Abstract
Previous studies on relatively small samples of individuals with trisomy 21 caused by paternally derived errors have shown that: (1) advanced paternal age is not a risk factor for chromosome 21 nondisjunction (NDJ), (2) absence of recombination, but not the location of recombination is associated with paternal NDJ and (3) there is an excess of males among live-births with paternally derived trisomy 21. An excess of males is also observed among all individuals with trisomy 21. Using 128 families that had a child with trisomy 21 due to a paternally derived error, we examined: paternal age, recombination and the male/female sex ratio. We genotyped STRs along 21q to identify the origin of the error and the location of recombination on the paternal chromosome. Results showed that 32% of paternal meiotic errors occurred in meiosis I (MI) and 68% in meiosis II (MII). We confirmed the lack of a paternal age association with either type of error (mean paternal age for controls, MI, and MII errors: 31.3 ± 6.6, 32.2 ± 6.3, 30.6 ± 6.5, respectively). However, contrary to previous findings, we did not find altered patterns of recombination among paternal MI or MII errors. We found an increased male/female sex ratio among paternal (1.28, 95% CI: 0.68–1.91) and maternal (1.16, 95% CI: 1.02–1.33) meiotic errors. While the sex ratio among individuals with paternal errors was not statistically significant, these findings suggest that selection against female fetuses with trisomy 21 may contribute to the excess of males observed among all individuals with trisomy 21.
Keywords: paternal nondisjunction, altered recombination, sex ratio, meiosis, Down syndrome, spermatogenesis
INTRODUCTION
Nondisjunction is defined as the failure of chromosomes to segregate during meiosis; this results in the production of aneuploid gametes. Aneuploidy is the leading cause of pregnancy loss, intellectual disabilities and birth defects [Hassold et al., 1996]. An estimated 10–35% of all human conceptions are estimated to involve an aneuploid gamete [Hassold et al., 1995]. Trisomy 21, caused by nondisjunction of human chromosome 21, has been estimated to occur in approximately 1 in 733 live births in the United States [Canfield et al., 2006]. It is one the few autosomal trisomies that survives gestation. Trisomy 21, also referred to as Down syndrome, is characterized by a host of symptoms, the hallmark being severe developmental delay and intellectual disabilities.
About 90% of chromosome 21 nondisjunction occurs within the oocyte; with less than half of the remaining 10% occurring in sperm [Takaesu et al., 1990; Antonarakis, 1991; Antonarakis et al., 1992; Yoon et al., 1996; Freeman et al., 2007]. As a result, most of the studies focused on understanding mechanisms underlying nondis-junction of chromosome 21 have been performed using maternal cases of trisomy 21. These studies have shown that meiotic nondis-junction of chromosome 21 occurs almost three times as frequently during the first meiotic division (MI) as during the second meiotic division (MII) [Antonarakis et al., 1992; Hassold et al., 1996; Freeman et al., 2007]. Increased maternal age, the absence of exchange and altered placement of exchange along chromosome 21 have been identified as risk factors for nondisjunction of chromo-some 21 in oocytes [Lamb et al., 1996, 1997a; Oliver et al., 2008].
With respect to nondisjunction of chromosome 21 occurring during spermatogenesis, few risk factors have been identified. We examined recombination profiles along the nondisjoined chromo-some 21 in a previous study [Savage et al., 1998] and found an increase in achiasmate tetrads (i.e., those with no exchanges). However, we found only weak evidence that the location of exchange played a role in nondisjunction. Importantly, our study was limited by sample size. In addition, we and others have noted an increased male to female sex ratio among all cases with trisomy 21 (1.18 [Huether et al., 1996]) and among offspring with trisomy 21 due to paternal nondisjunction [Mikkelsen et al., 1990; Petersen et al., 1993; Savage et al., 1998]. This excess of males with paternally derived errors is thought to contribute to the overall increase in sex ratio observed among infants born with trisomy 21.
Since our previous analysis [Savage et al., 1998], we have almost doubled our sample of paternal cases of trisomy 21. Thus the goal of the present study is to identify risk factors associated with nondisjunction of chromosome 21 in spermatogenesis. To do this, we genotyped short tandem repeat markers (STRs) along 21q to examine the origin of the error and the recombination profile along the nondisjoined chromosome. Unlike our previous studies, we did not find evidence that the absence of exchange was a risk factor for nondisjunction. We did find an increased male/female sex ratio among paternal and maternal meiotic cases of trisomy 21. While these differences were not statistically significantly different from the normal ratio of 1.05, these findings suggest there may be selection against female fetuses with trisomy 21. This could potentially contribute to the excess of males that is observed among all cases of trisomy 21.
METHODS
Trisomic Samples
Study population
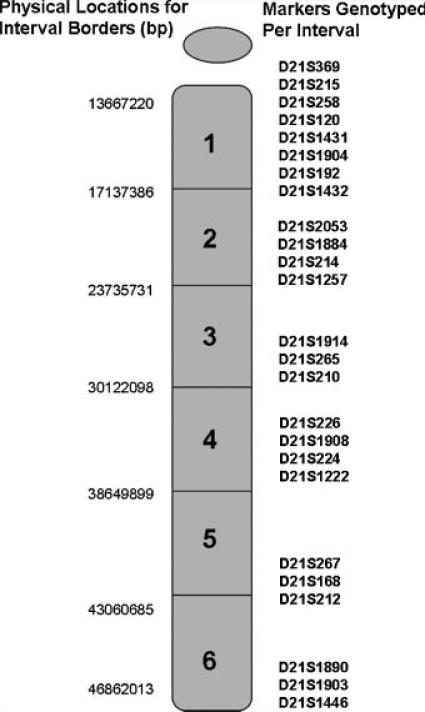
DNA was collected from individuals with Down syndrome and their parents as a part of several studies focused on understanding the etiology of trisomy 21 [Petersen et al., 1993; Yoon et al., 1996; Freeman et al., 2007]. Each site that participated in the recruitment of individuals in this study obtained the necessary Institutional Review Board approvals from their institutions. DNA was extracted from blood or tissue samples and STR markers specific to 21q were genotyped (Fig. 1). For the analysis of paternal cases of trisomy 21, families in which DNA was available from (1) both parents and the child with trisomy 21, henceforth referred to as “trios” and (2) the mother and the child with trisomy 21, henceforth referred to as “duos,” were eligible for inclusion. For the analysis of maternal cases of trisomy 21, we used only trios, as we had many more of these cases available for study.
FIG. 1.
Short tandem repeat markers along 21q genotyped in DNA collected from families interval and physical location of markers genotyped on 21q to determine the origin of the error and the location of recombination.
In all, the present study consists of 128 paternal cases of free trisomy 21 and 909 maternal cases of free trisomy 21 (Table I). Fifty-seven of the 128 paternal cases were included in a sample of 67 paternally derived cases of trisomy 21 reported by Savage et al. [1998]. Of the 909 maternal cases depicted in Table I, the sex of the proband was only known for 862. These 862 maternal cases were included in the study of maternal nondisjunction reported by Oliver et al. [2008] and are used to examine the sex ratio in Table V.
TABLE I.
Stage of Origin for Nondisjunction of Chromosome 21
| Paternal |
Maternal Trio |
||
|---|---|---|---|
| Meiotic outcome group | Duo | Trio | |
| Meiosis I | 13 | 26 | 616 |
| Meiosis II | 2 | 56 | 253 |
| Unknown meiotic origin | 12 | 1 | – |
| Mitotic | – | 20 | 40 |
| Normal | – | 300 | 295 |
TABLE V.
Male/Female Sex Ratio and 95% Confidence Intervals of Cases of Trisomy 21 by Meiotic Error
| Type of error | Males | Females | Sex ratio | Lower limit | Upper limit |
|---|---|---|---|---|---|
| Paternal MIa | 21 | 17 | 1.24 | 0.65 | 2.46 |
| Paternal MIIa | 33 | 22 | 1.50 | 0.79 | 2.70 |
| All paternal meiotica | 59 | 46 | 1.28 | 0.68 | 1.91 |
| Maternal MI | 329 | 283 | 1.16 | 0.99 | 1.36 |
| Maternal MII | 134 | 116 | 1.16 | 0.90 | 1.49 |
| Maternal meiotic | 463 | 399 | 1.16 | 1.02 | 1.33 |
Duos and trios included in calculation of sex ratio.
DNA studies
The parental origin of the nondisjoining error was determined by establishing the contribution of parental alleles to the child with trisomy 21. Amongst “trios,” parental origin was considered established when at least two markers were informative and all other markers were consistent with that origin of nondisjunction. To establish paternal origin amongst “duos,” at least two markers had to exclude maternal origin of the error.
Only trios can provide unbiased information about the type of error (MI, MII, or mitotic). Thus, duos were excluded from any analyses requiring this characterization. We used highly polymorphic STR markers located in the pericentric region (D21S369–D21S1432, Fig. 1) of 21q to infer the stage of nondis-junction. These markers are similar to those used in our previous studies of trisomy 21 [Savage et al., 1998; Oliver et al., 2008]. If parental heterozygosity at the most proximal marker in the above set was retained in the trisomic offspring (“nonreduced”), we concluded a MI error. If parental heterozygosity was “reduced” to homozygosity, we concluded a MII error. If there was no informative pericentric marker, but at least one marker in the trisomic offspring was “nonreduced” we concluded that the case was of unknown meiotic origin (i.e., either MI or MII).
When all informative markers in the parent of origin were reduced to homozygosity, we concluded a post-zygotic, mitotic error. Such cases could also be MII errors with no recombination. Since we have no method to further categorize these inferred mitotic cases, we excluded them from our analysis.
Characterizing the recombination profile
Our analysis of the number and location of recombination was restricted to 21q and was based on the genotyped STRs (Fig. 1). This set of STRs was similar to that used in our previous analysis [Savage et al., 1998], with the addition of six markers. The long arm of chromosome 21 was divided into six relatively equal physical intervals with interval 1 comprising the most centromeric region of 21q and interval 6 comprising the most telomeric region (Fig. 1). The presence of a recombinant event was identified by changes in the status of adjacent informative markers from “reduced” to “nonreduced” (or vice versa). In most cases, the location of recombination was scored as belonging to one of six distinct intervals along 21q. When one of the six intervals was uninformative, but markers defining the two flanking intervals were informative, we included the case. Those with two or more adjacent uninformative intervals were excluded from our analysis. In some instances, the recombinant event could not be located to one specific interval, but instead to one of two adjacent intervals (e.g., interval 1 or interval 2). The location of such events was treated as occurring at the midpoint of the two intervals (e.g., represented as interval 1.5) in most of our analyses (see Statistical Analyses Section). It is not possible to identify “reduced” loci among paternal cases in which the father’s DNA is missing (“duos”); thus we could not develop a recombination profile for these cases. As a result, only “trios” were analyzed in determining the recombination profile for paternal meiotic cases of trisomy 21. Our final analysis of recombination in paternal cases of trisomy 21 included a total of 22 paternal MI cases and 46 paternal MII cases.
Euploid Samples
Study population
Our control population consisted of 300 male and 295 female normal meioses taken from 23 CEPH Utah families previously genotyped with 133 chromosome 21-specific SNPs [Cheung et al., 2007].
Data used to determine the average paternal age in the normal population were from the fathers of children without Down syndrome who participated in the National Down Syndrome Project, a population-based study focused on understanding the etiology of Down syndrome and Down syndrome-associated birth defects [Freeman et al., 2007].
DNA studies
The most centromeric SNP on 21q was located at 15,009,674 bp (rs990141) and the most telomeric SNP located at 46,902,239 bp (rs2839337). It is important to point out that the marker set used to genotype these 23 CEPH families was different from that used to genotype the trisomic cases. Specifically, the most centromeric marker used for genotyping trisomic cases was located at 13678220 bp (D21S369) and the most telomeric marker was located at 46862013 bp (D21S1446). However, the criteria used to identify the location of recombination were the same for cases and controls.
Characterizing the recombination profile
In order to determine the location of recombination along 21q in normal paternal meiotic events, the transmission of paternal grandparental SNP genotypes to the paternal offspring was analyzed. A paternal recombinant event was noted when the sharing of SNPs identical by descent switched from one paternal grandparent to the other [Cheung et al., 2007]. Our final analysis included 300 informative paternal meioses: 180 had zero recombinants, 117 had one recombinant and three had two recombinants.
Statistical Analyses
Comparing paternal ages between meiotic outcome groups
Analysis of variance was used to determine whether the average paternal ages of mitotic, MI, MII, and controls were different from each other.
Estimating the exchange profile
Detailed methods for estimating exchanges from observed recombination profiles are described elsewhere [Lamb et al., 1997b]. Briefly, tetrad analysis was used to infer the frequency of exchange that occurred in each of six predefined intervals on 21q and this was done separately for paternal MI, MII, and control samples. Sample sizes were too small to conduct statistical comparisons of the spatial distribution of the location of exchange between normal and nondisjoining cases, thus we only provide a descriptive analysis.
In order to determine if the absence of exchange was a risk factor for paternal MI nondisjunction of chromosome 21, the 95% confidence intervals for the frequency of achiasmate tetrads were computed for normal meioses and for paternal MI errors as previously described [Lamb et al., 1997b].
Comparing sex ratios
In order to determine whether there was a difference between the sex ratio in cases of trisomy 21 and the normal sex ratio, the 95% confidence interval was calculated for the sex ratio among maternal and paternal cases of trisomy 21 due to various types of errors (i.e., MI, MII, mitotic, and free trisomy 21). This estimate was then compared to the normal sex ratio. The expected normal sex ratio, 1.05, was estimated from the male-female ratio of individuals born in the United States during the year 2002 [Mathews and Hamilton, 2005].
RESULTS
Stage of Origin
Of the 128 paternal cases, 103 had DNA available from the child with trisomy 21 and both parents. Of these, 26 were consistent with MI nondisjunction, 56 were consistent with MII nondisjunction, 20 were consistent with mitotic origin and one had an unknown but meiotic origin (Table I). The ratio of paternal MI/MII meiotic errors was 0.46 and was significantly different from that in maternal errors (maternal MI/MII ratio = 2.43; P < 0.0001).
Paternal Age
Using the analysis of variance we found no significant difference between the average paternal age of any of the meiotic outcome groups (F = 0.86, df = 3 P > 0.46, Table II).
TABLE II.
Mean Paternal Age and Standard Deviation of Each Meiotic Outcome Group
| Meiotic outcome group | n | Paternal age | Standard deviation |
|---|---|---|---|
| Meiosis I | 25 | 32.2 | 6.3 |
| Meiosis II | 54 | 30.6 | 6.5 |
| Mitotic | 20 | 29.5 | 5.8 |
| Normal | 1,308 | 31.3 | 6.6 |
Recombination Profile
Among 22 informative cases with paternal MI errors, 14 had no observed recombinants, seven had only one recombinant and one had a double recombinant (Table III). In order to determine if the absence of exchange was a risk factor for paternal nondisjunction, we used tetrad analysis to estimate the frequency of cases with zero exchanges. The frequency of achiasmates among normal paternal meiotic events was 22% (95% confidence interval (CI): 11–33%) and among MI errors was 32% (95% CI: 7–83%).
TABLE III.
Distribution of the Number of Recombination and the Number of Exchanges
| Frequency of observed recombinants |
Inferred exchanges |
||||||
|---|---|---|---|---|---|---|---|
| Meiotic outcome group | n | 0 | 1 | 2 | 0 | 1 | 2 |
| Meiosis I | 22 | 0.63 | 0.32 | 0.05 | 0.32 | 0.59 | 0.09 |
| Meiosis II | 46 | n/aa | 0.93 | 0.07 | n/a | 0.87 | 0.13 |
| Normal | 300 | 0.60 | 0.39 | 0.01 | 0.22 | 0.74 | 0.04 |
n/a reflects our assumption that MII errors with no recombination are post zygotic mitotic events, see Methods Section.
We then examined the location of recombination among the paternal MI cases, although there were too few cases with observed recombinants to conduct statistical tests (n = 8). Among those with one observed single event, the recombinants were located in the following intervals: two in interval 1.5, one in interval 2.5, one in interval 5 and three in interval 5.5. One case had two observed recombinant events, one in interval 3.5 and the other in interval 6.
Amongst our paternal population of MII cases, 43 had a single recombinant and three had two recombinants (Table III). We examined the spatial distribution of inferred single exchanges based on these data and found that they were similar to that in normal paternal meiotic events (Table IV). There was some suggestion of an increase in exchanges within the most proximal one-half of 21q (i.e., intervals 1–3), although no test of significance was conducted because of the small numbers used to infer these frequencies.
TABLE IV.
Spatial Distribution of the Frequency of Single Exchanges in MII and Normal Cases
| Interval | Meiosis II | Normal |
|---|---|---|
| 1 | 0.070 | 0.030 |
| 2 | 0.128 | 0.081 |
| 3 | 0.209 | 0.171 |
| 4 | 0.198 | 0.222 |
| 5 | 0.279 | 0.312 |
| 6 | 0.116 | 0.184 |
Sex Ratios of Cases of Trisomy 21
We examined the male/female sex ratios among maternal and paternal cases of trisomy 21 stratified by type of error and compared them to the sex ratio among live births in the United States estimated in 2002 taken from Mathews and Hamilton [2005]. Although the point estimates of the sex ratios for both maternal and paternal outcome groups were greater than 1.05, the 95% confidence intervals each included 1.05 (Table V).
DISCUSSION
In our previous analysis of risk factors associated with nondisjunction of chromosome 21 in spermatogenesis [Savage et al., 1998], we were limited in our analyses in several ways. First, although our sample size was the largest to date, it still was small. Second, we included cases that only had DNA from the proband and mother. Doing this compromised our ability to determine the recombination profile and identify the stage of the error. Lastly, we did not have extensive genotyping of loci along 21q. To circumvent these problems in the present analysis, we significantly increased our sample size and restricted analyses of paternal age, the ratio of MI/MII cases and the number and location of recombinant events to cases with both parent samples available. We also increased the number of loci genotyped.
In the present study, we were able to confirm two important characteristics of nondisjunction of chromosome 21 in spermato-genesis. First, the majority of errors occur during MII. This is in contrast to maternal cases of trisomy 21 where only one-fourth of cases exhibit MII errors [Antonarakis et al., 1992; Lamb et al., 1996]. Second, we found no evidence for a paternal age effect. As the time line for spermatogenesis differs significantly from that in oogenesis, these two findings are not surprising. Spermatogenesis in the human male begins at puberty and cells entering meiosis move from one stage to the next with no delay. The absence of an extended MI arrest in spermatogenesis may explain the absence of a paternal age effect as well as the reduction of MI errors in paternal cases of trisomy 21.
Previously we found that the absence of exchange was associated with a paternal MI error [Savage et al., 1998]. In our current analysis we found that 32% of paternal MI errors have no exchange compared to 22% of normal meiotic events. As the 95% confidence intervals overlap, we cannot say that these estimates are statistically different; however, the small sample size of paternal MI cases limits this analysis.
The altered location of exchanges has been associated with nondisjunction of chromosome 21 in oocytes, thus we examined the spatial distribution of exchange along chromosome 21 in our paternal meiotic cases. Although the numbers were too small to infer the exchange pattern among paternal MI errors, those for MII errors were sufficient (Table IV). The spatial distribution of exchanges did not appear to be different between paternal MII errors and controls. This observation is in agreement with that of our previous analysis.
Lastly, we examined the sex ratio in paternal cases of trisomy 21. Several previous studies have examined the sex ratio among paternally derived cases of trisomy 21 [Mikkelsen et al., 1990; Petersen et al., 1993; Savage et al., 1998] and results are summarized in Table VI. While the sample sizes in previous studies were smaller than those of our present analysis, an excess of males among paternally derived cases of trisomy 21 was found. We also found an increase in the frequency of males among our population of paternally derived cases of trisomy 21 (sex ratio of paternal meiotic cases = 1.28), however this sex ratio was not statistically different from controls (sex ratio among live-borns in the US in 2002 = 1.05). In our previous report [Savage et al., 1998], we found evidence to suggest that the excess of males was among paternal MII only. With increased number of paternal cases, we could not confirm this finding. We found that the point estimates for both MI and MII were increased, but neither was statistically different from that among normal outcomes (Table V). We suggest that our previous study and others were compromised by the small number of paternal errors. In our current dataset with 105 informative cases, the smallest difference from a sex ratio of 1.05 that we could detect at 5% significance level with 80% power using a two-sided test would be 1.86 and using a one-sided test would be 1.70. Detecting smaller differences between the normal sex ratio and the sex ratio among paternal errors would require a larger sample size, one that exceeds 600 cases.
TABLE VI.
Sex Ratios in Paternal Cases of Trisomy 21 Reported by Previous Studies
| Studies | Males | Females | Sex ratio | Lower limit | Upper limit |
|---|---|---|---|---|---|
| Mikkelsen et al. [1990] | 21 | 11 | 1.91 | 0.97 | 4.58 |
| Petersen et al. 1993 | 21 | 7 | 3.00 | 1.44 | 10.16 |
| Savage et al. [1998] | 34 | 20 | 1.70 | 1.00 | 3.14 |
| Current study | 59 | 46 | 1.28 | 0.88 | 1.91 |
While we did not detect a statistically significant excess in the number of live-born males among paternally derived cases of trisomy 21, this does not mean there is not one. Other evidence suggests there is an excess of males among paternal trisomy 21 cases. For example, Griffin et al. [1995] found that among 300,000 sperm taken from 24 normal, healthy males, the frequency of disomy 21 was 0.17%. Of sperm disomic for chromosome 21, the ratio of Y-bearing to X-bearing sperm was 1.57. Thus assuming that male and female trisomic fetuses have the same chance of being live-born, this observation would suggest an excess of males among paternally derived live-born cases of trisomy 21.
If there is a true excess of males among paternally derived trisomy 21 errors, we and others have asked whether this one contribution could explain the overall excess of males born with trisomy 21. Huether et al. [1996] estimated the sex ratio among all live-born cases with free trisomy 21, mosaic trisomy 21 and tranlocation trisomy 21 to be 1.18, 1.41, and 1.53, respectively in a relatively large study. Others have found estimates in the same range of 1.18 [Bernheim et al., 1979; Nielsen et al., 1981; Mikkelsen et al., 1990]. Based on this sex ratio and assuming that paternal cases account for at most 10% of all trisomy 21, we estimated that the sex ratio among paternal cases would have to be approximately 2.8 in order to account for the excess of males observed among all cases of trisomy 21. This estimate is considerably higher than our point estimates and that of Griffin et al. [1995]. Thus we conclude that other factors must explain the overall excess of males among infants born with trisomy 21.
To further examine possibilities, we calculated the sex ratio among maternal-derived cases of trisomy 21. We again observed an excess of males, although not statistically significant from that among normal outcomes: the sex ratio was 1.16 among all maternal meiotic cases. One simple explanation for the excess of males observed among all infants with trisomy 21 may be selection against female fetuses with trisomy 21.
In conclusion, we were able to confirm several of our previous findings with the increased sample size; for example, there is no evidence for a paternal age effect on nondisjunction of chromosome 21. However, identification of risk factors related to recombination remains inconclusive. There is no strong evidence for absence of recombination to be a risk factor for MI errors. Also, we were unable to detect any evidence that altered placement of exchange is associated with paternal MII errors. As it relates to the sex ratio among paternal cases of trisomy 21, we concluded that paternal cases of free trisomy 21 cannot solely account for the excess of males that is observed among all cases of trisomy 21. We suggest that selection may play a role in the excess of males among individuals with trisomy 21.
Footnotes
How to Cite this Article:
Oliver TR, Bhise A, Feingold E, Tinker S, Masse N, Sherman SL. 2009. Investigation of factors associated with paternal nondisjunction of chromosome 21.
Am J Med Genet Part A 149A:1685–1690.
REFERENCES
- Antonarakis SE. Parental origin of the extra chromosome in trisomy 21 as indicated by analysis of DNA polymorphisms. Down Syndrome Collaborative Group. N Engl J Med. 1991;324:872–876. doi: 10.1056/NEJM199103283241302. [DOI] [PubMed] [Google Scholar]
- Antonarakis SE, Petersen MB, McInnis MG, Adelsberger PA, Schinzel AA, Binkert F, Pangalos C, Raoul O, Slaugenhaupt SA, Hafez M, et al. The meiotic stage of nondisjunction in trisomy 21: Determination by using DNA polymorphisms. Am J Hum Genet. 1992;50:544–550. [PMC free article] [PubMed] [Google Scholar]
- Bernheim A, Chastang C, de Heaulme M, de Grouchy J. Excess of males in trisomy 21 (author's transl). Ann Genet. 1979;22:112–114. [PubMed] [Google Scholar]
- Canfield MA, Honein MA, Yuskiv N, Xing J, Mai CT, Collins JS, Devine O, Petrini J, Ramadhani TA, Hobbs CA, Kirby RS. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Burdick JT, Hirschmann D, Morley M. Polymorphic variation in human meiotic recombination. Am J Hum Genet. 2007;80:526–530. doi: 10.1086/512131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SB, Allen EG, Oxford-Wright CL, Tinker SW, Druschel C, Hobbs CA, O'Leary LA, Romitti PA, Royle MH, Torfs CP, Sherman SL. The National Down Syndrome Project: Design and implementation. Public Health Rep. 2007;122:62–72. doi: 10.1177/003335490712200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DK, Abruzzo MA, Millie EA, Sheean LA, Feingold E, Sherman SL, Hassold TJ. Non-disjunction in human sperm: Evidence for an effect of increasing paternal age. Hum Mol Genet. 1995;4:2227–2232. doi: 10.1093/hmg/4.12.2227. [DOI] [PubMed] [Google Scholar]
- Hassold T, Sherman S, Hunt PA. The origin of trisomy in humans. Prog Clin Biol Res. 1995;393:1–12. [PubMed] [Google Scholar]
- Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, Millie E, Saker D, Shen J, Zaragoza M. Human aneuploidy: Incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–175. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Huether CA, Martin RL, Stoppelman SM, D'Souza S, Bishop JK, Torfs CP, Lorey F, May KM, Hanna JS, Baird PA, Kelly JC. Sex ratios in fetuses and liveborn infants with autosomal aneuploidy. Am J Med Genet. 1996;63:492–500. doi: 10.1002/(SICI)1096-8628(19960614)63:3<492::AID-AJMG15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Freeman SB, Savage-Austin A, Pettay D, Taft L, Hersey J, Gu Y, Shen J, Saker D, May KM, Avramopoulos D, Petersen MB, Hallberg A, Mikkelsen M, Hassold TJ, Sherman SL. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. doi: 10.1038/ng1296-400. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Feingold E, Savage A, Avramopoulos D, Freeman S, Gu Y, Hallberg A, Hersey J, Karadima G, Pettay D, Saker D, Shen J, Taft L, Mikkelsen M, Petersen MB, Hassold T, Sherman SL. Characterization of susceptible chiasma configurations that increase the risk for maternal nondisjunction of chromosome 21. Hum Mol Genet. 1997a;6:1391–1399. doi: 10.1093/hmg/6.9.1391. [DOI] [PubMed] [Google Scholar]
- Lamb NE, Feingold E, Sherman SL. Estimating meiotic exchange patterns from recombination data: An application to humans. Genetics. 1997b;146:1011–1017. doi: 10.1093/genetics/146.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews TJ, Hamilton BE. Trend analysis of the sex ratio at birth in the United States. Natl Vital Stat Rep. 2005;53:1–17. [PubMed] [Google Scholar]
- Mikkelsen M, Poulsen H, Nielsen KG. Incidence, survival, and mortality in Down syndrome in Denmark. Am J Med Genet. 1990;37:75–78. doi: 10.1002/ajmg.1320370714. [DOI] [PubMed] [Google Scholar]
- Nielsen J, Jacobsen P, Mikkelsen M, Niebuhr E, Sorensen K. Sex ratio in Down syndrome. Ann Genet. 1981;24:212–215. [PubMed] [Google Scholar]
- Oliver TR, Feingold E, Yu K, Cheung V, Tinker S, Yadav-Shah M, Masse N, Sherman SL. New insights into human nondisjunction of chromo-some 21 in oocytes. PLoS Genet. 2008;4:e1000033. doi: 10.1371/journal.pgen.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MB, Antonarakis SE, Hassold TJ, Freeman SB, Sherman SL, Avramopoulos D, Mikkelsen M. Paternal nondisjunction in trisomy 21: Excess of male patients. Hum Mol Genet. 1993;2:1691–1695. doi: 10.1093/hmg/2.10.1691. [DOI] [PubMed] [Google Scholar]
- Savage AR, Petersen MB, Pettay D, Taft L, Allran K, Freeman SB, Karadima G, Avramopoulos D, Torfs C, Mikkelsen M, Hassold TJ, Sherman SL. Elucidating the mechanisms of paternal non-disjunction of chromosome 21 in humans. Hum Mol Genet. 1998;7:1221–1227. doi: 10.1093/hmg/7.8.1221. [DOI] [PubMed] [Google Scholar]
- Takaesu N, Jacobs PA, Cockwell A, Blackston RD, Freeman S, Nuccio J, Kurnit DM, Uchida I, Freeman V, Hassold T. Nondisjunction of chromosome 21. Am J Med Genet. 1990;37:175–181. doi: 10.1002/ajmg.1320370735. [DOI] [PubMed] [Google Scholar]
- Yoon PW, Freeman SB, Sherman SL, Taft LF, Gu Y, Pettay D, Flanders WD, Khoury MJ, Hassold TJ. Advanced maternal age and the risk of Down syndrome characterized by the meiotic stage of chromosomal error: A population-based study. Am J Hum Genet. 1996;58:628–633. [PMC free article] [PubMed] [Google Scholar]