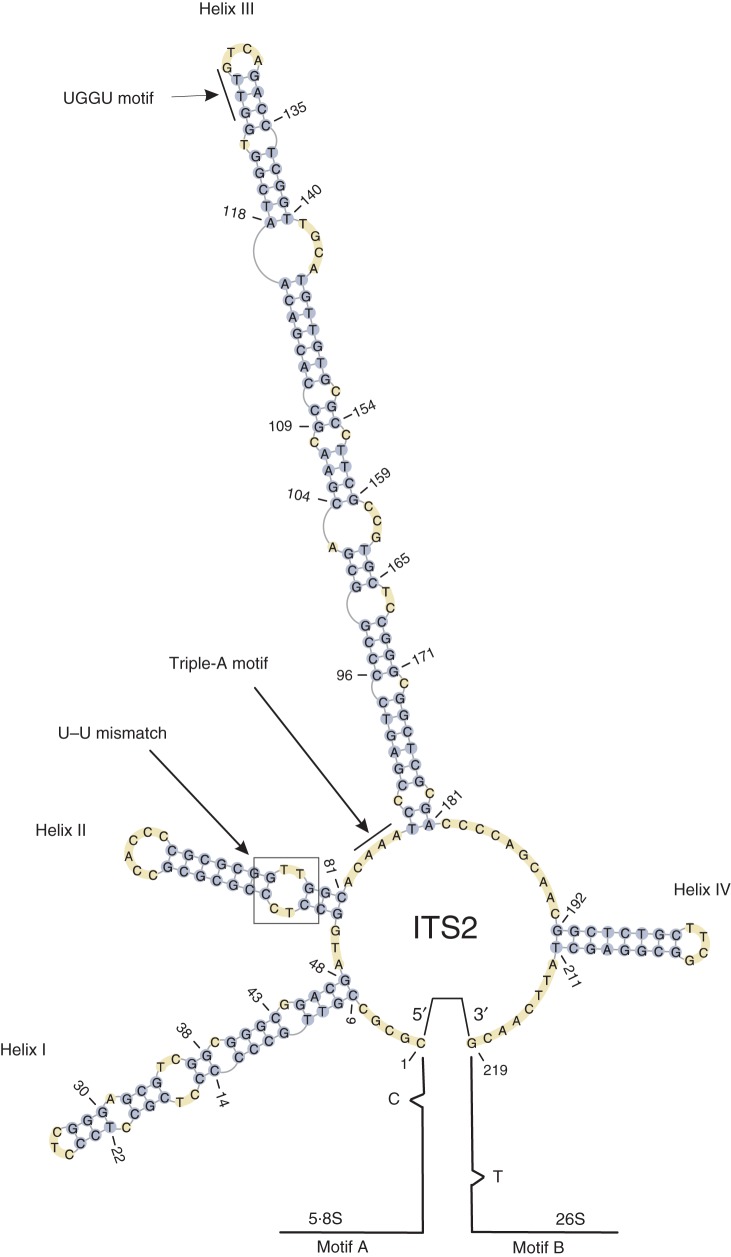
All analysed ITS2 sequences recovered from each accession are considered to be functional because they all returned the predicted secondary structure (Fig. 2) as characterized for Rosaceae (Mai and Coleman, 1997). The proximal stem of ITS2 structure is conserved across the entire range of ribotypes studied here. The 24-nucleotide motifs (A and B, Fig. 2) at the 3′ end of 5.8S and the 5′ end of 26S are also each conserved across all ribotypes. Only one private mutation in 0·08 % of sequences for motif B is observed without any particular pattern. This point mutation is probably due to PCR error (Eyre-Walker et al., 1998) as evidenced by the lack of a compensatory mutation in motif A. The fourth nucleotide from the 5′ end of 5.8S (equal to position 63 in the alignment) was mutated from G to C in all ribotypes belonging to C. series Cerrones ribotypes. These mutations are not located in the hybridized segment of the stem and hence they do not alter the structure of the proximal stem of ITS2. A cytosine (C)-free nucleotide at position 21 of 5.8S and a thymine (T)-free nucleotide at position 11 of 26S are conserved in all ribotypes investigated here (Fig. 2). The highly conserved UGGU motif (preceding the 5′ side to the apex of the third helix) and the triple-A motif (between helices II and III) are present in all ribotypes. Positions 152 and 153 at the 5′ side of helix II and positions 175 and 176 at the 3′ side of helix II correspond to the U–U mismatch. Position 176 is conserved, whereas position 174 had mutated to C in 10 % of ribotypes.
Fig. 2.
The predicted secondary structure of ITS2 for one accession of ribotypes of triploid C. suksdorfii sampled from Haida Gwaii (2010-42; GenBank accession no. KC173907). Annotation from HMMs of the 3′ 5.8S motif and 5′ 26S are displayed as solid lines in the proximal stem of ITS2. The free C nucleotide on the 5.8S subunit and the free T nucleotide on the 26S subunit are shown. The start of the structure and the end of structure are numbered. Four common helix structures, with the third as the longest, UGGU, triple-A motifs and the U–U mismatch were predicted for the entire data set studied here.
Phylogenetic analyses
The aligned ITS2 data matrix comprised 369 positions (including 23 coded characters for indels) of which 198 were variable, including outgroups (194 excluding outgroups), and 120 were potentially parsimony informative over the entire sample (93 for the ingroup alone). The second stage of parsimony analysis was stopped when it reached 100 000 trees, each with 458 steps, a consistency index (CI) of 0·55 (CI = 0·44, excluding uninformative characters), a rescaled consistency index (RC) of 0·50 and a retention index (RI) of 0·92. Poorly supported branches in the main clades are drawn as polytomies for discussion of ploidy evolution. The majority-rule consensus tree obtained from the Bayesian analysis, run under the GTR + I + G evolutionary model, was topologically congruent with the MP consensus tree (not shown). Bayesian PP estimations were superimposed on one of the most-parsimonious trees together with bootstrap values (Fig. 3).
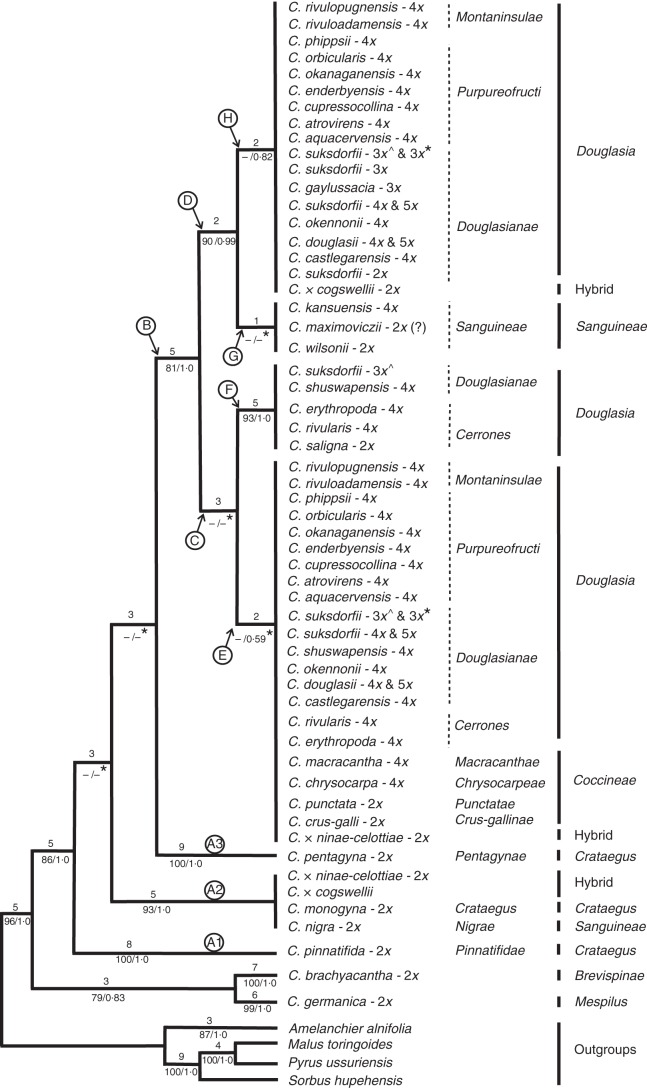
Fig. 3.
One of the most-parsimonious trees, randomly selected from >100 000 trees, and obtained from analysis of the data matrix of ITS2 sequences (299 OTUs). Tree length = 458, consistency index (CI) = 0·55, rescaled consistency index (RC) = 0·5, homoplasy index (HI) = 0·45 and retention index (RI) = 0·92. Branch lengths (ACCTRAN optimization) are indicated above branches and bootstrap percentages and posterior probabilities below branches (BS/PP). – indicates branches with BS <50 and collapsed in the 50 % majority-rule consensus tree of Bayesian inference. The branches in each main clade were collapsed into polytomies to simplify the tree visually. Ploidies are those reported in Talent and Dickinson (2005), Lo et al. (2007, 2013) or J. Coughlan and N. Talent (unpubl. data). ‘C. suksdorfii - 3x*’ indicates the accession sampled from Vancouver Island (JC385; Supplementary Data Table S1), and ‘C. suksdorfii - 3x̂’ represents the accession sampled from Haida Gwaii (2010-42; Table S1). An asterisk (*) after PP values below branches indicates the branches collapsed in the strict consensus of >100 000 most-parsimonious trees.
The four concatenated plastid markers comprised 2707 characters (including 26 coded characters from indels); 127 of these were variable over the entire sample, including outgroups plus the ingroup. Seventy-one characters were variable when outgroups were excluded, and 48 of these were potentially parsimony informative over the entire sample. Only 34 characters were potentially parsimony informative when outgroups were excluded. The second stage of parsimony analysis generated five trees, each with 183 steps, a CI of 0·87 an RC of 0·81 and an RI of 0·93. The majority-rule consensus tree obtained from the Bayesian analysis, run under the GTR + I evolutionary model, was topologically congruent with the MP consensus tree (not shown). Bayesian PP estimations were superimposed on one of the most-parsimonious trees together with bootstrap values (Fig. 4).
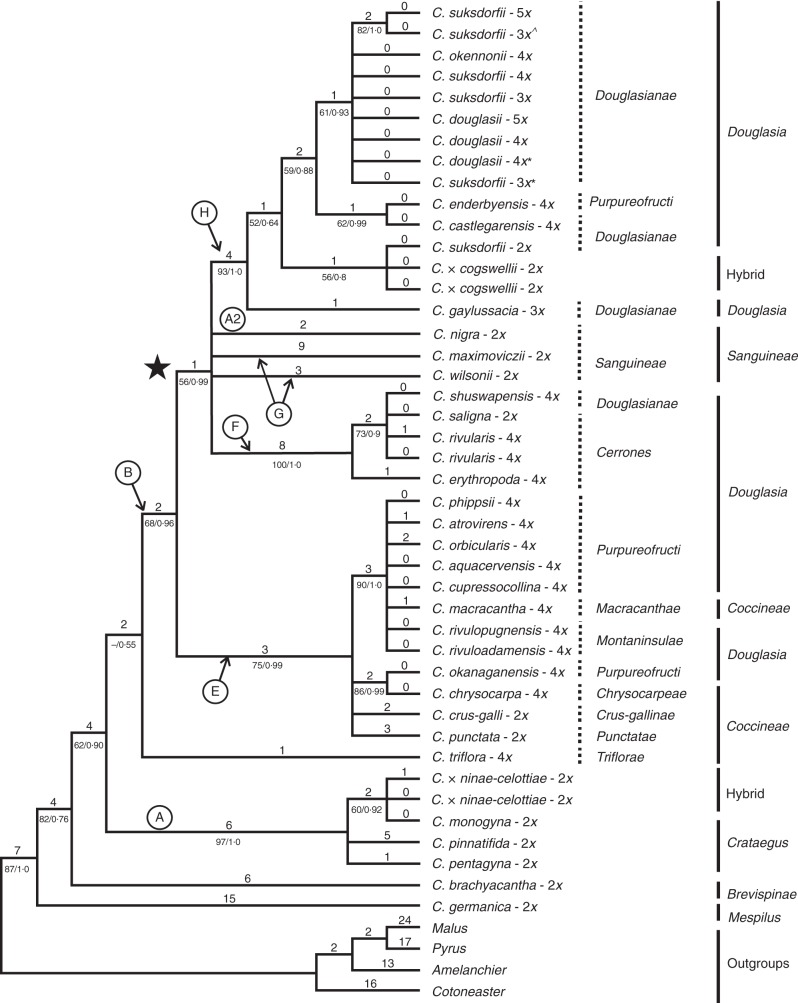
Fig. 4.
One of 25 equally most-parsimonious trees resulted from the analyses of four concatenated plastid markers (trnL-F, trnG-trnS, rpl2-trnH and rpl20-rps12; 47 OTUs, tree length = 182, consistency index (CI) = 0·87, rescaled consistency index (RC) = 0·81; homoplasy index (HI) = 0·12 and retention index (RI) = 0·9256). The clade coding and classification follow Fig. 3. An asterisk (*) after the species name indicates samples from Vancouver Island (JC385 and JC387), while ^ indicates samples from Haida Gwaii (2010-42; Supplementary Data Table S1).
Monophyly of the ingroup taxa is well supported (PP = 1·0) in both the ITS2 and plastid trees. The major clades (A, E, F and H) were well supported in the plastid tree (Fig. 4; PP 0·99–1·0). The PP support for the clades A1, A2, A3, B, D and F is high (PP = 0·99 and 1·0) in the ITS2 tree (Fig. 3), whereas support for clades E, and H is relatively weak (PP < 0·82). The overall topology of both trees is comparable with that seen earlier in analyses of nuclear and plastid loci (Lo et al., 2007, 2009a). In the ITS2 tree (Fig. 3), the unlabelled branch supporting C. brachyacantha and C. germanica plus the branches (A1, A2, A3) corresponding to clade A in the plastid tree (Fig. 4) were retained in the strict consensus tree of > 100 000 most-parsimonious trees and 27 000 Bayesian trees (clade H was not recovered in the Bayesian tree).
Both data sets support the taxonomic structure of the sample studied here, albeit with slightly different topologies, and in ways consistent with the matroclinal relationships and hybrid origins of most polyploids. All members of Eurasian C. section Crataegus formed a single clade (A) in the plastid tree (Fig. 4), whereas in the ITS2 tree this clade breaks down into three chained branches, A1 (C. pinnatifida), A2 (C. monogyna and its intersectional hybrids C. × cogswellii and C. × ninae-celottiae plus the individual of C. nigra from C. section Sanguineae) and A3 (C. pentagyna).
Clades C and D in the ITS2 tree (Fig. 3) collapse in the plastid tree (Fig. 4) so that the western North American black-fruited hawthorns in C. section Douglasia form two sister groups (clades F and H) in the unlabelled clade (star, Fig. 4) that together with clade E make up clade B. Clade F (Figs 3 and 4) comprises the members of C. series Cerrones, plus the individual of C. shuswapensis and, in Fig. 3, an allotriploid C. suksdorfii from Haida Gwaii (2010-42, Supplementary Data Table S1). Clade H comprises both diploid and polyploid members of C. section Douglasia that are found in C. series Douglasianae and the exclusively tetraploid members of C. series Purpureofructi (Figs 3 and 4). In the ITS2 tree, clade H also includes the two tetraploid Cypress Hills endemics with dark red fruit, C. rivuloadamensis and C. rivulopugnensis, that are placed in C. series Montaninsulae (Fig. 3). These two species are found in clade E in the plastid tree (Fig. 4). All of the sampled members of Eurasian C. section Sanguineae (including C. nigra, A2) are found in the unlabelled clade marked with an asterisk in the plastid tree (Fig. 4). In the ITS2 tree, with C. nigra in clade A2, the remaining Sanguineae form clade G, sister to the Douglasia in clade H (Fig. 3).
In both the ITS2 and plastid trees, members of red-fruited North American C. section Coccineae are found exclusively in clade E, with polyploid members of C. section Douglasia belonging to C. series Douglasianae, Montaninsulae and Purpureofructi (Figs 3 and 4). Note that in this account of how taxonomic structure is reflected by our molecular data, we have placed C. series Montaninsulae in section Douglasia (Table 1); this series was not placed in a section when first described (Phipps and O'Kennon, 2007), nor is it placed in a section in VASCAN (Brouillet et al., 2010 +).
As discussed elsewhere (Christensen et al., 2014), the hybrid taxa, C. × ninae-celottiae (C. monogyna × C. punctata) and C. × cogswellii (C. suksdorfii × C. monogyna) shared their ribotypes with both of their parents, i.e. each hybrid appears twice on the ITS2 tree (Fig. 3; C. × ninae-celottiae in clades A2 and E, and C. × cogswellii in clades A2 and H). However, as described below, the hybrids differed in terms of which species was the maternal parent (Fig. 4).
By cloning and sequencing ITS2 extensively and in proportion to ploidy, we see that almost all of the Crataegus section Douglasia polyploids are also found twice on the ITS2 tree, with series Cerrones tetraploids in clades E and F, and series Douglasianae, Montaninsulae and Purpureofructi polyploids in clades E and H (Fig. 3). Diploid C. saligna appears only once, in clade F (Fig. 3). Diploid C. suksdorfii (series Douglasianae) and putative autotriploids (C. gaylussacia and western Oregon triploids) are found only in clade H. The only exceptions to these patterns are the tetraploid individual of C. shuswapensis (clades E and F) and a triploid C. suksdorfii from Haida Gwaii (clades E, F and H; Fig. 3).
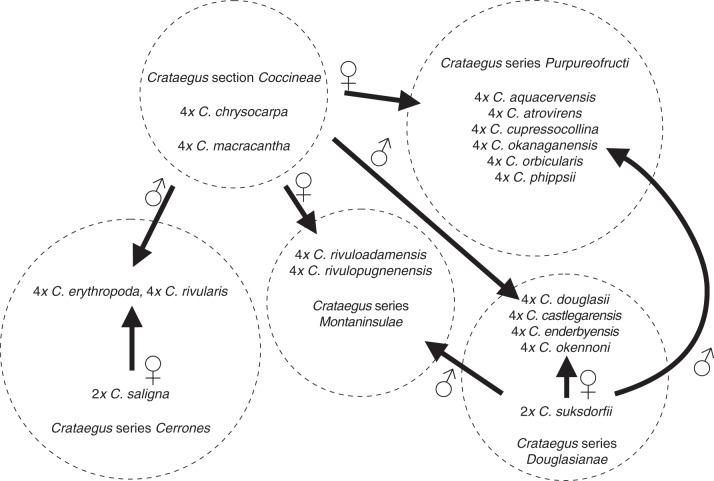
The plastid phylogenetic tree (Fig. 4) enables us to establish the maternal parentage of the recent hybrids and allopolyploids. Crataegus monogyna is the maternal parent of the hybrid with eastern North American C. punctata (clade A; Fig. 4). In contrast, C. suksdorfii appears to have been the maternal parent of the west coast hybrid with C. monogyna (clade H; Fig. 4). Tetraploid members of C. series Cerrones (and C. shuswapensis; clade F) appear to have C. saligna as their maternal parent, much as the tetraploids in C. series Douglasianae (and C. enderbyensis) ultimately have diploid C. suksdorfii as their maternal parent. Conversely, tetraploids assigned to C. series Montaninsulae and Purpureofructi (with the exception of C. enderbyensis) appear to have had one of the western members of C. section Coccineae as their maternal parent (clade G; Figs 4 and 5).
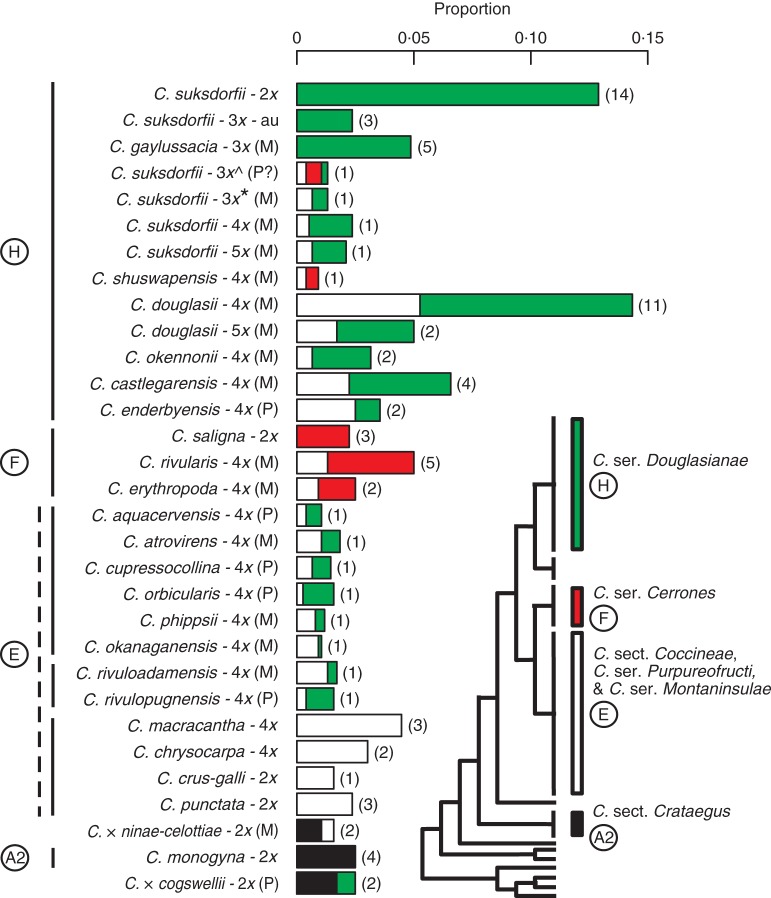
Fig. 5.
The distribution of ribotypes in C. sections Douglasia and Coccineae and C. monogyna, C. × ninae-celottiae and C. × cogswellii in clades A2 (black), E (white), F (red) and H (green) on the ITS2 phylogenetic tree (Fig. 3). All proportions sum up to one. Ploidy is shown for each entity studied. Letters inside parentheses beside each species refer to the directions of ITS2 homogenization (M, biased towards the maternal ribotypes; P, biased towards the paternal ribotypes). Numbers inside parentheses on the bars indicate the number of accessions analysed. Ploidies in C. suksdorfii are indicated as follows: 3x̂ = allotriploid C. suksdorfii sampled from Haida Gwaii (2010-42); 3x-au = autotriploid C. suksdorfii; 3x* = allotriploid C. suksdorfiî sampled from Vancouver Island (JC385).
In the ITS2 and the plastid phylogenetic trees, other ingroup taxa were associated with clades the same as or similar to those in which they were found in earlier studies (Lo et al., 2007, 2009a; Lo and Donoghue, 2012). Crataegus brachyacantha (section Brevispinae) and C. germanica (section Mespilus) are sister to the remainder of the genus (Figs 3 and 4). The Eurasian species C. pentagyna and C. pinnatifida are found with the other representative of C. section Crataegus in clade A (Figs 3 and 4). Crataegus nigra is the type of C. series Nigrae in C. section Sanguineae but is found with other members of section Sanguineae only in the plastid tree (Fig. 4). In our ITS2 tree, C. nigra appears together with members of C. section Crataegus (clade A2, Fig. 3), in contrast to earlier results (Lo et al., 2007). Crataegus triflora (series Triflorae in C. section Coccineae) is sister to clade B in the plastid tree. The remaining North American species (C. crus-galli and C. punctata) are found in clade E as part of an enlarged C. section Coccineae.
ITS2 composition in hybrids and polyploids
Results of the phylogenetic analyses of the ITS2 and plastid sequence data can be summarized graphically so as to contrast diploids and autotriploids with allopolyploids (Fig. 5). In this graph the behaviour of the two C. monogyna hybrids illuminates what is seen in the remainder of the sample: individuals of the three diploid parent species each contained only the ribotype found in ‘their’ clade (E, F, H), whereas individuals of each of the two hybrids contained ribotypes from the clades of both parents (A2 + E or A2 + H; Figs 3 and 4). The results thus suggest that hybridization between C. section Coccineae and C. series Cerrones is responsible for the tetraploids C. erythropoda, C. rivularis and C. shuswapensis (they contain ribotypes from clades E and F; Figs 3, 5 and 6). Similarly, hybridization between C. section Coccineae and C. series Douglasianae appears to be responsible for the allotetraploids C. aquacervensis, C. atrovirens, C. castlegarensis, C. cupressocollina, C. douglasii, C. enderbyensis, C. okanaganensis, C. okennonii, C. orbicularis, C. phippsii and C. rivuloadamensis (they contain ribotypes from clades E and H; Figs 3 and 5). The same appears to be true of pentaploid C. douglasii, some triploid C. suksdorfii and the tetraploid and pentaploid C. suksdorfii. On the other hand, C. gaylussacia and some other C. suksdorfii appear to be autotriploids (clade H ribotypes only; Figs 3, 5 and 6). A triploid C. suksdorfii from Haida Gwaii appears to contain ribotypes from all three North American clades (E, F, H; Figs 3 and 5). The relative proportions of maternal and paternal ribotypes recovered from each taxon or individual is used to infer the direction of ITS2 homogenization (Fig. 5).
Fig. 6.
A summary model of intersectional hybridization and polyploid formation in Crataegus section Douglasia, based on our analyses of ITS2 and plastid DNA sequences. This diagram thus does not include the way in which C. douglasii has contributed to the formation of polyploid C. suksdorfii (Lo et al., 2010). It also does not show the triploid bridges implicitly involved in the formation of tetraploids from crosses between diploid and tetraploid parents.
DISCUSSION
By extensively cloning ITS2, we have captured considerable ribotype diversity (Fig. 1B; Supplementary Data Table S1) from a sample that represents all of the major groups in Crataegus (Table 1). An examination of the secondary structure of the ITS2 sequences strongly suggests that the ribotype diversity we document is due to variation among orthologues and paralogues, not to the occurrence of pseudogenes. Visualized as a gene tree (Fig. 3), not all taxonomic groupings are well supported, but an NN graph (not shown) provided further evidence for the observed topological relationships. In addition, the topology of these groups (Fig. 3) corresponds to that seen with plastid data (Fig. 4) and is consistent with earlier studies based not only on ITS but also on additional nuclear (Lo et al., 2007, 2009a) and plastid (Lo and Donoghue, 2012) loci. This overall congruence encourages us to interpret our ITS2 data. We note that the ribotypes found in diploid individuals from Pacific Northwest and Rocky Mountain Crataegus section Douglasia occur only in clades F and H (Fig. 3), whereas those found in polyploid individuals from section Douglasia occur in clades F and H (Fig. 3), with the diploids, and in clade E with ribotypes from diploids and polyploids of Crataegus section Coccineae (Fig. 3).
Clade B received high PP support (1·0 and 0·96 in the ITS2 and plastid trees, respectively) and comprises all of the taxa belonging to C. sections Coccineae, Sanguineae and Douglasia (Fig. 3). However, C. nigra of C. section Sanguineae is found in the well-supported clade A2 in the ITS2 tree (Fig. 3), whereas this species has an unsupported position in clade B of the plastid tree (Fig. 4). The reliability of clades E, F and H is high (PP = 0·99 and 1·0) in the plastid tree. However, only clade F has strong support in the ITS2 tree (BS = 93; PP = 1·0), whereas clades E and H received only weak support with these sequences (PP ≤ 82). Crataegus series Montaninsulae and Purpureofructi (except C. enderbyensis) are found in clade E in the plastid tree, whereas C. series Douglasianae, except C. shuswapensis, are found in clade H in the plastid tree.
Direct vs. cloned sequencing of ITS2
For every sample investigated here, the cloned sequences confirmed the presence of the APS that were apparent in electropherograms resulting from the direct sequencing. To investigate the likelihood of possible mistakes incorporated into the DNA strand by Taq polymerase (Smith et al., 1997; Eyre-Walker et al., 1998; Nieto Feliner et al., 2004), the direct sequencing and cloning were performed on the identical pool of amplicons (accessions EL-32, 2007-03 and 18445; Supplementary Data Table S1). Moreover, only potentially parsimony-informative single nucleotide polymorphisms (SNPs) were considered for this purpose because incorporating private mutational errors at a single base is more likely than at potentially parsimony-informative SNPs (Nieto Feliner et al., 2004). All of the samples showed APS. However, if the frequency of occurrence of one base in a particular site is <15 %, only direct sequencing identified the base with higher frequency in the amplicon pool. This result emphasizes the limitation of direct sequencing to detect rare ITS2 ribotypes (see Rauscher et al., 2002; Nieto Feliner et al., 2004; Koch et al., 2010; Baldwin et al., 2011; Gao et al., 2012; Gardner et al., 2012). Eight, 12, 16 and 20 clones for diploid, triploid, tetraploid and pentaploid accessions were sufficient to capture ribotype diversity if the accession arose from hybridization or if multiple ribotypes of ITS2 were present in the genome.
Incomplete concerted evolution
By documenting the heterogeneous origins of ITS2 paralogues found in an individual, our results also show how concerted evolution is incomplete (Hollingsworth et al., 2011) in Crataegus, much as it has been found to be in other Rosaceae, such as diploid and polyploid Amelanchier spp. (Campbell et al., 1997), Malus (Feng et al., 2007) and Polylepis (Kerr, 2004). At least three factors appear to be potentially responsible for this. The presence of several ribotypes in the genome of the diploid Crataegus taxa studied here suggests that the rate of mutation among ribotypes is greater than that of concerted evolution (Arnheim, 1983; Nieto Feliner et al., 2004; Nieto Feliner and Rosselló, 2007). In polyploids the rate of concerted evolution is probably reduced by asexual reproduction, and by sexual reproduction involving unreduced (apomeiotic) gametes. Finally, all Crataegus taxa, like those of related genera (Rosaceae tribe Maleae), probably have greater ribotype diversity than might otherwise be the case because of their ancient polyploid origin (Campbell et al., 1997, 2007; Evans and Campbell, 2002).
Allo- and autopolyploidy
We interpret the presence in the C. section Douglasia polyploids, but not in the diploids, of ribotypes of two kinds (Douglasia-Sanguineae and Coccineae; Fig. 5) as evidence of allopolyploidy (Figs 5 and 6). This interpretation is strengthened by the behaviour of the ribotypes found in the recent hybrids between introduced, diploid C. monogyna (section Crataegus) and two native diploid species (Fig. 3; Christensen et al., 2014), one each from section Coccineae (C. punctata) and section Douglasia (diploid C. suksdorfii). Fehrer et al. (2009) obtained similar evidence of hybridization in Hieracium using the external transcribed spacer region (nrETS).
The contribution of parents to the ribotype composition of diploid hybrids and allopolyploids appears to vary. When different ITS repeats co-occur in single genomes due to hybridization or introgression, the speed and the direction of homogenization cannot be predicted, and it is not consistent in different descendant lineages (discussed in Nieto Feliner and Rosselló, 2007). Preferential PCR amplification (discussed in Bellemain et al., 2010) was probably not the main cause of the differences observed (Fig. 5) because different accessions of each ploidy showed the same pattern. Moreover, preferential PCR amplification was substantially reduced by using highly denaturing PCR conditions (see Buckler et al., 1997; Rudnóy et al., 2011).
In C. × ninae-celottiae more maternal than paternal ribotypes were recovered, whereas in C. × cogswellii the situation is reversed (Fig. 5). This could be a genetic effect, in that C. monogyna is the dominant contributor in both cases, as the female parent in the hybrid with C. punctata and as the male parent in the hybrid with C. suksdorfii. To the extent that allopolyploids originated via fertilization of unreduced female gametes by meiotically reduced male gametes from diploids or tetraploids (Talent, 2009), a greater representation of maternal ribotypes might be expected, and in fact is seen in more than half the cases (Fig. 5). However, our sample sizes may be too small in most cases to distinguish between any pattern and stochastic variation (and incomplete recovery of all ribotypes present).
A further confounding factor is that in both C. series Cerrones and C. series Douglasianae, the maternal parent of the allopolyploids is ultimately one of the diploid taxa, C. saligna and diploid C. suksdorfii, respectively. This strongly suggests that some allotetraploids, perhaps those which are most widespread (e.g. C. castlegarensis, C. douglasii, C. okennonii and C. rivularis), arose via a triploid bridge (Ramsey and Schemske, 1998), with the initial triploid arising from the fertilization of reduced gametes from diploid and tetraploid parents (Fig. 6). The triploid hybrid would have to have produced unreduced female gametes that developed into individuals that then took part in backcrosses with the diploid parent in order to produce allotetraploids with at least equal representation of the ribotypes of the diploid.
Other allotetraploids, perhaps those with much more restricted distributions (e.g. C. aquacervensis, C. cupressocollina, C. rivuloadamensis and C. rivulopugnensis in the Cypress Hills; C. atrovirens, C. enderbyensis and C. orbicularis in the southern interior of British Columbia) may have arisen through crosses involving only the reduced female gametes of other allotetraploid parents such as the widespread C. douglasii and members of C. section Coccineae. Seeds from reduced female gametes are produced much less frequently than from unreduced gametes in most tetraploid Crataegus (N. Talent, unpubl. data 2013).
Although C. gaylussacia in California (Table 1) appears to consist exclusively of autotriploids (Figs 3 and 5), autopolyploids are otherwise poorly represented in our sample. The accession EL-65 (triploid C. suksdorfii; Supplementary Data Table S1) was collected and studied by Lo et al. (2010), and it has been shown to be an autotriploid, along with two other accessions identified as autotriploids (EL-172 and EL-188). In the study of Lo et al. (2010), however, the emphasis lay on demonstrating the way in which ‘east side’ triploid C. suksdorfii (i.e. from east of the Cascades) arose as a result of hybridization between diploid C. suksdorfii and tetraploid C. douglasii. This conclusion was based on the distribution in C. douglasii and C. suksdorfii individuals of variants of the short and (or) long copies of the PEPC and PISTILLATA gene regions. Individuals of species in Crataegus section Coccineae were not studied by Lo et al. (2010). Our failure to identify EL-172 and EL-188 as allotriploids, as Lo et al. did, probably stems from the fact that we were unable to distinguish between the ITS2 ribotypes that they obtained from each of their parent species. This would imply either that due to chance we failed to clone any of the Coccineae ribotypes that this cytotype received from its C. douglasii parent or that in these allotriploids ITS2 homogenization was biased toward their paternal C. suksdorfii parent.
A different problem of interpretation is presented by the occurrence, in one triploid C. suksdorfii individual from Vancouver Island (JC385; Supplementary Data Table S1) and in the one individual of C. shuswapensis in our sample (tetraploid 2007-11; Table S1), of ITS2 ribotypes indistinguishable from those recovered from individuals in C. series Cerrones (Figs 3 and 5). In the latter case, plastid sequences also suggest a relationship between this individual and series Cerrones (Fig. 4). We doubt that these results are due to hitherto undiscovered floristic connections between the central Rocky Mountains and southern British Columbia. Analyses incorporating additional plastid sequences obtained subsequent to the work described here (M. Zarrei et al., unpubl. data) suggest that the relationship between C. shuswapensis and the members of C. series Cerrones is considerably more tenuous than seen in Fig. 4. If instead this relationship is due to long-branch attraction, the ITS2 results might then be seen as resulting from our cloning a ribotype that has persisted unaltered by concerted evolution from the common ancestor of series Cerrones and series Douglasianae.
Hybridization and apomixis: ‘the Crataegus problem’
Elsewhere we will present analyses of leaf, flower and fruit morphometric data from most of the species studied here (T. A. Dickinson et al., unpubl. res.) to show that the morphological variation observed in the entities studied here is consistent with the occurrence of hybridization. Further, we note that our interpretation is consistent with flow cytometric data documenting the ploidy (Table 1) and breeding system (Talent and Dickinson, 2007a) of the taxa studied here (in many cases, based on the same individuals from which our molecular data were obtained). Seeds of all of the polyploid Crataegus individuals studied to date exhibit embryo–endosperm ratios of nuclear DNA content that are consistent with their origin predominantly from unreduced female gametes by means of pseudogamous gametophytic apomixis (Talent and Dickinson, 2007a). In addition, almost all of the tetraploid Crataegus studied to date produce highly fertile (i.e. stainable) pollen at a frequency comparable with that seen in diploids (Dickinson and Phipps, 1986; Dickinson et al., 1996). We surmise that this pollen developed from reduced microspores, as many of our data from seeds that prove to be biparental are consistent with this origin (Talent and Dickinson, 2007a).
Our interpretation of the results presented here (and earlier, regarding the origins of polyploid C. suksdorfii; Lo et al., 2009b, 2010) runs counter to recent suggestions that intersectional hybridization should be rare in Crataegus (Phipps, 2005). This latter conclusion was based on qualitative analyses of morphological variation in the well-collected Crataegus flora of Missouri. In other work (e.g. Dickinson and Phipps, 1985, 1986; Phipps, 2013), however, Phipps emphatically recognized the roles played by gametophytic apomixis and hybridization in creating the ‘Crataegus problem’, as did earlier workers including Camp (1942), Rickett (1936) and Palmer (1932). Nevertheless, entities ascribed to C. section Douglasia (see above and Table 1), with limited geographical distributions, that can only with difficulty be distinguished from each other, with some more widely distributed entities (C. castlegarensis, C. douglasii, C. erythropoda and C. rivularis; Table 1) have all proved to be tetraploid, apomictic and demonstrably of hybrid origin. Apart from the autotriploids we have found, the polyploid segregates of C. suksdorfii (Table 1) are similar. These Pacific Northwest hawthorns thus provide a model for better understanding evolution in Crataegus.
As noted earlier (Dickinson et al., 2007), genera such as Sorbus sensu lato (s.l.) have already shown that this model of hybridization, gametophytic apomixis, pseudogamy and allopolyploidization may be widely applicable to other genera belonging to Rosaceae tribe Maleae, including Amelanchier, Aronia, Cotoneaster and Malus. In particular, results from molecular analyses of Sorbus s.l. parallel our documentation of the extent to which western North American Crataegus polyploids have arisen as intersectional hybrids. In Sorbus s.l., two distinct clades (recognizable as minimally two segregate genera, Aria and Sorbus; Lo and Donoghue, 2012) have given rise to numerous apomictic, polyploid hybrids within and between these clades, as documented by morphological, isozyme, RFLP (restriction fragment length polymorphism), microsatellite, plastid DNA sequence and flow cytometry data (Aas et al., 1994; Nelson-Jones et al., 2002; Chester et al., 2007; Robertson et al., 2010; Pellicer et al., 2012). As with some of the Crataegus taxa studied here, some of the hybrids of Sorbus s.l. have limited distributions, whereas others are more widely distributed (Chester et al., 2007; Ludwig et al., 2013). Coincidentally, Sorbus subgenus Sorbus sections Sorbus and Commixtae both show amphi-Beringean distributions (McAllister, 2005) comparable with that exhibited by clade D in Fig. 3 that supports clades G and H, i.e. C. sections Douglasia and Sanguineae, that have been recognized as Crataegus subgenus Sanguineae (Ufimov, 2013).
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
Nadia Talent and Jennifer Coughlan provided unpublished flow cytometric data on ploidy level in the Crataegus accessions studied here, as well as assistance in the field and comments on the manuscript (N.T.). In addition to those individuals acknowledged in Supplementary Data Table S1, the following also assisted us with fieldwork or made collections for us: Adam Dickinson, John Dickinson, Beth Dickson, Fannie Gervais, Marta Heckel, Sophie Nguyen, Jenny Shiller and Jurgena Tusha. James B. Phipps identified several of our 2007 collections. This work could not have been completed without the help of Anuar Rodrigues, Thomas Braukmann, Kristen Choffe, Oliver Haddrath, Maria Kuzmina, Samantha Beddington and Ionatan Waisgluss. We would also like to thank Mark Chase and Christopher Campbell for discussion of concerted evolution, and Matthias Wolf for discussion on predicting the secondary structure ITS2. We are indebted to the following institutions for allowing us to collect on their property: the Arnold Arboretum of Harvard University; Morton Arboretum; and the Point Reyes National Seashore (US Department of the Interior, National Park Service). This work was supported by a Natural Sciences and Engineering Research Council of Canada Strategic Research Project Grant [381073 to T.A.D., S.S., P. Shipley (UBC-Okanagan), S. Proctor (University of Alberta) and the Naturally Grown Herb and Spice Growers Co-operative (Edgewood BC, J. Lee, President)]. Paula Brown (British Columbia Institute of Technology) materially assisted us in obtaining this grant. The Canada Foundation for Innovation and the Ontario Research Fund provided funding through the Canadensys initiative for the equipment and personnel used to document voucher specimens for this study. Additional funding for fieldwork and other aspects of this research was provided by NSERCC Discovery Grants [A3430 to T.A.D.; 326439 to S.S.]. The generous support of the Louise Hawley Stone Charitable Trust enabled the ROM Green Plant Herbarium to accept the University of Western Ontario's gift of the J. B. Phipps Hawthorn Research Collection. Further funding from the Department of Natural History of the Royal Ontario Museum is gratefully acknowledged, as are an award from the Royal Ontario Museum Foundation to T.A.D. and Mark Engstrom for the purchase of the thermocycler used in part of this work, and the generous support of the ROM Reproductions Acquisitions and Research Fund.
LITERATURE CITED
- Aas G, Maier J, Baltisberger M, Metzger S. Morphology, isozyme variation, cytology, and reproduction of hybrids between Sorbus aria (L) Crantz and S. Torminalis (L) Crantz. Botanica Helvetica. 1994;104:195–214. [Google Scholar]
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Applied and Environmental Microbiology. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnarsson I, Miller JA. Is ACCTRAN better than DELTRAN? Cladistics. 2008;24:1–7. doi: 10.1111/j.1096-0031.2008.00229.x. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- Arnheim N. Concerted evolution of multigene families. In: Nei M, Koehn R, editors. Evolution of genes and proteins. Sunderland, MA: Sinauer; 1983. pp. 38–61. [Google Scholar]
- Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ. The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Annals of the Missouri Botanical Garden. 1995;82:247–277. [Google Scholar]
- Baldwin BG, Kalisz S, Armbruster WS. Phylogenetic perspectives on diversification, biogeography, and floral evolution of Collinsia and Tonella (Plantaginaceae) American Journal of Botany. 2011;98:731–753. doi: 10.3732/ajb.1000346. [DOI] [PubMed] [Google Scholar]
- Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiology. 2010;10:189. doi: 10.1186/1471-2180-10-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillet L, Coursol F, Meades SJ, et al. VASCAN, the Database of Vascular Plants of Canada. 2010 +. Crataegus gaylussacia A. Heller. http://data.canadensys.net/vascan/taxon/20019. (accessed on November 7, 2013) [Google Scholar]
- Bruen T, Phillipe H, Bryant D. A quick and robust statistical test to detect the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunsfeld SJ, Johnson FD. Cytological, morphological, ecological and phenological support for specific status of Crataegus suksdorfii (Sarg.) Kruschke. Madroño. 1990;37:274–282. [Google Scholar]
- Bryant D, Moulton V. Neighbor-Net: an agglomerative method for the construction of phylogenetic networks. Molecular Biology and Evolution. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Buckler ESI, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Huelsenbeck JP, Cunningham CW, Swofford DL, Waddell JP. Partitioning and combining data in phylogenetic analysis. Systematic Biology. 1993;42:384–397. [Google Scholar]
- Camp WH. The Crataegus problem. Castanea. 1942;7:51–55. [Google Scholar]
- Campbell CS, Wojciechowski MF, Baldwin BG, Alice LA, Donoghue MJ. Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier (Rosaceae) agamic complex. Molecular Biology and Evolution. 1997;14:81–90. doi: 10.1093/oxfordjournals.molbev.a025705. [DOI] [PubMed] [Google Scholar]
- Campbell CS, Evans RC, Morgan DR, Dickinson TA, Arsenault MP. Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): limited resolution of a complex evolutionary history. Plant Systematics and Evolution. 2007;266:119–145. [Google Scholar]
- Chase MW, Hills HG. Silica gel: an ideal desiccant for preserving field collected leaves for use in molecular studies. Taxon. 1991;40:215–220. [Google Scholar]
- Chen S, Yao H, Han J, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M, Cowan RS, Fay MF, Rich TCG. Parentage of endemic Sorbus L. (Rosaceae) species in the British Isles: evidence from plastid DNA. Botanical Journal of the Linnean Society. 2007;154:291–304. [Google Scholar]
- Christensen KI. Revision of Crataegus sect. Crataegus and nothosect. Crataeguineae (Rosaceae-Maloideae) in the Old World. Systematic Botany Monographs. 1992;35:1–199. [Google Scholar]
- Christensen KI, Zarrei M, Kuzmina M, Talent N, Lin C, Dickinson TA. Crataegus × ninae-celottiae and C. × cogswellii (Rosaceae, Maleae), two spontaneously formed intersectional nothospecies. Phytokeys. 2014;36:1–26. doi: 10.3897/phytokeys.36.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke T, Adams RP. The effect of plant polysaccharides and buffer additives on PCR. Biotechniques. 1992;12:332–334. [PubMed] [Google Scholar]
- Dickinson T, Phipps J. Studies in Crataegus L. (Rosaceae: Maloideae). XIII. Degree and pattern of phenotypic variation in Crataegus sect. Crus-galli in Ontario. Systematic Botany. 1985;10:322–337. [Google Scholar]
- Dickinson T, Phipps J. Studies in Crataegus (Rosaceae: Maloideae) XIV. The breeding system of Crataegus crus-galli sensu lato in Ontario. American Journal of Botany. 1986;73:116–130. doi: 10.1002/j.1537-2197.1986.tb09687.x. [DOI] [PubMed] [Google Scholar]
- Dickinson TA, Belaoussoff S, Love RM, Muniyamma M. North American black-fruited hawthorns: I. Variation in floral construction, breeding system correlates, and their possible evolutionary significance in Crataegus sect. Douglasii Loudon. Folia Geobotanica and Phytotaxonomica. 1996;31:355–371. [Google Scholar]
- Dickinson TA, Lo EYY, Talent N. Polyploidy, reproductive biology, and Rosaceae: understanding evolution and making classifications. Plant Systematics and Evolution. 2007;266:59–78. [Google Scholar]
- Dickinson TA, Lo EYY, Talent N, Love RM. Black-fruited hawthorns of western North America – one or more agamic complexes? Botany. 2008;86:846–865. [Google Scholar]
- Dickinson TA, Zarrei M, Kuzmina M, Stefanović S. Presented at Botany 2011. St. Louis, MO: Healing the planet; 2011. Mixed success with DNA barcoding of hawthorns (Crataegus L., Rosaceae) [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, et al. 2012. Geneious v5.6. Available from http://www.geneious.com . [Google Scholar]
- Edwards JE, Brown PN, Talent N, Dickinson TA, Shipley PR. A review of the chemistry of the genus Crataegus. Phytochemistry. 2012;79:5–26. doi: 10.1016/j.phytochem.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Efron B. Bootstrap methods: another look at the jackknife. Annals of Statistics. 1979;7:1–26. [Google Scholar]
- Evans RC, Campbell CS. The origin of the apple subfamily (Maloideae; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. American Journal of Botany. 2002;89:1478–1484. doi: 10.3732/ajb.89.9.1478. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to domestication of maize. Proceedings of the National Academy of Sciences; USA. 1998. pp. 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS. The retention index and the rescaled consistency index. Cladistics. 1989;5:417–419. doi: 10.1111/j.1096-0031.1989.tb00573.x. [DOI] [PubMed] [Google Scholar]
- Fehrer J, Krak K, Chrtek J. Intra-individual polymorphism in diploid and apomictic polyploid hawkweeds (Hieracium, Lactuceae, Asteraceae): disentangling phylogenetic signal, reticulation, and noise. BMC Evolutionary Biology. 2009;9:239. doi: 10.1186/1471-2148-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Feng TT, Zhou ZQ, Tang JM, Cheng MH, Zhou SL. ITS sequence variation supports the hybrid origin of Malus toringoides Hughes. Botany. 2007;85:659–666. [Google Scholar]
- Fitch WM. Towards defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–426. [Google Scholar]
- Gao Q, Zhang D, Duan Y, Zhang F, Li Y, Fu P, Chen S. Intraspecific divergences of Rhodiola alsia (Crassulaceae) based on plastid DNA and internal transcribed spacer fragments. Biological Journal of the Linnean Society. 2012;168:204–215. [Google Scholar]
- Gardner AG, Vaio M, Guerra M, Emshwiller E. Diversification of the American bulb-bearing Oxalis (Oxalidaceae): dispersal to North America and modification of the tristylous breeding system. American Journal of Botany. 2012;99:152–164. doi: 10.3732/ajb.1100152. [DOI] [PubMed] [Google Scholar]
- Gu C, Spongberg SA. Crataegus Linnaeus. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 9 (Pittosporaceae through Connaraceae). Beijing: Science Press; 2003. [Google Scholar]
- Guo R, Pittler MH, Ernst E. Hawthorn extract for treating chronic heart failure. Cochrane Database of Systematic Reviews. 2008;1:CD005312. doi: 10.1002/14651858.CD005312.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hamilton MB. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology. 1999;8:521–523. [PubMed] [Google Scholar]
- Han K, Lee Y, Kim K. PseudoViewer: automatic visualization of RNA pseudoknots. Bioinformatics. 2002;18:S321–S328. doi: 10.1093/bioinformatics/18.suppl_1.s321. [DOI] [PubMed] [Google Scholar]
- Heinze B. A database of PCR primers for the chloroplast genomes of higher plants. Plant Methods. 2007;3:4. doi: 10.1186/1746-4811-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller AA. Notes on plants from middle western California. Bulletin of the Southern California Academy of Sciences. 1903;2:65–70. [Google Scholar]
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S. Bootstrapping phylogenetic trees: theory and methods. Statistical Science. 2003;18:241–255. [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Keller A, Schleicher T, Schultz J, Müller T, Dandekar T, Wolf W. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Kerr MS. 2004. A phylogenetic and biogeographic analysis of Sanguisorbeae (Rosaceae) with emphasis on the pleistocene radiation of the high Andean genus Polylepis. PhD thesis, University of Maryland, College Park, MD. [Google Scholar]
- Koch MA, Karl R, Kiefer C, Al-Shehbaz IA. Colonizing the American Continent: systematics of the genus Arabis in North America (Brassicaceae) American Journal of Botany. 2010;97:1040–1057. doi: 10.3732/ajb.0900366. [DOI] [PubMed] [Google Scholar]
- Koetschan C, Hackl T, Müller T, Wolf M, Förster F, Schultz J. ITS2 Database IV: interactive taxon sampling for internal transcribed spacer 2 based phylogenies. Molecular Phylogenetics and Evolution. 2012;63:585–588. doi: 10.1016/j.ympev.2012.01.026. [DOI] [PubMed] [Google Scholar]
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology. 2001;50:913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lo EYY, Donoghue MJ. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae. Molecular Phylogenetics and Evolution. 2012;63:230–243. doi: 10.1016/j.ympev.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Lo EYY, Stefanović S, Dickinson TA. Molecular reappraisal of relationships between Crataegus and Mespilus (Rosaceae, Pyreae) – two genera or one? Systematic Botany. 2007;32:596–616. [Google Scholar]
- Lo EYY, Stefanović S, Christensen KI, Dickinson TA. Evidence for genetic association between East Asian and Western North American Crataegus L. (Rosaceae) and rapid divergence of the Eastern North American lineages based on multiple DNA sequences. Molecular Phylogenetics and Evolution. 2009a;51:157–168. doi: 10.1016/j.ympev.2009.01.018. [DOI] [PubMed] [Google Scholar]
- Lo EYY, Stefanovic S, Dickinson TA. Population genetic structure of diploid sexual and polyploid apomictic hawthorns (Crataegus; Rosaceae) in the Pacific Northwest. Molecular Ecology. 2009b;18:1145–1160. doi: 10.1111/j.1365-294X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- Lo EYY, Stefanović S, Dickinson TA. Reconstructing reticulation history in a phylogenetic framework and the potential of allopatric speciation driven by polyploidy in an agamic complex in Crataegus (Rosaceae) Evolution. 2010;64:3593–3608. doi: 10.1111/j.1558-5646.2010.01063.x. [DOI] [PubMed] [Google Scholar]
- Love R, Feigen M. Interspecific hybridization between native and naturalized Crataegus (Rosaceae) in western Oregon. Madroño. 1978;25:211–217. [Google Scholar]
- Ludwig S, Robertson A, Rich TCG, et al. Breeding systems, hybridization and continuing evolution in Avon Gorge Sorbus. Annals of Botany. 2013;111:563–575. doi: 10.1093/aob/mct013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR. The discovery and importance of multiple islands of most-parsimonious trees. Systematic Zoology. 1991;40:315–328. [Google Scholar]
- Mai JC, Coleman AW. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. Journal of Molecular Evolution. 1997;44:258–271. doi: 10.1007/pl00006143. [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister HA. The genus Sorbus – mountain ash and other rowans. Richmond, Surrey: Royal Botanic Gardens, Kew; 2005. [Google Scholar]
- Muniyamma M, Phipps JB. Studies in Crataegus L. (Rosaceae). I. Cytological proof of apomixis in Crataegus L. American Journal of Botany. 1979a;66:149–155. [Google Scholar]
- Muniyamma M, Phipps JB. Studies in Crataegus L. (Rosaceae). II. Studies of pollen meiosis and polyploidy with relation to Crataegus systematics in Ontario. Canadian Journal of Genetics and Cytology. 1979b;21:231–241. [Google Scholar]
- Müller K. SeqState – primer design and sequence statistics for phylogenetic DNA data sets. Applied Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nelson-Jones EB, Briggs D, Smith AG. The origin of intermediate species of the genus Sorbus. Theoretical and Applied Genetics. 2002;105:953–963. doi: 10.1007/s00122-002-0957-6. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G, Rossello JA. Better the devil you know? Guidelines for insightful utilization of nrDNA ITS in species-level evolutionary studies in plants. Molecular Phylogenetics and Evolution. 2007;44:911–919. doi: 10.1016/j.ympev.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Nieto Feliner GN, Larena BG, Aguilar JF. Fine scale geographic structure, intra-accession polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae) Annals of Botany. 2004;93:189–200. doi: 10.1093/aob/mch027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2.3 program. Distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- Palmer EJ. The Crataegus problem. Journal of the Arnold Arboretum. 1932;13:342–362. [Google Scholar]
- Pellicer J, Clermont S, Houston L, Rich TCG, Fay MF. Cytotype diversity in the Sorbus complex (Rosaceae) in Britain: sorting out the puzzle. Annals of Botany. 2012;110:1185–1193. doi: 10.1093/aob/mcs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps JB. Introduction to the red-fruited hawthorns (Crataegus, Rosaceae) of western North America. Canadian Journal of Botany. 1998;76:1863–1899. [Google Scholar]
- Phipps JB. The relationships of the American black-fruited hawthorns Crataegus erythropoda, C. rivularis, C. saligna, and C. brachyacantha to C. Ser. Douglasianae (Rosaceae) SIDA. 1999;18:647–660. [Google Scholar]
- Phipps JB. A review of hybridization in North American hawthorns – another look at ‘the Crataegus problem.’. Annals of the Missouri Botanical Garden. 2005;92:113–126. [Google Scholar]
- Phipps JB. Crataegus tenuior (Rosaceae) – an intriguing new species from the Okanagan of British Columbia and Washington and a new variety of C. Okanaganensis. Journal of the Botanical Research Institute of Texas. 2013;7:275–297. [Google Scholar]
- Phipps JB, O'Kennon RJ. Three new species of Crataegus (Rosaceae) from western North America: C. okennonii, C. okanagenensis, and C. phippsii. SIDA. 1998;18:169–191. [Google Scholar]
- Phipps JB, O'Kennon RJ. New taxa of Crataegus (Rosaceae) from the northern Okanagan–southwestern Shuswap diversity center. SIDA. 2002;20:115–144. [Google Scholar]
- Phipps JB, O'Kennon RJ. Hawthorns (Crataegus: Rosaceae) of the Cypress Hills, Alberta and Saskatchewan. Journal of the Botanical Research Institute of Texas. 2007;1:1031–1090. [Google Scholar]
- Phipps JB, Robertson KR, Smith PG, Rohrer JR. A checklist of the subfamily Maloideae (Rosaceae) Canadian Journal of Botany. 1990;68:2209–2269. [Google Scholar]
- Qiu X, Wu L, Huang H, et al. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Applied and Environmental Microbiology. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. 2007. Tracer v1.4. Available from http://tree.bio.ed.ac.uk/software/tracer/ .
- Ramsey J, Schemske DW. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology. 1998;29:467–501. [Google Scholar]
- Rauscher JT, Doyle JJ, Brown HD. Internal transcribed spacer repeat-specific primers and the analysis of hybridization in the Glycine tomentella (Leguminosae) polyploid complex. Molecular Ecology. 2002;11:2691–2702. doi: 10.1046/j.1365-294x.2002.01640.x. [DOI] [PubMed] [Google Scholar]
- Rickett HW. Forms of Crataegus pruinosa. Botanical Gazette (Crawfordsville) 1936;97:780–793. [Google Scholar]
- Robertson A, Rich TCG, Allen AM, et al. Hybridization and polyploidy as drivers of continuing evolution and speciation in Sorbus. Molecular Ecology. 2010;19:1675–1690. doi: 10.1111/j.1365-294X.2010.04585.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ronquist F, van de Mark P, Huelsenbeck JP. Bayesian phylogenetic analyses using MrBayes. In: Lemey P, Salemi M, Vandamme AM, editors. The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing. Cambridge: University Cambridge Press; 2010. pp. 2010–2066. [Google Scholar]
- Rudnóy S, Páldi E, Bratek Z, Lásztity D, Rácz I. Detection of genome-specific ribosomal DNA sequences from bread wheat by a modified PCR-based method. Journal of Applied Botany and Food Quality. 2011;84:200–206. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Smith DB, McAllister J, Casino C, Simmonds P. Virus ‘quasispecies’: making a mountain out of a molehill? Journal of General Virology. 1997;78:1511–1519. doi: 10.1099/0022-1317-78-7-1511. [DOI] [PubMed] [Google Scholar]
- Stefanović S, Kuzmina M, Costea M. Delimitation of major lineages within Cuscuta subgenus Grammica (Convolvulaceae) using plastid and nuclear DNA sequences. American Journal of Botany. 2007;94:568–589. doi: 10.3732/ajb.94.4.568. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), Verson 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Talent N. Evolution of gametophytic apomixis in flowering plants: an alternative model from Maloid Rosaceae. Theory in Biosciences. 2009;128:121–138. doi: 10.1007/s12064-009-0061-4. [DOI] [PubMed] [Google Scholar]
- Talent N, Dickinson TA. Polyploidy in Crataegus and Mespilus (Rosaceae, Maloideae): evolutionary inferences from flow cytometry of nuclear DNA amounts. Botany. 2005;83:1268–1304. [Google Scholar]
- Talent N, Dickinson TA. Apomixis and hybridization in Rosaceae subtribe Pyrineae Dumort.: a new tool promises new insights. In: Grossniklaus U, Horandl E, Sharbel T, van Dijk P, editors. Apomixis: evolution, mechanisms and perspectives. Liechtenstein: Verlag Ruggell; 2007a. pp. 301–316. [Google Scholar]
- Talent N, Dickinson TA. Endosperm formation in aposporous Crataegus L. (Rosaceae, Spiraeoideae, tribe Pyreae): parallels to Ranunculaceae and Poaceae. New Phytologist. 2007b;173:231–249. doi: 10.1111/j.1469-8137.2006.01918.x. [DOI] [PubMed] [Google Scholar]
- Talent N, Dickinson TA. The potential for ploidy level increases and decreases in Crataegus L. (Rosaceae, Spiraeoideae, tribe Pyreae) Botany. 2007c;85:570–584. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura Y, Yoshimura K, Tomaru N, Ohba K. Molecular phylogeny of conifers using RFLP analysis of PCR-amplified specific chloroplast genes. Theoretical and Applied Genetics. 1995;91:1222–1236. doi: 10.1007/BF00220933. [DOI] [PubMed] [Google Scholar]
- Ufimov RA. Notes on the genus Crataegus L. (Rosaceae) Novosti sistematiki vysshikh rastenii˘. 2013;44:113–125. [Google Scholar]
- Vaillancourt RE, Jackson HD. A chloroplast DNA hypervariable region in eucalypts. Theoretical and Applied Genetics. 2000;101:473–477. [Google Scholar]
- Wells TC, Phipps JB. Studies in Crataegus (Rosaceae: Maloideae). XX. Interserial hybridization between Crataegus monogyna (series Oxyacanthae) and Crataegus punctata (series Punctatae) in southern Ontario. Botany. 1989;67:2465–2472. [Google Scholar]
- White TJ, Bruns T, Lee T, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Whitton J, Sears CJ, Baack EJ, Otto SP. The dynamic nature of apomixis in the angiosperms. International Journal of Plant Sciences. 2008;169:169–182. [Google Scholar]
- Wilkinson M. Coping with missing entries in phylogenetic inference using parsimony. Systematic Biology. 1995;44:501–514. [Google Scholar]
- Winship PR. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Research. 1989;17:1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Estimating the pattern of nucleotide substitution. Journal of Molecular Evolution. 1994;39:105–111. doi: 10.1007/BF00178256. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Molecular Biology and Evolution. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- Yao H, Song J, Liu C, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010;5:e13102. doi: 10.1371/journal.pone.0013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, Wilkin P, Fay MF, Ingrouille MJ, Zarre S, Chase MW. Molecular systematics of Gagea and Lloydia (Liliaceae; Liliales): implications of analyses of nuclear ribosomal and plastid sequences for infrageneric classification. Annals of Botany. 2009;104:125–142. doi: 10.1093/aob/mcp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, Wilkin P, Ingrouille JM, et al. Speciation and evolution in the Gagea reticulata species complex (Tulipeae; Liliaceae) Molecular Phylogenetics and Evolution. 2012;62:624–639. doi: 10.1016/j.ympev.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Sodmergen Why does biparental plastid inheritance revive in angiosperms? Journal of Plant Research. 2010;123:201–206. doi: 10.1007/s10265-009-0291-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.