Abstract
Background and Aims
Both regional and local plant abundances are driven by species' dispersal capacities and their abilities to exploit new habitats and persist there. These processes are affected by clonal growth, which is difficult to evaluate and compare across large numbers of species. This study assessed the influence of clonal reproduction on local and regional abundances of a large set of species and compared the predictive power of morphologically defined traits of clonal growth with data on actual clonal growth from a botanical garden. The role of clonal growth was compared with the effects of seed reproduction, habitat requirements and growth, proxied both by LHS (leaf–height–seed) traits and by actual performance in the botanical garden.
Methods
Morphological parameters of clonal growth, actual clonal reproduction in the garden and LHS traits (leaf-specific area – height – seed mass) were used as predictors of species abundance, both regional (number of species records in the Czech Republic) and local (mean species cover in vegetation records) for 836 perennial herbaceous species. Species differences in habitat requirements were accounted for by classifying the dataset by habitat type and also by using Ellenberg indicator values as covariates.
Key Results
After habitat differences were accounted for, clonal growth parameters explained an important part of variation in species abundance, both at regional and at local levels. At both levels, both greater vegetative growth in cultivation and greater lateral expansion trait values were correlated with higher abundance. Seed reproduction had weaker effects, being positive at the regional level and negative at the local level.
Conclusions
Morphologically defined traits are predictive of species abundance, and it is concluded that simultaneous investigation of several such traits can help develop hypotheses on specific processes (e.g. avoidance of self-competition, support of offspring) potentially underlying clonal growth effects on abundance. Garden performance parameters provide a practical approach to assessing the roles of clonal growth morphological traits (and LHS traits) for large sets of species.
Keywords: Clonal plant growth, species abundance, botanical garden collections, LHS traits, leaf-specific area, plant height, seed mass, lateral expansion, seed reproduction, Ellenberg indicator values
INTRODUCTION
For more than a decade, identifying the determinants of plant species abundance has been recognized as a major challenge for ecology (May, 1999; McGill, 2006; Comita et al., 2010; Gaston, 2011). Indeed, it has been the subject of extensive investigation (e.g. Murray et al., 2002; Espeland and Emam, 2011). This has been stimulated by the need to discern causes of species rarity, both for conceptual and for practical/conservation purposes.
Abundance of a species is constrained both by the extent of habitats that match its niche requirements, and by its ability to reach and exploit these habitats (Boulangeat et al., 2012b). The extent and spatial arrangement of these potential habitats depend on the particular region under study and provide the stage on which the population biology processes (dispersal, establishment and persistence) play out (Hanski, 1982; Ozinga et al., 2005; Boulangeat et al., 2012b). Indeed, in heterogeneous landscapes, habitat requirements are often the best predictor of species' regional abundance (Ozinga et al., 2005; Soons and Ozinga, 2005; Kolb et al., 2006; Moore and Elmendorf, 2006; Lososová et al., 2008; Knapp et al., 2009; Boulangeat et al., 2012a, b).
Within the constraints of the landscape structure, a species' abundance results from its dispersal capacity and its ability to exploit new habitat and persist there. However, identification of the particular drivers that determine relative abundances among species of similar habitat requirements has been difficult (Duralia and Reader, 1993; Craine et al., 2001; McGill, 2006; Öster et al., 2009). Nevertheless, reproductive traits (Kunin and Shmida, 1997) and dispersal capacity are known to be important for colonization of new habitats and have often been identified as predictors of species abundance at the regional level (Soons and Ozinga, 2005; van der Veken et al., 2007; Römermann et al., 2008; Ozinga et al., 2009; Saar et al., 2012; but see e.g. Öster et al., 2009). It is more difficult to generalize about processes determining species persistence at any given site, as they are likely to be more habitat-specific than is dispersal (Craine et al., 2001; Murray et al., 2002; Espeland and Emam, 2011).
The attempt to find abundance predictors across a large number of species is necessarily limited by the availability of comparative data, which in practice are restricted to readily quantified traits (‘soft’ or ‘easy’ traits, Weiher et al., 1999; Lavorel and Garnier, 2002) that are assumed to approximate important field processes and for which data collection is relatively easy (see e.g. Westoby, 1998). However, the soft traits that have traditionally been used in examinations of trait–abundance relationships have not included any that represent the capacity for lateral spreading by clonal growth. This capacity is a key attribute for many perennial herbaceous plants (Klimeš and Klimešová, 1999; Klimešová and de Bello, 2009), and thus these analyses have omitted an important aspect of life history. Indeed, clonal growth clearly plays roles both in short-distance spread and in persistence within habitats (Benot et al., 2010), while also contributing to dispersal; however, we lack adequate information on these roles for many species.
Identification of a set of easily measurable traits that describe clonal growth is essential to discerning its effects on these processes, and, ultimately, on species abundance. However, identifying such traits is especially challenging because clonal growth is highly environmentally dependent (Sammul, 2011) and cannot easily be captured in a few parameters. Nevertheless, considerable data have been amassed characterizing clonal growth morphology in terms of particular parameters (Klimeš and Klimešová, 1999; Tamm et al., 2001; Klimešová and de Bello, 2009). The necessary next step is to use such comparative data sets to examine the ecological role of these parameters across many species and different habitats (for similar approaches, see e.g. Klimeš and Klimešová, 2000; Gough et al., 2012). In addition, we need to examine to what extent the morphological parameters of clonal growth are good approximations of actual clonal growth of the plant in the field. Indeed, the relatively straightforward examination of the community effects of seed dispersal and growth/competition has revealed the limitations of soft traits, with comparative studies showing that population biology processes, and not necessarily easily measurable traits, determine species' performance and success (Lavergne et al., 2004; Münzbergová, 2005; Moora and Jõgar, 2006; Miller et al., 2007; Matesanz et al., 2009). Thus, one cannot assume a priori that easily measurable clonal growth traits would have predictive power for species abundance, especially in the case of morphologically defined traits, which are not necessarily reliable predictors of actual clonal growth in the field (Klimeš, 1999).
In this study, we had two goals. First, we aimed to assess, for a large set of species, the effects of clonal growth on species abundance in comparison with those of seed reproduction, growth-related traits (specific leaf area and plant height) and habitat requirements. Because the definition of abundance is scale-dependent (Kunin et al., 2000; Hartley et al., 2004), and different drivers can operate at different levels (Murray et al., 2002, 2005; Hartley et al., 2004; Cornwell and Ackerly, 2010; Mokany and Roxburgh, 2010), we examined the effects of these potential predictors both at a regional scale (number of records of the species in the Czech Republic) and at a local scale (mean species cover in individual vegetation records in the Czech National Phytosociological Database).
Second, we aimed to compare the ability to predict abundance based on morphologically defined clonal growth traits versus that based on data recorded on actual vegetative growth performance. Because collecting comparable vegetative reproduction data for a large set of species would be a daunting task, we used data from existing growth records of plants in a botanical garden. We relied on the assumptions that: (1) species in the garden are maintained in conditions reasonably close to their natural habitats; and (2) therefore, their garden growth and reproduction provide approximations of their potential vegetative and seed reproduction in favourable (non-competitive) conditions. This approach provides a standardized way to assess directly, across many species, potential reproduction, rather than just measuring morphological traits that may or may not influence reproductive potential. Although there can be a number of issues regarding data from these records (e.g. reproduction assessments not necessarily fully quantitative, small sample sizes, weeding eliminating constraints imposed by interspecific competition, potentially non-representative genetic composition of cultivated populations), these disadvantages are easily outweighed by the large number of species that can be compared (for further discussion of caveats in using such data see Herben et al., 2012a, 2013).
We used two different approaches to treat habitat effects on species abundance. First, we used Ellenberg indicator values as covariates. Although this does not address differences in niche widths among species, it removes the other major source of environmental effects, namely differences in the spatial extent of suitable habitat among species in the region. Second, we also analysed the data based on classification of the vegetation records according to seven major habitat types.
METHODS
Trait and other database data for species
We used four traits from the CLO-PLA3 database (Klimešová and de Bello, 2009): shoot lifespan (1 or 1+ years, available for 831 species, and called cyclicity by Klimešová and de Bello, 2009); persistence of connections between shoots (1, 2, >2 years; 834 species); multiplication rate (number of offspring shoots per mother shoot per year: <1, 1, >1; 834 species); and lateral spread (distance from the mother shoot at which offspring shoots are formed (<0·01, 0·01–0·25, >0·25 m; 834 species). For non-clonal species, persistence, multiplication rate and lateral spread were assigned the value of zero. We refer to these four traits as CLOPLA traits or morphological parameters. Data on seed mass (available for 593 species), plant height (850 species) and specific leaf area (SLA; 603 species) were taken from the LEDA traitbase (Kleyer et al., 2008). If several records were available for a species, the simple (unweighted) arithmetic mean value was used. Plant height data missing from LEDA for a species were calculated as the mean of the range values given by Kubát et al. (2002). Following Westoby (1998), we further refer to these traits as to LHS (leaf–height–seed) traits.
Ellenberg indicator values (EIVs) were taken from the BiolFlor database (Klotz et al., 2002). Species with a value of x for the particular EIV (i.e. no specific response to the given factor) were assigned the means of the endpoint values of the EIV's entire possible range (i.e. 4·5, 5 or 5·5 depending on the range).
Phylogenetic data were obtained from Durka (2002), with dated branch lengths updated by Stefan Michalski (Helmholtz Centre for Environmental Research-UFZ Halle, Germany, unpubl. data). If no data were available for a given species in Durka (2002), data on congeneric species were taken from it; species for which data on no congener were available in the phylogenetic dataset were excluded from the phylogenetic analysis. This approach yielded independent phylogenetic information for 628 species; including species for which congener data were used yielded a total of 752 species for which we had phylogenetic information.
Vegetative and seed reproduction
Data on plant performance came from the collection of native plants of the Central European Flora, the Botanical Garden of the Faculty of Science, Charles University in Prague (http://www.bz-uk.cz; see also Herben et al., 2012a). This collection houses about 1200 Central European plant species, collected mainly from the Czech Republic and Slovakia. The garden's habitats range from open, dry, sandy habitats and limestone, rocky habitats through mesic, open habitats and shaded forest stands to moist (shaded and unshaded) habitats. Plants are grown in open soil and are subject to weeding to keep stands of individual species separate. Each species is maintained in habitat and conditions that, based on field knowledge, match as closely as possible those in which it typically occurs; however, conditions in which the plants grow cannot fully match their field conditions owing to weeding, thinning and occasional irrigation. For all native, non-woody, perennial plant species that have been growing in the garden for at least for 10 years (836 species; for the list of species see Herben et al., 2012a), we scored the reproduction for the last 10 years of cultivation. Seed and vegetative reproduction were scored separately using the same five-degree ordinal scale (for further details see Herben et al., 2012a). In most cases, seedlings could be distinguished from vegetative offspring; however, for some plants with vigorous vegetative reproduction, assessment of seed reproduction was impossible because individuals that probably were seedlings were mixed with vegetative progeny. For these plants (43 species), seed reproduction was treated as a missing value. We further refer to these scores as seed reproduction and vegetative reproduction (together referred to as garden performance parameters). We used all perennial species for which reproduction data from the garden were available in addition to the LHS and CLOPLA data. Annuals were left out because they are underrepresented in the garden, rarely show clonal growth and their drivers of abundance differ from those of perennials (Herben et al., 2012b).
Regional and local abundance data
Two sources of regional abundance data were used. First, we obtained the number of each species' records from a stratified subset of the Czech National Phytosociological Database (Chytrý and Rafajová, 2003) containing 20 468 plots sampled after 1970 (see Chytrý et al., 2005 for the stratification procedure). Second, we also obtained the number of each species' records from the floristic database of the Institute of Botany, Academy of Sciences of the Czech Republic (FLDOK; see http://www.ibot.cas.cz/index.php?p=databazeandsite=default). These two measures of regional abundance were subject to principal components analysis after a log(x + 1) transformation. The first axis in this analysis (accounting for 93·6 % of the total variance) was used as the compound measure of regional abundance and is referred to hereafter as regional abundance. We also used the set of 20 468 plots (described above) from the Czech National Phytosociological Database to determine the local abundance of each species. This was assessed using mean cover values calculated over all database plots in which the species was present. In contrast to regional abundance, which is based on non-replicated data, these values are estimated using a large number of replicates for each species (although not independent across species).
We also used the Czech National Phytosociological Database to assess species' regional and local abundances classified by habitat type. Plots in the stratified set (see above) were assigned to seven major EUNIS habitat types (see Chytrý et al., 2005): grasslands (6702 plots); forests (3391 plots); scrub (354 plots); water habitats (4173 plots); rocky habitats (286 plots); peatlands and mires (531 plots); and synanthropic habitats (5030 plots). These are further referred to as habitat types. Plots not assigned to any habitat type by Chytrý et al. (2005) were excluded from all habitat-based analyses. Within each habitat type, we calculated the regional abundance of each species as the natural logarithm of the number of database plots in the given habitat type in which the species occurred (with a given species potentially included in the datasets of several habitat types). To avoid trivial cases of absence or low abundance of species that could not occur appreciably in a habitat type, we excluded all species that had less than 1 % of all their occurrences in that habitat. Local abundance was calculated for each habitat type in which a species occurred as the mean of the species' cover in all the database plots in which it occurred in that habitat.
Data analysis
Two different approaches were used to identify relationships between abundance and its potential predictors. In the first, we examined individual predictors separately by using the Akaike information criterion (AIC) to build the best regression model for the given variable. In the second, we examined the effects of whole predictor groups.
For the model-building procedure, we first assessed the relationships of regional and local abundance to species' ecological preferences by building linear models with EIVs as the only predictors. We used untransformed EIVs as well as their squares as predictors, because relationships between some EIVs and both local and regional abundances were markedly non-linear. We chose the best-fitting model by using the AIC in a bidirectional stepwise procedure implemented through the ‘step’ function in R ver. 2·15·1 (R Development Core Team, 2012). We built models both for the whole dataset and for the individual habitat types (for the final models, see Table 1).
Table 1.
Best models (with lowest AIC) of regional and local abundance in relationship to Ellenberg indicator values
| Regional abundance |
Local abundance |
Correlation of local and regional abundance |
|||||
|---|---|---|---|---|---|---|---|
| Habitat type | Model | Residual d.f. | Model | Residual d.f. | R | n | P |
| Whole dataset | Temp + Temp2 + pH2 + Moisture + Moisture2 + N + N2 | 614 | Light2 + N + pH + pH2 + Temp2 | 580 | –0·063 | 673 | 0·103 |
| Water habitats | pH2 + Moisture + N + N2 + Cont + Cont2 | 193 | Moisture + Moisture2 + pH2 + Temp2 | 195 | 0·102 | 214 | 0·137 |
| Mire habitats | pH + pH2 | 118 | Moisture2 | 119 | 0·181 | 143 | 0·029 |
| Grassland habitats | Light + Light2 + Temp + Temp2 + Moisture + N + N2 + pH2 | 504 | N + Moisture2 + Cont + Cont2 + Light + Temp | 505 | 0·17 | 585 | 0 |
| Rocky habitats | pH + pH 2 | 95 | Light2 + Temp + Temp2 | 94 | –0·047 | 99 | 0·641 |
| Scrub habitats | pH + Moisture2 + Temp + Temp2 | 209 | Moisture2 | 212 | 0·178 | 266 | 0·003 |
| Forest habitats | Light + pH2 + Temp + Temp2 + Moisture2 + Cont + Cont2 | 415 | Light2 + pH + pH2 + Moisture2 + Temp + Temp2 | 415 | 0·152 | 493 | 0·001 |
| Synanthropic habitats | pH2 + Temp + Temp2 + N + N2 + Moisture2 + Light + Light2 | 504 | Moisture2 | 511 | 0·027 | 566 | 0·52 |
Temp, temperature; Cont, continentality. 2 indicates second power of the given Ellenberg value. Significant values of Pearson correlations are in bold.
We also built linear models of regional and local abundance using two different sets of predictors: (1) garden performance parameters only, and (2) garden performance parameters along with CLOPLA and LHS traits. Using different predictor sets allowed us to maximize the number of cases (species) available for the analysis, as particular species were missing values for some potential predictors, but not for others. In all analyses, we used only cases which had complete data for all candidate predictor variables. All LHS traits were log-transformed before analysis. Each of these predictor sets was used for model building both with and without using EIVs (shown in Table 1) as covariates. The best models were selected by implementing a stepwise procedure using the AIC; for the final models, the significance of each individual term was checked by using the F-statistic to compare the models with and without the term included. Identical analyses were done across the set of all plots and separately within each of the seven EUNIS habitat types. The latter analyses considered only species that had more than 1 % of all their occurrences in the given habitat type.
In the second approach, we assessed the relative effects of different groups of predictors by comparing the R2 values of models containing all the predictors from each of the given groups. We grouped the predictors in different ways to answer different questions. To examine the roles of particular life-history components, we divided the predictors into the following groups: (1) seed reproduction in the garden and seed size (seed reproduction predictors), (2) vegetative reproduction in the garden and CLOPLA traits (clonal growth predictors), (3) height and SLA, and (4) EIVs. To assess the relative effects of traits (LHS and morphological traits) vs. performance, we used the following groupings: (1) seed and vegetative reproduction in the garden (performance predictors); (2) all clonal growth traits, including shoot life span (CLOPLA predictors); (3) height, seed mass and SLA (LHS predictors); (4) CLOPLA and LHS predictors combined; and (5) EIVs. The overall effect of each group was assessed in terms of the R2 of the model including all predictors from that group without any other predictors; the net effect of each group was assessed as the difference between the R2 values of the full model (containing all predictors) and the model containing all groups except the group being tested. Adjusted R2 was used as a safeguard against possible model overparameterization. All calculations were done in R ver. 2·15·1 (R Development Core Team, 2012).
All further analyses were based on the set of all perennial species (herbaceous and dwarf shrubs; life span data taken from Kubát et al., 2002) native to the Czech Republic. To correct for phylogenetic relatedness in the regressions we used the approach of (Diniz-Filho et al., 1998; see also Desdevises et al., 2003), which is well suited to phylogenetic trees involving polytomies. We calculated phylogenetic distances contained in the source tree (Durka, 2002) by using the ‘cophenetic.phylo’ function from R package ‘ape’ (Paradis et al., 2012). The matrix of phylogenetic distances was summarized by non-standardized principal coordinates analysis (PCoA) using the ‘ade4’ package for R (Dray and Dufour, 2007). Scores along the first 17 PCoA axes (accounting for 90·0 % of the total phylogenetic variation) were used as covariates in the regressions to capture phylogenetic relatedness of the taxa. In all analyses that employed phylogenetic information, species with no phylogenetic information were treated as missing.
RESULTS
Local and regional abundances were uncorrelated at the level of the whole dataset (Table 1). In contrast, at the level of the EUNIS habitat types, local and regional abundances were (positively) correlated in grasslands, scrub and (weakly) mire habitats, and showed no correlation in water, rocky and synanthropic habitats.
When comparing across the whole dataset, the effect of EIV was very strong on regional abundance and much weaker on local abundance (Table 2). Within habitat types, its effect on regional abundance was generally large; it was smaller at rocky and synanthropic habitats. Its effect on local abundance was high in some habitat types (water, rocky, and partly forest and scrub) and rather small in mire, grassland and synanthropic habitats (Table 3).
Table 2.
Best models of regional and local abundance for the whole dataset
| Potential predictors | Regional abundance |
Local abundance |
||||||
|---|---|---|---|---|---|---|---|---|
| Garden performance parameters | Garden performance parameters and traits | Garden performance parameters | Garden performance parameters and traits | Garden performance parameters | Garden performance parameters and traits | Garden performance parameters | Garden performance parameters and traits | |
| Covariate | None | None | EIV | EIV | None | None | EIV | EIV |
| R2 due to covariates | 0 | 0 | 0·2976 | 0·2609 | 0 | 0 | 0·0277 | 0·0306 |
| R2 of the model | 0·02439 | 0·06113 | 0·3041 | 0·284 | 0·05809 | 0·1206 | 0·09249 | 0·158 |
| Adjusted R2 of the model | 0·0218 | 0·05433 | 0·2952 | 0·2677 | 0·05528 | 0·1117 | 0·08172 | 0·1336 |
| Residual d.f. | 753 | 414 | 626 | 394 | 672 | 393 | 590 | 379 |
| Seed reproduction | 0·0156(+)*** | 0·0162(+)** | 0·008(–)* | 0·0145(–)* | 0·0166(–)** | 0·015(–)* | ||
| Vegetative reproduction | 0·0146(+)*** | 0·0093(+)* | 0·0381(+)*** | 0·0436(+)*** | 0·0337(+)*** | 0·0303(+)*** | ||
| Shoot lifespan | NA | NA | NA | 0·0048(+) + | NA | 0·0063(+)+ | ||
| Lateral spread | NA | 0·0507(+)*** | NA | 0·026(+)** | NA | NA | ||
| Persistence | NA | NA | NA | NA | 0·008(+)+ | |||
| Multiplication | NA | NA | NA | NA | ||||
| Seed mass | NA | NA | NA | NA | 0·0068(+)+ | |||
| SLA | NA | NA | NA | 0·0169(–)** | NA | 0·0146(–)* | ||
| Height | NA | 0·0048(+)+ | NA | 0·0065(+)+ | NA | NA | ||
Partial R2 values due to the individual terms are shown. Symbols in parentheses indicate the sign of the relationships. ***P < 0·001, **P < 0·01, *P < 0·05, +P < 0·1. Empty cells indicate terms not included in the best model; NA indicates terms not available for use in the given analysis. Ellenberg indicator values (EIVs) used as covariates in particular models are listed in Table1.
Table 3.
Best models of regional and local abundance for individual habitat types
| Water habitats |
Mires |
Grasslands |
Rocky habitats |
Scrub |
Forests |
Synanthropic |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Covariate | None | EIV | None | EIV | None | EIV | None | EIV | None | EIV | None | EIV | None | EIV |
| Regional abundance | ||||||||||||||
| R2 due to covariates | 0 | 0·3031 | 0 | 0·2457 | 0 | 0·2978 | 0 | 0·1985 | 0 | 0·2241 | 0 | 0·4179 | 0 | 0·2205 |
| R2 of the model | 0·1412 | 0·3614 | 0·07447 | 0·3002 | 0·05968 | 0·3345 | 0·2092 | 0·313 | 0·1416 | 0·2835 | 0·1918 | 0·4772 | 0·104 | 0·2812 |
| Adjusted R2of the model | 0·1189 | 0·3212 | 0·05614 | 0·2638 | 0·05162 | 0·3108 | 0·1683 | 0·2506 | 0·1241 | 0·2479 | 0·1839 | 0·4541 | 0·09164 | 0·2564 |
| Residual d.f. | 154 | 143 | 101 | 96 | 350 | 336 | 58 | 55 | 147 | 141 | 309 | 294 | 361 | 347 |
| Seed reproduction | 0·0573(+)* | 0·0391(+)* | 0·0312(+) + | 0·0369(+) + | 0·0066(+) + | |||||||||
| Vegetative reproduction | 0·0143(+)* | 0·0078(+) + | 0·0113(+)* | 0·01(+) + | ||||||||||
| Shoot lifespan | 0·0214(+)** | 0·0224(+)** | 0·0066(+) + | |||||||||||
| Lateral spread | 0·0586(+)** | 0·0261(+)* | 0·0292(+) + | 0·0203(+) + | 0·0202(+)** | 0·0252(+)** | 0·0261(+)** | 0·0244(+)** | 0·0147(+)* | |||||
| Persistence | 0·0464(+)*** | |||||||||||||
| Multiplication | 0·1143(+)** | 0·0669(+)* | 0·0534(+)** | 0·0326(+)* | ||||||||||
| Seed mass | 0·0397(–)** | 0·0337(–)* | 0·021(–) + | 0·0182(–)** | 0·0067(–) + | 0·0471(–) + | 0·0352(–) + | 0·0389(–)* | 0·0198(–) + | 0·0111(–) + | 0·0145(–)* | 0·0076(–) + | ||
| SLA | 0·0202(+) + | 0·1067(+)*** | 0·0263(+)** | |||||||||||
| Height | 0·0232(+)* | 0·0264(+)* | 0·0125(+)* | 0·0234(–)* | 0·0134(–) + | 0·0191(+)* | 0·0255(+)** | 0·0284(+)** | ||||||
| Local abundance | ||||||||||||||
| R2 due to covariates | 0·3406 | 0·0737 | 0·0651 | 0·2532 | 0·1129 | 0·1308 | 0·0167 | |||||||
| R2 of the model | 0·1496 | 0·4231 | 0·07419 | 0·1383 | 0·08634 | 0·1269 | 0·05241 | 0·3393 | 0·05601 | 0·1578 | 0·09394 | 0·2105 | 0·04028 | 0·05648 |
| Adjusted R2 of the model | 0·1275 | 0·3952 | 0·05585 | 0·1119 | 0·07587 | 0·1011 | 0·03662 | 0·2921 | 0·04325 | 0·1403 | 0·09102 | 0·1894 | 0·03235 | 0·04315 |
| Residual d.f | 154 | 145 | 101 | 98 | 349 | 338 | 60 | 56 | 148 | 145 | 311 | 299 | 363 | 354 |
| Seed reproduction | 0·0379(–)** | 0·0396(–)* | 0·019(–) + | 0·0124(–)* | 0·0082(–) + | 0·0085(–) + | ||||||||
| Vegetative reproduction | 0·0186(+) + | 0·0511(+)** | 0·0284(+)* | 0·0231(+) + | 0·0939(+)*** | 0·0601(+)*** | 0·0181(+)** | 0·0089(+) + | ||||||
| Shoot lifespan | 0·0231(+)* | 0·0364(+)* | 0·013(–)* | |||||||||||
| Lateral spread | ||||||||||||||
| Persistence | 0·0298(+) + | 0·0449(+)* | 0·0216(+)** | 0·0254(+)** | ||||||||||
| Multiplication | 0·0524(+) + | 0·1152(+)** | ||||||||||||
| Seed mass | 0·0365(+)* | 0·0354(+)* | 0·0054(+) + | |||||||||||
| SLA | 0·0156(–) + | 0·025(–)* | 0·0247(–)** | 0·0136(–)* | 0·0086(–) + | 0·0173(–)* | ||||||||
| Height | 0·0165(–)* | 0·0136(–)* | 0·0149(+)* | |||||||||||
Partial R2 values due to the individual terms are shown. Symbols in parentheses indicate the signs of the relationships. ***P < 0·001, **P < 0·01, *P < 0·05, +P < 0·1. Empty cells indicate terms not included in the best model. Ellenberg indicator values (EIVs) used as covariates for the individual habitat types are listed in Table 1.
Considering the dataset as a whole, regional abundance was predicted, apart from the above-mentioned effect of EIV, by seed reproduction in the garden and by CLOPLA lateral spread values (Table 2). In cases where CLOPLA traits were not available for selection, both seed and vegetative reproduction positively affected regional abundance. The effect of seed reproduction disappeared when EIVs were used as covariates. Within habitat types, seed reproduction in the garden was a predictor of regional abundance in mire and rocky habitats, with its effect remaining stable when EIVs were used as covariates (Table 3). Vegetative reproduction in the garden was a weak predictor of regional abundance in forests and synanthropic habitats. In almost all habitat types, lateral spread or clonal multiplication were predictors of regional abundance (Table 3). Seed mass was a predictor (negative effect) on regional abundance in a number of habitat types – often together with seed reproduction (positive effect).
Local abundance was consistently predicted by vegetative reproduction in the garden, both across the whole dataset and within individual habitat types (Tables 2 and 3). Of other predictors, only SLA contributed more than 1 % to the variance (negative effect) in local abundance when considering the entire dataset. The effect of seed reproduction in the garden, although weak, was significant and negative. Within habitat types, local abundance was predicted also by seed reproduction (negative effect, namely in water and mire habitats). Additionally, local abundance was predicted by either persistence (mires, grasslands) or multiplication (rocky habitats). SLA was a negative predictor for water, grassland and synanthropic habitats. Predictors of both regional and local abundance across the whole dataset remained qualitatively unchanged when examined with phylogenetic signal removed (results not shown).
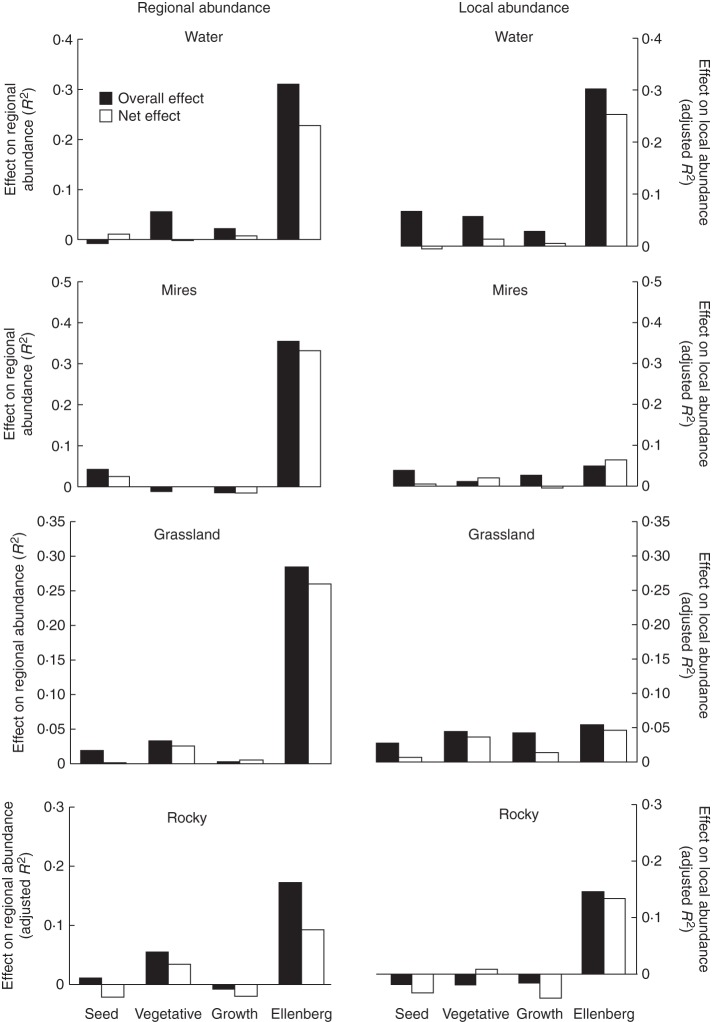
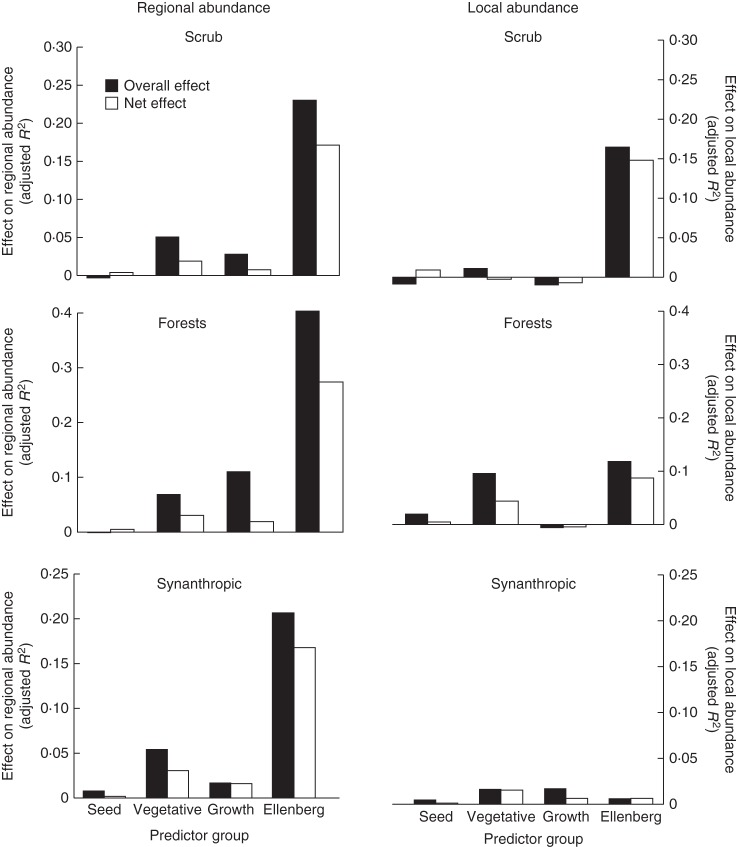
Analysis of entire predictor groups showed a similar pattern: EIVs were the dominant source of variation in regional abundance when considering the dataset as a whole, whereas garden and LHS predictors explained much less variation (Fig. 1). The biggest contribution was from clonal growth, in terms of both its overall and its net effects (Fig. 2). A similar pattern was found within habitat types, with EIVs playing the largest role, followed by vegetative reproduction. For regional abundance in mires, seed reproduction was more important than clonal growth, whereas the opposite was true of regional abundance in forests (Fig. 3).
Fig. 1.
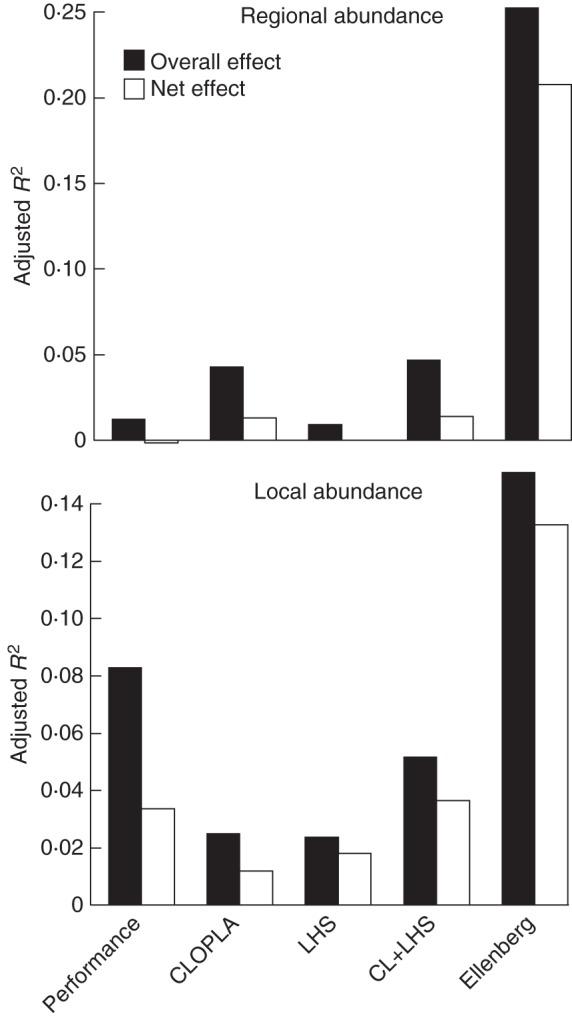
Effects of individual sets of predictors of species abundance, showing total (overall) effect of the predictor set and the net effect after effects of all other predictor sets have been partialled out (as indicated in the key). Note that the effects of seed reproduction on local abundance have a negative sign (see Table 2).
Fig. 2.
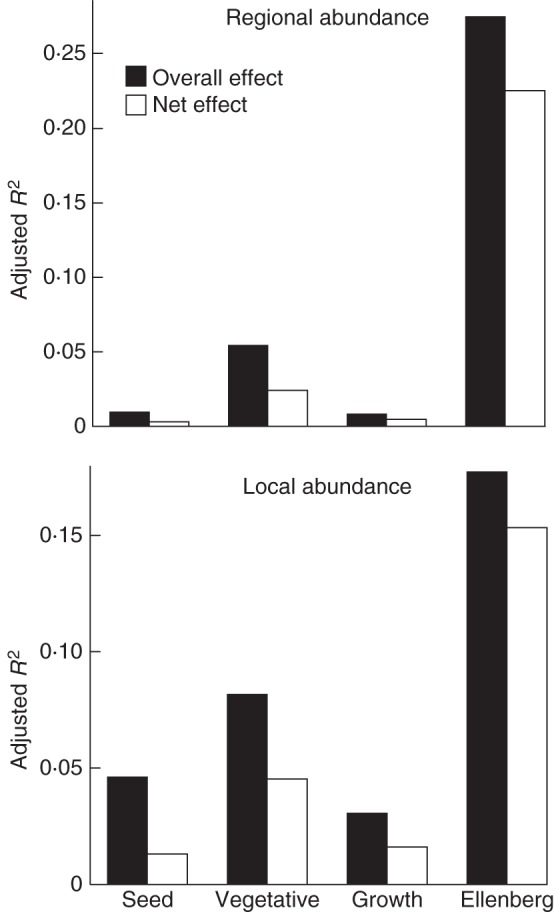
Effects of individual aspects of species life history on species abundance, showing total (overall) effect of the predictor set and the net effect after effects of all other predictor sets have been partialled out (as indicated in the key). Note that the effects of seed reproduction on local abundance have a negative sign (see Table 2). Life-history aspects are indicated as follows (see Methods): seed – seed size and garden seed reproduction; vegetative – garden vegetative reproduction and morphological traits from CLOPLA; growth – SLA and height; Ellenberg – Ellenberg indicator values.
Fig. 3.
Effects of individual components of life history on species abundance in individual habitat types, showing total (overall) effect of the predictor set and the net effect after effects of all other predictor sets have been partialled out. Life-history aspects are indicated as follows (see Methods): seed – seed size and garden seed reproduction; vegetative – garden vegetative reproduction and morphological traits from CLOPLA; growth – SLA and height; Ellenberg – Ellenberg indicator values.
In contrast, variation in local abundance was accounted for both by EIVs and by clonal growth predictors (Figs 1 and 2). The effect of seed reproduction parameters was negative. Within habitat types, EIVs were often important for local abundance in habitat types in which their effect on regional abundance was minor, i.e. in water, rocky and forest habitats. Clonal growth predictors, considered as a group, were important for local abundance in forest, grassland and water habitats.
DISCUSSION
Clonal growth and species abundance
While differences in habitat factors are clearly the best predictors of species abundance, clonal growth predictors (both morphological and vegetative reproduction in the garden) account for a significant amount of variation in plant abundance after differences in habitat factors are taken out. This is true both at the regional and (especially) at local levels. The amount of variation explained by clonal growth was larger than that explained by the parameters of seed reproduction used, but it must be kept in mind that these seed reproduction parameters do not incorporate explicit parameters of dispersal distance. Although both garden reproduction and seed mass are negatively correlated with dispersal distance, they provide only an indirect measure of seed reproduction, and this is largely limited to wind-dispersed species
The proximate effects of clonal growth traits (such as lateral spread) occur only at the scale of individual plants. There are number of candidate processes through which these clonal growth traits may affect local abundance, but patterns identified in the data can help us to identify those that are particularly important. Specifically, the capacity for lateral expansion and/or garden vegetative reproduction was a much better predictor of local abundance (i.e. a proxy of success of individual plants) than multiplication rate (number of offspring per plant) and persistence of connections (see also Sammul, 2011). These findings are likely to reflect the pervasive effect of self-competition in clonal plants, which may seriously limit their success in some habitats (Oborny et al., 2007), in that lateral spread reduces intragenet (and hence intraspecific) contact. If self-competition is very strong, the potential number of offspring per mother plant would be largely irrelevant, as the offspring would be unlikely to have a chance to develop (except in rocky habitats with abundant open space). The importance of self-competition in our study system is probably due to the prevailing ecological conditions experienced by Central European vegetation, where productivity is often high enough to lead to self-competition, and there are no strong selective forces against lateral spread (for discussion of a different system, see Klimešová et al., 2012). Some clonal traits were relevant only in particular habitats. Persistence of connections was favourable only in grassland habitats (see also Klimešová et al., 2011), although the explanation for this is hard to infer. It may have been due to limited nutrient availability in these habitats (Jónsdóttir and Watson, 1997; Klimešová et al., 2011), although the role of asymmetric light competition there cannot be ruled out.
The effects of clonal traits are not restricted to the local level, however, as our data show that they (especially lateral spread) are important for regional abundance as well. Several other studies have shown that traits assumed to act locally are also important for regional abundance (Klimeš and Klimešová, 2000; Ozinga et al., 2007; Tremlová and Münzbergová, 2007; Lososová et al., 2008; Kolb et al., 2006). The mechanisms of their action at the regional level resist easy elucidation. High persistence and capacity to attain dominance at the local level can translate into higher numbers of diaspores that would increase regional abundance (Hanski, 1982). This mechanism would yield a positive correlation between the abundances at measured at both scales; however, this relationship was generally weak in the studied dataset. In fact, positive effects of clonal traits on regional abundance were found even in habitat types such as synanthropic or water habitats for which there was no or negative correlation between abundance at the two levels. However, positive correlations between local and regional abundance can be absent from non-equilibrium systems driven by extinction debt, if traits enable persistence at sites. Indeed, clonal traits have been shown to extend the time to extinction (Ozinga et al., 2007; Saar et al., 2012), which, if site occupancy has an important non-equilibrium component, could explain their effect on regional abundances.
Alternatively, the regional effects of clonal traits could be due to habitat preferences not captured by EIV characterization, different abundances of habitats themselves, associated mass effects or similar processes. Vegetative reproduction has an important phylogenetic component (van Groenendael et al., 1996; J. Klimešová and T. Herben, unpubl. data), which may constrain effects of EIVs, some of which are known to be phylogenetically conservative (Prinzing et al., 2001). This phylogenetic mechanism, however, would not explain the regional effects we found, as our phylogeny-independent analysis yielded similar results.
LHS traits, morphological parameters and performance in the garden
In addition to assessing effects of clonal growth, our study also enabled examination of the predictive power of individual proxy variables for it. In particular, we were able to assess the explanatory power of functional parameters provided by garden reproduction data. Garden performance had good predictive power mainly at the local level, and provided information partly independent of morphological and LHS traits. Vegetative reproduction in the garden was by far the strongest predictor of species local abundance (apart from habitat information assessed by EIVs). Seed reproduction was a predictor at the regional level, but its effect disappeared when EIVs are taken into account.
The effects of performance parameters covaried with the effects of CLOPLA and LHS traits to an important degree (see also Herben et al., 2012a), but they also explained a portion of variation in abundance that was unexplained by these traits. This demonstrates that a species' potential performance in favourable conditions, even though it does not take into account environmental variability in demography, is an independent predictor of its local abundance (see also Tremlová and Münzbergová, 2007). In contrast, for regional abundance, the traits were more important than garden performance parameters, and generally remained significant predictors even after these parameters were partialled out. Thus, the trait values also affected species regional abundance through processes not captured by measuring vegetative and seed reproduction in favourable conditions. This could have been due to differences between realized and fundamental niches (for which garden conditions essentially represented proxies) or to hidden habitat drivers, i.e. niche attributes for which we did not account, but which are correlated with traits.
A key question implicit in trait-based studies regards the mechanisms of action of ‘soft’ traits. The likely mechanism of some traits is clear: for example, the negative correlation of seed mass with dispersal distance, in conjunction with the effect that dispersal distance can have on regional abundance (Soons and Ozinga, 2005; Ozinga et al., 2009; but see also Öster et al., 2009). Indeed, this mechanism could well explain the relevant findings of our study. In particular, at the regional scale, the effect of seed reproduction was positive, probably because it enabled colonization of new habitat. This aspect could also underlie its strong effect in habitat types such as mires/peat bogs and rocky habitats that tend to be fragmented in Central Europe; in both these habitat types, of the predictors examined, seed reproduction and/or seed mass was the best predictor of regional abundance (see also e.g. Ozinga et al., 2009). By contrast, on the local scale, its effect was generally negative, due either to increased emigration because of small seeds or, in dense vegetation, poor establishment of species that rely on seed reproduction (see also Murray et al., 2005; Kolb et al., 2006). A negative effect of seed reproduction on local abundance could also be due to ecological trade-offs between vegetative and seed reproduction. While such trade-offs may be difficult to detect at the trait level, they can be discerned when using demographic data instead of traits per se (Eriksson, 1992; Boedeltje et al., 2008; Herben et al., 2012a).
In contrast to seed traits, the mechanisms by which clonal growth traits influence plant functions are much less readily apparent. Clonal traits can enable local dispersal as a safeguard against local extinction, but are involved in trade-offs with clonal and other traits (possibly also including habitat preferences, see e.g. Klimeš and Klimešová, 1999), complicating inference. This necessitates further research into the mechanisms of action of predictive clonal growth traits, in particular how they affect local spreading and persistence, and how these processes scale up to affect regional abundance.
Effects of habitat factors
The strong effect of habitat factors on regional abundance corresponds well with studies that show a prevailing role of habitat requirements at relatively large, including regional, scales (e.g. Soons and Ozinga, 2005; Kolb et al., 2006). The significant EIV effects on regional species abundance probably reflected differing spatial extents of suitable habitat. Indeed, non-linearity in habitat abundance might underlie the frequently found non-linear effects of EIV on species abundance, as habitat abundance need not change linearly, or even monotonically, along a given gradient. Such non-linear relationships between EIVs (such as pH) and species abundance are well known from Central Europe (e.g. Chytrý et al., 2003; Ewald, 2003). In contrast, the significant EIV effects on local abundance indicate that the cover of a species (in habitats in which it occurs) depends on its EIV preferences. From this we can infer that there is a systematic relationship between species' optimal EIVs, and the proportions of their populations occurring in suboptimal habitats, where their cover is necessarily lower, due to poor performance or interspecific competition. This relationship would account for both the much weaker effects of EIV on local abundance (especially in some habitat types) and the poor correspondence between EIV effects on local and regional abundance.
The strong differences we found among habitat types in the roles of individual predictors, both for regional and for local abundance, were probably due to the prevailing ecological mechanisms differing among these habitats (e.g. dispersal in fragmented habitats such as mires, and vegetative growth in forests). Also, the importance of habitat preference varied strongly, such as the relative breadth of some habitat types (mires and grasslands), probably resulted in differences within these habitats, making the role of demography less clear.
CONCLUSIONS
We demonstrated that clonal growth is an important predictor of species local abundance, while also contributing to regional abundance. Although such a finding does not directly address causal relationships, detecting these patterns is important, as it generally has been difficult to identify predictors of abundance (May, 1999; McGill, 2006). In particular, we showed that morphologically defined clonal growth traits can predict species abundance. Simultaneously examining several such morphologically defined parameters and their relationships to habitat and other relevant variables can help develop hypotheses on the contributions of specific processes (e.g. avoidance of self-competition, or role of offspring support) to the effects of clonal growth in particular habitat types. We also showed that the use of garden performance parameters provides a practical approach to assessing the roles of morphologically defined clonal growth traits (and also of LHS traits) for large sets of species.
Our findings also provide the basis for further investigations. In particular, we must learn more about the relative influence of the recruitment and established phases of plant life cycles (Silvertown et al., 1993; Ehrlén et al., 2006; Öster et al., 2009) on patterns of local and regional abundance. Moreover, we must assess the degree to which these roles can truly be proxied by the simple traits that have been widely used. We believe that using performance parameters such as those obtained from the garden provide a viable approach to assessing trait–performance relationships. Comparison of performance and soft trait data can be used to infer mechanisms of action of the traits in question, and could also prompt searches for other simple correlates of performance and abundance.
ACKNOWLEDGEMENTS
The research reported here would not have been possible without the vision and long-term generous support of the garden collections of the Faculty of Science, Charles University. Members of the Plant Ecology Discussion Group at the University of Michigan greatly helped to shape ideas presented here. Ingolf Kühn kindly provided the updated BiolFlor phylogenetic tree. We thank Tomáš Koubek and Martin Weiser for comments on an earlier draft of the manuscript. The research was partially supported by the Grant Agency of the Czech Republic (P505/12/1007, GA 526/09/0963), by the Academy of Sciences of the Czech Republic (RVO 67985939) and by the Ministry of Education (institutional resources for the support of research).
LITERATURE CITED
- Benot ML, Bittebiere AK, Ernoult A, Clément B, Mony C. Fine-scale spatial patterns in grassland communities depend on species clonal dispersal ability and interactions with neighbours. Journal of Ecology. 2010;101:626–636. [Google Scholar]
- Boedeltje G, Ozinga WA, Prinzing A. The trade-off between vegetative and generative reproduction among angiosperms influences regional hydrochorous propagule pressure. Global Ecology and Biogeography. 2008;17:50–58. [Google Scholar]
- Boulangeat I, Gravel D, Thuiller W. Accounting for dispersal and biotic interactions to disentangle the drivers of species distributions and their abundances. Ecology Letters. 2012a;15:584–593. doi: 10.1111/j.1461-0248.2012.01772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangeat I, Lavergne S, Van Es J, Garraud L, Thuiller W. Niche breadth, rarity and ecological characteristics within a regional flora spanning large environmental gradients. Journal of Biogeography. 2012b;39:204–214. [Google Scholar]
- Chytrý M, Rafajová M. Czech National Phytosociological Database: basic statistics of the available vegetation-plot data. Preslia. 2003;75:1–15. [Google Scholar]
- Chytrý M, Tichý L, Roleček J. Local and regional patterns of species richness in Central European vegetation types along the pH/calcium gradient. Folia Geobotanica. 2003;38:429–442. [Google Scholar]
- Chytrý M, Pyšek P, Tichý L, Knollová I, Danihelka J. Invasions by alien plants in the Czech Republic: a quantitative assessment across habitats. Preslia. 2005;77:339–354. [Google Scholar]
- Comita LS, Muller-Landau HC, Aguilar S, Hubbell SP. Asymmetric density dependence shapes species abundance in a tropical tree community. Science. 2010;329:330–332. doi: 10.1126/science.1190772. [DOI] [PubMed] [Google Scholar]
- Cornwell WK, Ackerly DD. A link between plant traits and abundance: evidence from coastal California woody plants. Journal of Ecology. 2010;98:814–821. [Google Scholar]
- Craine JM, Froehle J, Tilman DG, Wedin DA, Chapin FS. The relationships among root and leaf traits of 76 grassland species and relative abundance along fertility and disturbance gradients. Oikos. 2001;93:274–285. [Google Scholar]
- Desdevises Y, Legendre P, Azouzi L, Morand S. Quantifying phylogenetically structured environmental variation. Evolution. 2003;57:2647–2652. doi: 10.1111/j.0014-3820.2003.tb01508.x. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, de Sant'Ana CER, Bini LM. An eigenvector method for estimating phylogenetic inertia. Evolution. 1998;52:1247–1262. doi: 10.1111/j.1558-5646.1998.tb02006.x. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour AB. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software. 2007;22:1–20. [Google Scholar]
- Duralia TE, Reader RJ. Does abundance reflect competitive ability? A field test with three prairie grasses. Oikos. 1993;68:82–90. [Google Scholar]
- Durka W. Phylogenie der Farn- und Blütenpflanzen Deutschlands. In: Klotz S, Kühn I, Durka W, editors. BIOLFLOR – Eine Datenbank mit biologisch-ökologischen Merkmalen zur Flora von Deutschland. Vol. 38. Bonn: Schriftenreihe für Vegetationskunde; 2002. pp. 75–91. [Google Scholar]
- Ehrlén J, Münzbergová Z, Diekmann M, et al. Long-term assessment of seed limitation in plants: results from an 11-year experiment. Journal of Ecology. 2006;94:1224–1232. [Google Scholar]
- Eriksson O. Evolution of seed dispersal and recruitment in clonal plants. Oikos. 1992;63:439–448. [Google Scholar]
- Espeland EK, Emam TM. The value of structuring rarity: the seven types and links to reproductive ecology. Biodiversity and Conservation. 2011;20:963–985. [Google Scholar]
- Ewald J. A critique for phytosociology. Journal of Vegetation Science. 2003;14:291–296. [Google Scholar]
- Gaston K. Common ecology. BioScience. 2011;61:354–362. [Google Scholar]
- Gough L, Gross KL, Cleland EE, et al. Incorporating clonal growth form clarifies the role of plant height in response to nitrogen addition. Oecologia. 2012;169:1053–1062. doi: 10.1007/s00442-012-2264-5. [DOI] [PubMed] [Google Scholar]
- Hanski I. Dynamics of regional distribution: the core and satellite species hypothesis. Oikos. 1982;38:210–221. [Google Scholar]
- Hartley S, Kunin WE, Lennon JJ, Pocock M JO. Coherence and discontinuity in the scaling of specie's distribution patterns. Proceedings of the Royal Society, London B. 2004;271:81–88. doi: 10.1098/rspb.2003.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herben T, Nováková Z, Klimešová J, Hrouda L. Species traits and plant performance: functional trade-offs in a large set of species in a botanical garden. Journal of Ecology. 2012a;100:1522–1533. [Google Scholar]
- Herben T, Suda J, Klimešová J, Mihulka S, Říha P, Šímová I. Ecological effects of cell-level processes: genome size, functional traits and regional abundance of herbaceous plant species. Annals of Botany. 2012b;110:1357–1367. doi: 10.1093/aob/mcs099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herben T, Březina S, Hadincová V, Krahulec F, Skálová H. Mutual replacement of species in space in a grassland community: is there an evidence for functional complementarity of replacement groups? Oikos. 2013;122:111–121. [Google Scholar]
- Jónsdóttir IS, Watson MA. Extensive physiological integration: an adaptive trait in resource-poor environments? In. In: de Kroon H, van Groenendael J, editors. The ecology and evolution of clonal plants. Backbuys Publishers: Leiden; 1997. pp. 109–136. [Google Scholar]
- Kleyer M, Bekker RM, Knevel IC, et al. The LEDA Traitbase: a database of life-history traits of the Northwest European flora. Journal of Ecology. 2008;96:1266–1274. [Google Scholar]
- Klimeš L. Small-scale plant mobility in a species-rich grassland. Journal of Vegetation Science. 1999;10:209–218. [Google Scholar]
- Klimeš L, Klimešová J. CLO-PLA2 – a database of clonal plants in central Europe. Plant Ecology. 1999;141:9–19. [Google Scholar]
- Klimeš L, Klimešová J. Plant rarity and the type of clonal growth. Zeitschrift für Ökologie und Naturschutz. 2000;9:43–52. [Google Scholar]
- Klimešová J, de Bello F. CLO-PLA: the database of clonal and bud bank traits of Central European flora. Journal of Vegetation Science. 2009;20:511–516. [Google Scholar]
- Klimešová J, Doležal J, Sammul M. Evolutionary and organismic constraints on the relationship between spacer length and environmental conditions in clonal plants. Oikos. 2011;120:1110–1120. [Google Scholar]
- Klimešová J, Doležal J, Prach K, Košnar J. Clonal growth forms in Arctic plants and their habitat preferences: a study from Petuniabukta, Spitsbergen. Polish Polar Research. 2012;33:421–442. [Google Scholar]
- Klotz S, Kuehn I, Durka W. BIOLFLOR – Eine Datenbank zu biologisch-oekologischen Merkmalen der Gefaesspflanzen in Deutschland. Schriftenreihe fuer Vegetationskunde. 2002;38:75–91. [Google Scholar]
- Knapp S, Kühn I, Bakker JPC, et al. How species traits and affinity to urban land use control large-scale species frequency. Diversity and Distributions. 2009;15:533–546. [Google Scholar]
- Kolb A, Barsch F, Diekmann M. Determinants of local abundance and range size in forest vascular plants. Global Ecology and Biogeography. 2006;15:237–247. [Google Scholar]
- Kubát K, Hrouda L, Chrtek J, Kaplan Z, Kirschner J, Štěpánek J. Klíč ke Květeně České republiky (Key to the flora of the Czech Republic) Praha: Academia; 2002. [Google Scholar]
- Kunin WE, Shmida A. Plant reproductive traits as a function of local, regional, and global abundance. Conservation Biology. 1997;11:183–192. [Google Scholar]
- Kunin WE, Hartley S, Lennon JJ. Scaling down: on the challenge of estimating abundance from occurrence patterns. American Naturalist. 2000;156:560–566. doi: 10.1086/303408. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Thompson JD, Garnier E, Debussche M. The biology and ecology of narrow endemic and widespread plants: a comparative study of trait variation in 20 congeneric pairs. Oikos. 2004;107:505–518. [Google Scholar]
- Lavorel S, Garnier E. Predicting the effects of environmental changes on plant community composition and ecosystem functioning: revisiting the Holy Grail. Functional Ecology. 2002;16:545–556. [Google Scholar]
- Lososová Z, Chytrý M, Kühn I. Plant attributes determining the regional abundance of weeds on central European arable land. Journal of Biogeography. 2008;35:177–187. [Google Scholar]
- Matesanz S, Escudero A, Valladares F. Impact of three global change drivers on a Mediterranean shrub. Ecology. 2009;90:2609–2621. doi: 10.1890/08-1558.1. [DOI] [PubMed] [Google Scholar]
- May R. Unanswered questions in ecology. Philosophical Transactions of The Royal Society B: Biological Sciences. 1999;354:1951–1959. doi: 10.1098/rstb.1999.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill BJA. Renaissance in the study of abundance. Science. 2006;314:770–772. doi: 10.1126/science.1134920. [DOI] [PubMed] [Google Scholar]
- Miller MT, Antos JA, Allen GA. Demographic differences between two sympatric lilies (Calochortus) with contrasting distributions, as revealed by matrix analysis. Plant Ecology. 2007;191:265–278. [Google Scholar]
- Mokany K, Roxburgh SH. The importance of spatial scale for trait–abundance relations. Oikos. 2010;119:1504–1514. [Google Scholar]
- Moora M, Jõgar Ü. Competitive response of the rare Viola elatior in comparison with a common V. mirabilis. Plant Ecology. 2006;184:105–110. [Google Scholar]
- Moore KA, Elmendorf SC. Propagule vs. niche limitation: untangling the mechanisms behind plant species' distributions. Ecology Letters. 2006;9:797–804. doi: 10.1111/j.1461-0248.2006.00923.x. [DOI] [PubMed] [Google Scholar]
- Münzbergová Z. Determinants of species rarity: population growth rates of species sharing the same habitat. American Journal of Botany. 2005;92:1987–1994. doi: 10.3732/ajb.92.12.1987. [DOI] [PubMed] [Google Scholar]
- Murray BR, Thrall PH, Gill AM, Nicotra AB. How plant life-history and ecological traits relate to species rarity and commonness at varying spatial scales. Austral Ecology. 2002;27:291–310. [Google Scholar]
- Murray BR, Kelaher BP, Hose GC, Figuera WF. A meta-analysis of the interspecific relationship between seed size and plant abundance within local communities. Oikos. 2005;110:191–194. [Google Scholar]
- Oborny B, Szabó G, Meszéna G. Survival of species in patchy landscapes: percolation in space and time. In: Storch D, Marquet P, Brown J, editors. Scaling biodiversity. Cambridge: Cambridge University Press; 2007. pp. 409–440. [Google Scholar]
- Öster M, Ask K, Römermann C, Tackenberg O, Eriksson O. Plant colonization of ex-arable fields from adjacent species-rich grasslands: the importance of dispersal vs. recruitment ability. Agriculture, Ecosystems and Environment. 2009;130:93–99. [Google Scholar]
- Ozinga WA, Schaminée JHJ, Bekker RM, et al. Predictability of plant species composition from environmental conditions is constrained by dispersal limitation. Oikos. 2005;108:555–561. [Google Scholar]
- Ozinga W, Hennekens SM, Schaminée JHJ, et al. Local above-ground persistence of vascular plants: Life-history trade-offs and environmental constraints. Journal of Vegetation Science. 2007;18:489–497. [Google Scholar]
- Ozinga WA, Römermann C, Bekker RM, et al. Dispersal failure contributes to plant losses in NW Europe. Ecology Letters. 2009;12:66–74. doi: 10.1111/j.1461-0248.2008.01261.x. [DOI] [PubMed] [Google Scholar]
- Paradis E, Bolker B, Claude J, et al. Package ape. Analyses of Phylogenetics and Evolution. R package version, 2012·04-04. 2012 http://www.cran.r-project.org/package=ape/ [Google Scholar]
- Prinzing A, Durka W, Klotz S, Brandl R. The niche of higher plants: evidence for phylogenetic conservatism. Proceedings of the Royal Society, Series B: Biological Sciences. 2001;268:2383–2389. doi: 10.1098/rspb.2001.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012. Available at: http://www.R-project.org . [Google Scholar]
- Römermann C, Tackenberg O, Jackel AK, Poschlod P. Eutrophication and fragmentation are related to species' rate of decline but not to species rarity! Results from a functional approach. Biodiversity and Conservation. 2008;17:591–604. [Google Scholar]
- Saar L, Takkis K, Pärtel M, Helm A. Which plant traits predict species loss in calcareous grasslands with extinction debt? Diversity and Distributions. 2012;18:808–817. [Google Scholar]
- Sammul M. Length of the spacer rather than its plasticity relates to species distribution in various natural habitats. Folia Geobotanica. 2011;46:137–153. [Google Scholar]
- Silvertown J, Franco M, Pisanty I, Mendoza A. Comparative plant demography – relative importance of life-cycle components to the finite rate of increase in woody and herbaceous perennials. Journal of Ecology. 1993;81:465–476. [Google Scholar]
- Soons MB, Ozinga WA. How important is long-distance seed dispersal for the regional survival of plant species? Diversity and Distributions. 2005;11:165–172. [Google Scholar]
- Tamm A, Kull K, Sammul M. Classifying clonal growth forms based on vegetative mobility and ramet longevity: a whole community analysis. Evolutionary Ecology. 2001;15:383–401. [Google Scholar]
- Tremlová K, Münzbergová Z. Importance of species traits for species distribution in fragmented landscapes. Ecology. 2007;88:965–977. doi: 10.1890/06-0924. [DOI] [PubMed] [Google Scholar]
- van der Veken S, Bellemare J, Verheyen K, Hermy M. Life-history traits are correlated with geographical distribution patterns of western European forest herb species. Journal of Biogeography. 2007;34:1723–1735. [Google Scholar]
- van Groenendael JM, Klimeš L, Klimešová J, Hendriks RJJ. Comparative ecology of clonal plants. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences. 1996;351:1331–1339. [Google Scholar]
- Weiher E, van der Werf A, Thompson K, Roderick M, Garnier E, Eriksson O. Challenging Theophrastus: a common core list of plant traits for functional ecology. Journal of Vegetation Science. 1999;10:609–620. [Google Scholar]
- Westoby M. A leaf-height-seed (LHS) plant ecology strategy scheme. Plant and Soil. 1998;199:213–227. [Google Scholar]