Abstract
Cells respond to a variety of stimuli, including biochemical, topographical and mechanical signals originating from their micro-environment. Cell responses to the mechanical properties of their substrates have been increasingly studied for about 14 years. To this end, several types of materials based on synthetic and natural polymers have been developed. Presentation of biochemical ligands to the cells is also important to provide additional functionalities or more selectivity in the details of cell/material interaction. In this review article, we will emphasize the development of synthetic and natural polymeric materials with well-characterized and tunable mechanical properties. We will also highlight how biochemical signals can be presented to the cells by combining them with these biomaterials. Such developments in materials science are not only important for fundamental biophysical studies on cell/material interactions but also for the design of a new generation of advanced and highly functional biomaterials.
1. Introduction
Our body contains several types of tissues (skin, bone, cartilage…) whose mechanical and biochemical properties depend on their composition. Tissues are composed of cells embedded within an extracellular matrix (ECM) made of proteins, polysaccharides, and other bioactive molecules such as growth factors. The field of tissue engineering, which consists in recreating new tissues by means of a combination of engineering, cell biology and materials, was pioneered about 18 years ago by Langer and colleagues from MIT.1 A goal of biomaterials scientists is to design biocompatible scaffolds in which cells can adhere, proliferate, differentiate and synthesize their own matrix to regenerate tissue. Molecules promoting cell adhesion have already been included in the design of biomaterials, as it is known that many cells need to adhere for their survival.2
More recently, other parameters like mechanical properties of biomaterials3,4 and delivery of growth factors5 have also been taken into account. On the other hand, biophysicists have long been studying the process of cell adhesion6,7 and the cell’s mechanical properties. More recently, cell aggregates and tissues have been studied.8 To this end, several characterization techniques have been adapted to soft biological materials, including micromanipulation, microrheology9 and nano-indentations.
How the cells exert forces on to a substrate and how these forces are transmitted at the molecular level inside the cells are key questions, which have been and are still being investigated. Such questions are tackled by a wide range of investigators, from a cell biology point of view to a mechanical point of view.7,10
This has also led to the development of new materials that would, ideally, make possible independent variation in mechanical and biochemical properties. If surface properties of the materials are taken into consideration, they are viewed as 2D materials and cells will interact with them from their basal side. In the case of hydrogel materials, their bulk (volumic) properties are important, as the cells embedded in the hydrogel are fully surrounded by it.
We are now entering a new era where the 3rd dimension is more and more taken into account. In this context, measurements of forces in 3D are starting to be measured. However, it is important to underline that both 2D and 3D studies of cell/material interactions are required, as these studies will provide complementary information.
Although the two scientific communities of biomaterials scientists and biophysicists have different goals and different experimental approaches, they nevertheless share a common interest in designing materials with well-defined mechanical and biochemical properties. For biomaterials scientists, these may serve as new scaffolds to control cell fate and tissue regeneration. For biophysicists, they may be used as a toolbox to decorticate and understand the specific effects of different environmental signals on the cell. Until recently, cell biologists commonly used glass substrates or tissue culture polystyrene substrates to investigate cell behavior. Commercial products of model basement membrane-like ECM such as Matrigel are also used and have become popular in cancer cell biology. Matrigel is composed of mainly laminin-111, collagen IV, heparan sulfate proteoglycan, entactin/nidogen, and various growth factors (fibroblast growth factor, transforming growth factor beta, epidermal growth factor, etc.) but is poorly defined. Even though it contains natural biomolecules, it cannot be used to identify the role of specific parameters on cell behavior and to modulate them in a controlled manner.
In this review, we will be writing from a materials point of view. First, we will give an overview of the different types of materials, including synthetic and natural ones, which have been developed for their tunable physical/mechanical and biochemical properties (Table 1).
Table 1.
Summary of the main properties of natural and synthetic materials, from 2D to 3D materials, which are used in mechano-sensitivity studies. This includes their physical/mechanical and biochemical properties. Their main disadvantages and advantages are also given
| PROPERTIES | NATURAL | SYNTHETIC |
|---|---|---|
| 2D ▲ 3D |
PEM-films | PA gels PDMS |
| IPN Composites | ||
| Fibrin Collagen Hyaluronan Alginate |
PEG | |
| Physical/mechanical properties | - Viscoelasticity - Physical architecture - Porosity (nm to μm scale) - Degradability (proteases) |
- Pure elasticity - No physical architecture - Small porosity - Non biodegradabile (unless grafted with MMP peptides) |
| Biochemical properties | - Non spectfic interactions (electrostatic, H-bonds) - Specific (natural ligands) |
- Inertness - Need grafting with ligands |
| Main disadvantage | - Difficulty to decouple mechanics and chemistry | - High swellability (for PEG) - Stability over time |
| Main advantage | - Biomimetism (natural presence in tissues) | - Versatility of the control |
We will focus on the advances made in the design of 2D and 3D polymeric materials with well-defined mechanical and biochemical properties (Fig. 1). We will discuss the range of mechanical properties, depending on type and composition of the material.
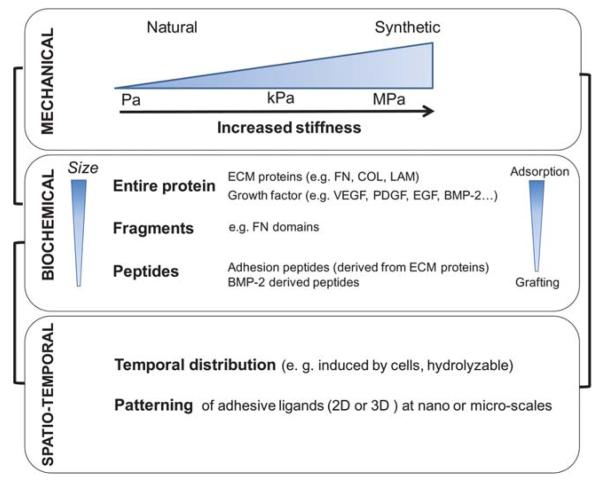
Fig. 1.
Scheme presenting the possibilities of control of the cell micro-environment using engineered materials: mechanical properties with typical variation in elastic moduli from a few Pa to tens of MPa; biochemical properties obtained by adsorbing or grafting entire proteins, protein fragments as well as peptides; spatio-temporal properties, e.g. hydrolytically degradable materials or controlled presentation of ligands by nano and micropatterning.
We will also present different ways of providing them a biochemical functionality. Two main strategies of functionalization are usually employed: covalent coupling or physical adsorption of the bioactive molecules (entire proteins, fragments or peptides). The coupling strategy is often required for synthetic materials, which do not have any natural interaction with biomolecules. Conversely, natural materials that exhibit low and high affinity interactions with ECM proteins and growth factors, can be favorably exploited to present these stimuli.
The third important aspect concerns spatio-temporal properties of the materials, especially spatial control of stiffness or of ligand presentation. For more information on these aspects, the reader is referred to very interesting recent reviews, which adopt either a “cell point of view”11 or a “biomaterials point of view”.12
Finally, the paper will end with some concluding remarks and a short outlook.
2. 2D and 3D materials used for mechano-sensitivity studies
Polymeric materials have been developed both by biophysicists and biomaterial scientists. Controlling and modulating their biochemical and mechanical properties is one of the current challenges, ideally aimed to achieve simultaneous and independent control of each of these properties. First, we should mention that polymeric materials have mechanical properties that are somewhat difficult to compare, due to the various methods used to measure them. Each of these methods, including dynamic shear rheology, dynamic compression for hydrogels and nano-indentations for films, is well-suited to a given type of material. The Young’s modulus (E0) is most often measured by traction tests or nano-indentations. The elastic and viscous moduli (G′, G″) of soft materials are instead measured by oscillatory shear rheology. However, a close look at all values measured for various materials indicates that E0 or G′ lie in the range of a few Pa to hundred MPa, depending on the material (Fig. 2A). Indeed, this is in the physiological range of cell and tissue stiffnesses (Fig. 2B). It has to be noted that, very often, the strategies employed for modulating mechanical properties also involve changes in the nature or density of the chemical bonds within the materials. Unfortunately, it is simply impossible to fully decouple both. The main strategies for creating ionic or/and covalent crosslinks in polymeric materials are summarized in Fig. 3. Incorporation of nano-objects has also been shown to stiffen a polymeric material. However, this method has never been applied in the context of the 2D or 3D materials used for cell mechano-sensitivity studies. Here, we will distinguish between synthetic materials and natural materials, which are made of naturally occurring biomolecules. The main physical/mechanical and biochemical properties, advantages and drawbacks of these two types of materials are summarized in Table 1.
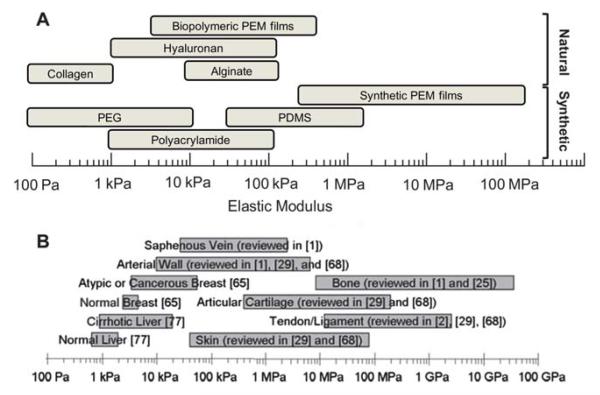
Fig. 2.
(A) Range of stiffnesses of the different synthetic and natural materials that are currently employed for mechanosensitivity studies, which are presented in this review. These include both synthetic and natural 2D and 3D materials. (B) Range of stiffnesses found in selected human tissues (from ref. 12).
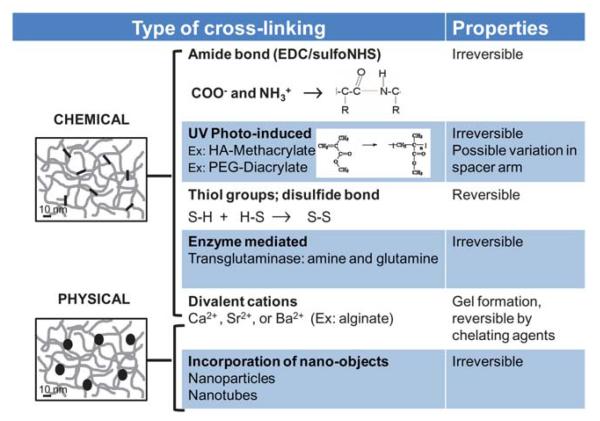
Fig. 3.
Overview of the main strategies used to modulate mechanical properties of synthetic and natural materials. The methods are essentially based on chemical cross-linking, as physical cross-linking is so far barely employed for biomaterials. We have classified cross-linking by divalent cations at the border between chemical and physical cross-linking, as addition of cations changes the film chemistry but, at the same time, induces a physical gelation (no need for covalent crosslinks).
2.1 Synthetic polymeric materials
Synthetic polymers can be tuned in terms of composition, rate of degradation, mechanical and chemical properties. There are four major types of polymers that are used in mechano-sensitivity studies (Fig. 2 and Table 1). Three of them are mostly employed as 2D culture substrates, e.g. polyacrylamide (PA), polydimethylsiloxane (PDMS) and polyelectrolyte multilayer films made of synthetic polyelectrolytes, whereas the fourth, poly (ethylene glycol) (PEG), is used as a 3D hydrogel with cells embedded in it. We will present below the design strategies for each of these.
2.1.1 Polyacrylamide hydrogels
PA gels, initially used by biologists for protein electrophoresis, have been used for about 14 years for mechano-sensitivity studies.3 Their stiffness can be adjusted by varying the molar fraction of the bis-acrylamide cross-linker. Cross-linking can be induced by chemicals such as ammonium persulfate to trigger a free radical-dependent polymerization of double bonds (vinyl groups) in the otherwise stable acrylamide and bis-acrylamide monomers. Alternately, cross-linking can be photo-induced (Irgacure is commonly used), leading to E0 in the range of 10 to 100 kPa. PA hydrogels are relatively simple in their mechanics, and have been extensively characterized by other traditional techniques, including bulk tensile loading, microindentation, and rheology.3,13 Classic theory of rubber elasticity would predict that the elastic modulus of a polymer gel scales linearly with cross-linker concentration, which is fairly well validated by experimental measurements.14 For this reason, PA gels are very popular and have been extensively used to investigate the mechanical effects on cell morphology, adhesion, migration and differentiation in 2D cell cultures.4,15,16
The chemical inertness of polyacrylamide is one of its greatest advantages and disadvantages. PA doesn’t promote any specific cell adhesion when used as a culture substrate, and ligands have to be grafted to control adhesive interactions, which is a clear advantage when demonstrating the mechanical contribution of a given receptor or adhesive protein. Unfortunately, the degree of inertness is also a limitation, as the chemical nature of the PA does not allow easy covalent attachment.
There are additional possibilities offered by PA gels. First, in case of photo-polymerized gels, gradients in mechanical properties can be created by illuminating the gel using photomasks.17 Second, as cells cannot be entrapped in 3D in PA gels due to the toxicity of acrylamide, a simple approach that involves sandwiching cells between two polyacrylamide hydrogels has been proposed.18,19 This method does not fully embed the cells in the environment but it does engage at least part of the dorsal cell receptors, thus mimicking the native 3D environment. Furthermore, it makes it possible to manipulate compliance and measure traction forces. Indeed, microparticles of well-defined sizes can be inserted into PA gels, allowing cell traction forces to be measured, provided that the Young’s modulus and the deformation regime of the gels are known.20 For this reason, PA gels are by far the most commonly used substrate for quantification of traction forces.21
2.1.2 PDMS
PDMS has arisen from the development of soft lithography. As PDMS is elastically deformable, non toxic and exhibits excellent optical properties, it has become a material of choice to stretch cells in controlled conditions. PDMS is always used as a 2D culture substrate (i.e with cells grown at its surface): this material it too dense for the cells to migrate through in 3D. In addition, it is a non-degradable material and cannot be remodeled by cells. To prepare PDMS, a “base” and a “curing agent”, which contains monomers and is also named “cross-linker”, are typically mixed in a 10 : 1 w : w ratio. Thus, to prepare substrates with different elastic moduli, the silicone elastomer base and the cross-linker can be mixed at various ratios, forming gels from 50 kPa to 1.7 MPa.22–24 However, PDMS exhibits uncontrolled protein adsorption and can sometimes cause non-specific cell adhesion, depending on its surface properties and cell type. Thus, PDMS surface needs to be chemically modified by various cell adhesion molecules to induce more reproducible adhesion.22,23 Various strategies have been developed for this purpose (see paragraph 3.1).
2.1.3 Polyelectrolyte multilayer films made of synthetic polymers
Polyelectrolyte multilayer films are a new kind of self-assembled material that emerged almost 20 years ago25 and has versatile properties depending on the assembly conditions and post-assembly treatments. The thickness of these films can be adjusted in the range of few nm to several μm by varying the deposition conditions (pH, ionic strength and concentration of the polyelectrolytes) and the polyelectrolyte pairs. Using pH-dependent assembly of poly(acrylic acid)/poly(allylamine) (PAA/PAH), Van Vliet et al. evidenced that such films can exhibit elastic moduli E from 200 kPa to 142 MPa (measured by nano-indentation), which is as much as one thousand-fold more compliant than tissue-culture polystyrene.26 Extremely stiff films with a high degree of ionic cross-links are obtained at neutral pH whereas soft films are obtained when films are built in acidic pH.27,28
A different strategy was proposed by Senger et al.,29 who prepared a composite film made of a first (PLL/HA)24 stratum capped by a second (PSS/PAH)n stratum (n varying between 0 and 12). As the (PSS/PAH) films were much stiffer, the progressive deposition of these layers rendered the composite film stiffer, from roughly 50 to 500 kPa. An apparent elastic modulus was estimated from elasticity measurements by modeling the different strata. These films were recently used to investigate whether substrate elasticity has an effect on nuclear processes such as replication and transcription.30
2.1.4 PEG-based
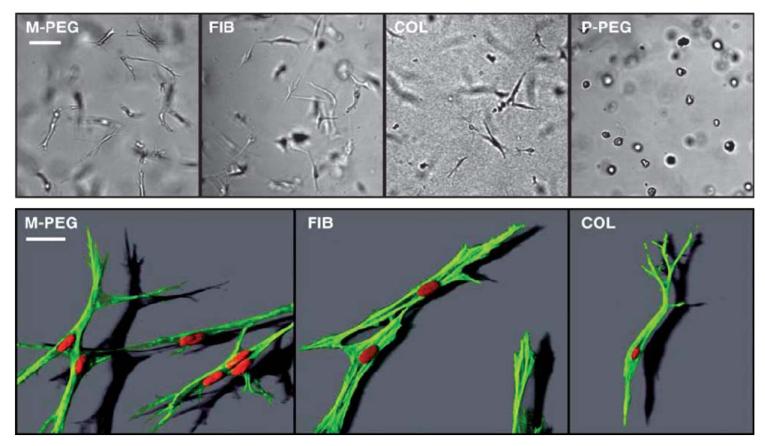
To date, there are very few synthetic materials with controlled mechanical properties that can be used for 3D cell studies, because of their high density, low porosity and lack of biodegradability. PEG-based hydrogels with well-controlled molecular properties have been developed for this purpose. These synthetic hydrogels are biologically inert and, as such, they often require the insertion of adhesion peptides during polymerization.31 Typically, the hydrogels are formed by Michael-type addition of PEG-diacrylate (PEG-DA) and of thiol-containing peptides on to multiarm PEG-Vinyl sulfone (VS). These gels are very sensitive to their preparation conditions including pH, stoichiometry, precursor concentration, chain length and number of arms of the macromers.31 PEG gels are known to swell greatly when introduced in solution, with the equilibrium swelling ratio ranging from 10 to 70 and elastic moduli from 0 to 6 kPa as determined by small strain oscillatory shear measurements.31 These parameters were found to be connected by a power law, with more swelling for softer gels. Of note, these gels have a very low viscous component G″ and very low porosity (of around 25 nm) as compared to physical hydrogels such as fibrin or collagen.32 This low porosity renders them resistant to cell migration, as incorporated cells essentially have an isotropic continuum without sensible physical architecture. Subsequent developments of these gels have included the grafting of protease-sensitive peptides (sensitivity to matrix metallo-proteases) with bifunctional groups to be grafted at both of their extremities.33 Thus, these materials are locally degraded in response to cell–surface proteases, allowing cells to create paths for 3D migration (Fig. 4).
Fig. 4.
Representative images of human neonatal foreskin fibroblasts (HFFs) cultured within the four materials: two biopolymers, namely collagen I (COL, at 2 mg mL−1) and fibrin (FIB, at 2 mg mL−1) and PEG hydrogels that possess matrix metalloprotease sequence (MMP-sensitive PEG hydrogels, M-PEG) or plasmin-sensitive PEG hydrogels (P-PEG). Upper row: brightfield images; Lower row: confocal images obtained after staining for F-actin (rhodamine-phalloidin, green) and nuclei (DAPI, red). Scale bars, 100 μm (upper row) and 30 μm (lower row). Of note, the MMP sensitive cross-linker allows HFFs to spread and attain cell shapes in synthetic M-PEG gels (M-PEG) very similarly to HFFs in biopolymers (FIB, COL) (images of the bottom row). In contrast, HFFs are not able to form a spindle-shaped morphology in plasmin-sensitive PEG hydrogels (P-PEG) as seen by the increased compactness and a decrease in projected cell area (from ref. 32, copyright Cell Press).
2.2. Natural materials
Natural biopolymers have the advantage of being components of native ECM matrices, i.e. they provide compositional uniqueness such as stimulating a specific cellular response and serve both as mechanical as well as biochemical signals. Natural materials are also particularly interesting due to their unique structural properties (Table 1). Their nano and microstructure are similar to that of native tissues in terms of functional groups, backbone (presence of neutral and charged groups, chirality) and structural organization (coils, fibers…). Conversely, natural materials have also some drawbacks. They are more fragile, polydisperse, and not always pure. Moreover, their mechanical properties are often difficult to measure mechanically or rheologically as they can exhibit non linear behaviors. In addition, their natural bioactivity makes it fastidious to fully decouple the effect of mechanics from chemistry. The main biopolymers used to study the effect of substrate stiffness on cell behavior are collagen, alginate, fibrin, and agarose (Fig. 2 and Table 1). 2D PEM coatings made by self-assembly of polypeptides and polysaccharides are emerging as a new class of materials with well-defined properties. The methods employed to cross-link natural materials are similar to those employed for synthetic materials. They are summarized in Fig. 3.
2.2.1 Collagen
Type I collagen is a major protein component of fibrous connective tissues, which provides mechanical support and frameworks for the other tissues in the body. Collagen is a natural ligand for several integrin receptors. Collagen gels exhibiting different mechanical properties can be prepared by varying the pH during hydrogel formation34 or by varying its concentration.35 Thus, porosity as well as density of ligands, which are changed simultaneously, are coupled to the material’s mechanical properties. Elastic modulus G′ of such gels can vary between 5 and 1000 Pa as measured by oscillation rheometry. Collagen gels can be prepared as 2D culture surfaces or 3D matrices.35 They exhibit a rather organized physical architecture characterized by the presence of fibers.
Grinnel et al. recently investigated the effect of 2D and 3D collagen matrices on cell adhesion and migration. They quantified the effects of matrix stiffness and porosity on collagen translocation, fibroblast cell spreading and cell migration for collagen gels with average pore diameter varying from 1.1 to 2.2 μm. Drying collagen fibrils appears to have an impact on cell spreading and proliferation. Plant et al.36 showed that thin films of collagen fibrils can be dehydrated, and when seeded on these dehydrated fibrils, smooth muscle cells spread and proliferate extensively. Indeed, the dehydrated collagen gels were found to be mechanically stiffer than their hydrated counterparts. Tanishita et al. found that in vitro formation of microvessel networks by endothelial cells was also affected by the mechanical properties of collagen gels.34
Microbial transglutaminase, an enzyme that catalyzes the formation of a covalent bond between a free amine group (e.g., protein- or peptide-bound lysine) and the gamma-carboxamid group of protein- or peptide-bound glutamine, can also be employed to covalently crosslink collagen I.37 This resulted in a 6-fold increase in G″ (1.3 kPa versus 210 Pa). In terms of cell behavior, these authors showed a significant reduction in the level of cell-mediated contraction of scaffolds with increased concentrations of enzymes.
2.2.2 Alginate
Alginate is a linear polysaccharide of (1–4)-linked β-mannuronic acid and α-guluronic acid monomers, which forms a gel in the presence of certain divalent cations (calcium, strontium, or barium) (Fig. 3).38 The block structure of alginate dictates the structure of ionic cross-links, and covalent cross-links can also be formed.39–41 Due to their biocompatibility, alginate gels have long been used for biomedical purposes, particularly in the manufacture of surgical dressings for exuding wounds. More recently, they were employed as scaffolds for the immunoprotection of transplanted cells. Elastic modulus and toughness can be modulated from 2 to 70 kPa by controlling the parameters for gel cross-linking. However, alginate needs to be chemically modified to interact specifically with mammalian cells, which is usually achieved by grafting RGD (arginine–glycine–aspartic acid)-containing cell adhesion ligands.42 In this context, alginate gels were used to investigate the substrate mechanics effect on chondrocyte adhesion.43 Very recently, the same group demonstrated that the commitment of mesenchymal stem cell populations changes in response to the rigidity of 3D alginate gels, with osteogenesis occurring predominantly at 11–30 kPa.44 Matrix stiffness was found to regulate integrin binding as well as reorganization of adhesion ligands at the nanoscale. Both were traction-dependent and correlated with osteogenic commitment of mesenchymal stem cell populations.
2.2.3 Hyaluronan and other biopolymers
Hyaluronan (HA) is a non-sulfated glycosaminoglycan (GAG) that is present in different types of tissues and fluids, including synovial fluid, cartilage, tendon and skin. It plays a role in tissue viscoelasticity and hydration, due to its ability to interact with water molecules and to establish multiple hydrogen bonds.45 Hyaluronan is also present in the pericellular coat (also called glycocalyx) of different cell types, chondrocytes being a prominent example with a thick coat of ~5 μm.46 Despite their biocompatibility, native HA gels have poor mechanical properties. Although HA can be cross-linked using carbodiimide 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)47 before serving as a soft substrate for cell biology experiments, such hydrogel preparations lack long-term stability and have very low elasticity (3 to 250 Pa). To increase its mechanical properties, the Prestwich group has proposed a method that consists in grafting thiol groups to HA (HA–SH)48 to form disulfide (S–S) bonds in the presence of an oxidizing agent. Adding PEG-DA to the mixture can provide additional cross-links. AFM nano-indentations have been performed on S–S crosslinked HA gels, and moduli were found in the range of 1 to 100 kPa.14 One of the drawbacks of such gels is the dissociation of the S–S bond over several days. Another is that PEG-DA addition leads to the formation of hydrolytically degradable esters, which may balance the stiffening effect of the cross-links. Engler et al. recently compared the growth of pre-cardiac cells on HA and PA gels of similar stiffness.49 They showed that pre-cardiac cells grown on collagen-coated HA hydrogels exhibit a 3-fold increase in mature cardiac specific markers and form up to 60% more maturing muscle fibers than they do when grown on compliant PA hydrogels over 2 weeks.
Other biomolecules are being developed to investigate the effects of substrate mechanics. Bellamkonda et al. used agarose gels at different concentrations (ranging from 0.75 to 2% wt/vol) to investigate the rate of neurite extension,50 which was found to be inversely correlated to the mechanical stiffness of the gels. Soichet et al. have developed cross-linkable forms of chitosan by grafting methacrylate groups.51 These hydrogels have been used for the regeneration of neuronal tissue.
2.2.4 Polyelectrolyte multilayer films made of biopolymers as 2D coatings
As mentioned above, polyelectrolyte multilayer films are a new type of self-assembled coating that has found applications for cell studies in the past 10 years.52 PEM films made of polysaccharides and polypeptides have been engineered and studied.53,54 Their mechanical properties can be modulated by several methods. A now popular method is to covalently cross-link carboxylic with amine groups in the films to form covalent amide bonds. This was first applied to poly(l-lysine)/hyaluronan (PLL/HA) films55 for a fixed cross-linker concentration and subsequently to the same films on a large range of cross-linker concentration.56 The apparent Young’s modulus of the films, as probed by AFM-nano-indentations, could thus be modulated over a range of a few kPa to ~500 kPa. More recently, this strategy has been applied to several other types of multilayer films to obtain mechanical properties that depend on the type of polyelectrolyte pairs, deposition conditions and cross-linker concentrations.57 Recent developments include the investigation of the effect of film cross-linking on hepatocyte adhesion,58 on the differentiation of myoblast cells into myotubes59 and of selective cross-linking on the outer region of the films. This results in a rigid outer “skin” to promote cell attachment, while leaving the film’s interior unaffected.60
Another strategy relies on the use of a natural cross-linking agent such as genipin.61 The viscoelastic properties of chitosan/hyaluronan (CHI/HA) and chitosan/alginate (CHI/ALG) multilayer films without cross-linking or after cross-linking with genipin have been investigated using quartz crystal microbalance with dissipation monitoring (QCM-D). (CHI/HA) cross-linked films proved to be highly non-adhesive for pre-osteoblasts and fibroblastic skin cells. Conversely, cross-linking (CHI/ALG) films dramatically improved pre-osteoblast and rat fibroblastic skin cell adhesion, especially for high bi-layer numbers and using high concentrations of cross-linker. Finally, photo-crosslinking can be employed to modulate the Young’s modulus of (PLL/HA) films that contain a photosensitive derivative of HA (HA-vinyl benzyl) grafted at various percentages.62
2.3. Mixtures of synthetic/natural
As both synthetic and natural biomaterials have advantages and drawbacks, efforts have also been made to develop composite biomaterials made of mixtures of synthetic and natural materials. In this respect, Putnam et al. developed PEG-conjugated fibrinogen gels,63 by coupling PEG-DA to full-length fibrinogen. These gels can be additionally cross-linked by exogenous cross-linkable PEG-DA (typical range from 0 to 2 wt%) and make possible the simultaneous manipulation of mechanical properties and adhesion ligand density presented to cells. Their bulk compressive moduli ranged from 450 to 5.2 kPa.63 However, their mechanical properties decreased over a seven-day immersion period in phosphate buffered saline, and it was probably due to the combined effects of hydrolysis and proteolysis. Adding soluble factors such as ascorbic acid to the gels was found to stimulate matrix remodeling by modulating smooth muscle cells phenotype (induction of contractility), which led to an increase in elastic modulus.64
Semi-interpenetrating hydrogels (IPNs) are an emerging class of hydrogels, which make it possible to combine the advantages of each component. For instance, photo-cross-linkable hyaluronic acid (HA) and semi-interpenetrating collagen components were found to exhibit superior mechanical properties.65 The inclusion of the semi-interpenetrating collagen chains provided a synergistic mechanical improvement over unmodified HA hydrogels. These semi-IPNs supported fibroblast adhesion and proliferation and were shown to be suitable for cell encapsulation at high levels of cell viability. They were also employed to fabricate cell-laden microstructures and microchannels. Another example is that of fibroin/collagen hybrid hydrogels,66 which were prepared by cross-linking a fibroin/collagen solution using the water soluble EDC. G′ of these gels varied between 3 and 10 kPa. Some mobility of fibroin molecules inside the gels was noticed. These composite gels allowed vascular smooth muscle cells to grow.
3. Biochemical functionalization
A common approach in the field of biomaterials is to start from a “blank slate”,67 i.e. a substrate or material preventing protein adsorption and cell adhesion, and to add a biochemical functionality to the material in a controlled fashion. As mentioned above, cell attachment on many synthetic polymers is very poor, due to their inertness and lack of specific adhesive motifs. Such a low background attachment has been observed for PA, PDMS and PEG hydrogels. In the case of natural materials, although cells may possess specific receptors recognizing the material, their naturally high hydration, softness and the possible lack of accessibility for functional groups often render them poorly adhesive. Such low cell attachment has been observed for hydrated polysaccharides such as HA and ALG.
Researchers have thus designed strategies for giving additional biochemical functionality to different types of synthetic and natural materials. We will distinguish here between three types of biochemical functionality (Fig. 1): i) full length ECM proteins, ii) fragments and peptides derived from these proteins and iii) growth factors (GF). Among the important biomolecules are ECM proteins like fibronectin, collagen and laminin as well as GAGs. These GAGs are negatively charged polysaccharides that can interact with proteins by non covalent and covalent interactions.68 In this latter case, they form what is called a proteoglycan. In addition, growth factors are an important class of signaling molecules playing a key role in cellular processes including growth, proliferation, differentiation, adhesion and migration.69
Biochemical functionality can be provided either by grafting or by physically adsorbing the bioactive molecule. Notably, presentation of a biochemical signal from a biomaterial or a substrate in a “matrix-bound” manner is important for mimicking physiological conditions, as many bioactive molecules are bound to the ECM matrix in vivo.70 Indeed, in the ECM, glycosaminoglycans are found immobilized by ionic or covalent cross-links. We will present below the different strategies (grafting versus physical adsorption), how they are achieved and what types of biomolecules have been grafted or adsorbed to date.
3.1. Biochemical functionalization by grafting
The advantage of grafting is that it provides good control of surface composition, a stable link and limits release of the functional group into the culture medium. Covalent grafting of short bioactive peptides or protein fragments is more frequently performed than that of full length ECM proteins, which is more difficult to handle. A key issue is to preserve the bioactivity of the grafted molecules, especially entire proteins, because their activity depends on their 3D conformation. Moreover, using harsh solvents and/or high temperatures often leads to the denaturation of biologically active molecules. Thus, selecting the appropriate conjugation strategy and using spacer arms are essential to retain the bioactivity of grafted molecules and provide them with sufficient flexibility and accessibility by the cell receptors.
3.1.1 Major grafting strategies
Grafting of proteins/peptides can be performed in solution on hydrogel components prior to formation of the 2D or 3D biomaterial or directly at the surface of a biomaterial. This latter strategy can only be performed on 2D biomaterials. As mentioned earlier, a key requirement is to preserve the bioactivity of the biomolecules.
3.1.1.1 Targeting amino and carboxylic groups
Proteins can be coupled to polymers via their amino-groups. To this end, sulfo-SANPAH (sulfosuccinimidyl-6-[40-azido-20-nitrophenylamino]hexanoate) can be employed. It is a heterobifunctional cross-linker containing a photosensitive phenyl azide group on one end and an amine-reactive N-hydroxysuccinimide on the other end. Proteins can thus react via their amine group with sulfo-SANPAH, which can itself react with the gel71 upon exposure to UV light. However, the limited solubility, stability and shelf-life of sulfo-SANPAH have urged researchers to look for alternative grafting strategies. Alternatively, the water soluble carbodiimide coupling chemistry can be employed to create a covalent amide bond, which is formed between activated carboxylic groups and ammonium groups.72 This is, in principle, straightforward coupling chemistry, but several side reactions are known to complicate the subject. These are especially present when the polymer contains a large amount of water, as in the case of HA.73 Therefore, sulfo-N-hydroxysulfosuccinimide ester is often employed to catalyze the reaction. This strategy has been applied to alginate gels, which contain carboxylic groups42 to covalently attach a G4RGDSP oligopeptide so as to promote cell-matrix interactions.74 It has also been used to graft different proteins or fragments (extracted human fibronectin, collagen I or collagen IV; recombinant fragments of fibronectin and vitronectin) on to polyelectrolylte multilayer films containing hyaluronan.75
3.1.1.2 Targeting cysteine residues
Thiol groups of proteins or peptides are another target for coupling reactions. Maleimides linked to PEG are often used as flexible linker molecules to attach whole proteins, protein fragments or peptides to surfaces. The double bond of maleimide readily reacts with the thiol group found on cysteine to form a stable carbon-sulfur bond. Attaching the other end of the polyethylene chain to a bead or solid support, or to a polyelectrolyte76 allows one to separate the protein from other molecules in solution, provided that molecules do not also possess thiol groups.
Acrylate groups are often used in Michael addition, which is a conjugate reaction based on the nucleophilic addition of a carbanion or another nucleophile to an alpha, beta unsaturated carbonyl compound. This is one of the most useful methods for the mild formation of C–C bonds and thus for covalently cross-linking acrylated polymers, usually by light activation. Acrylates are also known to react with thiols of cysteines (on peptides or proteins) under defined experimental conditions. This reaction, which belongs to the thiol-ene family of reactions, involves the addition of an S–H bond across a double or triple bond by either a free radical or ionic mechanism. Thus, acrylates are often employed to graft either peptides or full-length proteins. For example, RGD sequences have been incorporated into PEG hydrogel networks through the acrylation of the peptide sequence at the N-terminus, followed by copolymerization of the acrylated peptides with PEG-DA via photocross-linking in aqueous solution.77–79 On similar PEG hydrogels, bis-cysteine peptides that contain an additional acrylate group and that are MMP-sensitive (e.g., Ac-GCRD-GPQGYIWGQ-DRCG) or plasmin sensitive (e.g., Ac-GC-YKYNRD-CG) have been prepared.32 Burdick et al.80 used an acrylate derivative of HA to graft two peptide components: one to support cell adhesion and the other for proteolytic degradability. Full-length fibrinogen was also coupled to PEG-DA at room temperature but in the presence of a strong denaturating agent (urea).81
The potentiality of acrylate to serve for cross-linking polymeric chains but also for peptide coupling has recently been shown for acrylated HA hydrogels. These were subjected to two step experimental protocol: the first step was designed to couple peptides to the acrylate groups and the second to initiate free radical polymerization of the remaining acrylate groups by exposure to UV light.80 The resulting UV-HA hydrogels were expected to prevent remodeling due to the incorporation of non-degradable covalent cross-links from kinetic chain formation and thus to confine encapsulated cells to a rounded morphology.
3.1.2 Grafting of different types of molecules
As mentioned above, we will distinguish here the three types of molecules – full length ECM proteins, protein fragments or shorter peptides (typically from 4 to 20 amino acids) and GF – that can be grafted. Grafting sequences has great advantages over grafting full length molecules. In entire proteins (ECM proteins or GF), many different active sequences there can be recognized by cell surface receptors. Using a bioactive fragment makes it possible to enhance the specificity of the interaction and to target one particular partner to better control cellular processes. The problem is that such short sequences are usually less bioactive than entire molecules because of the loss of active site spatial architecture owing the protein’s specific conformation.82
3.1.2.1 Peptides
The most common grafted peptides are derived from ECM proteins, mainly fibronectin,83 collagen,84 laminin85,86 and vitronectin87 (Table 2). More recently, peptides that exhibit protease sensitive sequences have been grafted to the biomaterials to add biodegradability in response to cellular activity.88
Table 2.
Peptide sequences used for targeting adhesion receptors of four main ECM proteins (collagen, fibronectin, laminin and vitronectin) as well as for providing degradability (matrix-metalloprotease sequence). The targeted receptor (or receptor family) as well as cell type used in the study are indicated
| Ecm protein | Peptide sequence | Targeted receptor | Cell type | Reference |
|---|---|---|---|---|
| COLLAGEN (Type I) | GFOGER | Integrin α2β1 | Primary bone m arrow stromal cells | Reyes et al., Biomaterials, 200794 |
| CGPKGDRGDAGPKGA | Integrins α1β1,α2β1 | Primary human osteoblasts | Picart et al., Adv. Funct. Mater., 200576 | |
| DGEA | Integrin α2β1 | MIN6 b-cells | Weber et al., Biomaterials, 200795 | |
| FIBRONECTIN (FN) | rhFN fragment FNIII7-10 (with RGD and PHSRN) | Integrin α5β1 | Osteoblasts | Petrie et al., Biomaterials, 200683 |
| rESC, rMSC | Doran et al., Biomaterials, 201075 | |||
| RGD-PHSRN | Integrin α5β1 | Osteoblasts | Benoit and Anseth, Biomaterials, 200592 | |
| LAMININ (LAM) | RKRLQVQLSIRT (α1 chain LAM-1, LG 4 module) | Syndecans | Human dermal fibroblasts, neural PC12 | Hozumi et al., Biomaterials, 200985 |
| ATLQLQEGRLHFX FDLGKGR, X: Nle (α1 chain, LG4 module) | Integrin α2β1 | |||
| PPFLMLLKGSTRFC (LG3 of the lam-5 α3 chain) | Integrins α6β4,α3β1 | Oral keratinocycle cell line TERT-2OKF-8 |
Werner et al., Biomaterials, 200996 | |
| IKLLI (LAM α1 chain) | Integrins α3β1 | MIN6 b-cells | Weber et al., Biomaterials, 200795 | |
| IKVAV (LAM α1 chain) | 110 kDa laminin receptor protein | |||
| YIGSR (LAM β1 chain) | 67 kDa lamminin receptor protein | |||
| VITRONECTIN | rhVN, N-terminal Somatomedin B and RGD domain |
Plasminogen activator inhibitor-1 (PAI-1), integrin receptors | hESC | Doran et al., Biomaterials, 201075 |
| Multiple ECM proteins | RGDSPC | Integrins | MC3T3-E1 preostoblasts | Zouani et al., Biomaterials, 2010101 |
| Human foreskin fibroblasts | Lutolf et al., Nat. Biotechnol., 200384 | |||
| G4RGDSP | Integrins | Primary human bone marrow stromal cells, MC3T3-E1 preosteoblasts, mouse bone marrow stromal D1 cell line | Hsiong et al., Tissue Eng., 200990 | |
| Cyclic RGD: G4CRGDSPC | Integrin receptors, higher speciality for αVβ3 | |||
| MMP-sensitive peptide: Ao-GCRD-GPQGIWGQDRCG-NH2 | Matrix metaloproteinases (MMP) | Human foreskin fibroblasts | Lutolf et al., Nat. Biotechnol., 200384 |
The tripeptide sequence RGD is very popular, as it is present in many ECM proteins, including fibronectin, vitronectin, fibrinogen, von Willebrand factor, thrombospondin, laminin, osteopontin, bone sialo protein, and some collagen isoforms.82 It binds to a wide range of integrin receptors in a non selective manner, i.e. not specific to a given integrin receptor. The literature about the various forms of RGD peptides is rich and the reader is referred to more specialized reviews.89 To achieve better selectivity and/or target only one type of integrin receptor, several strategies have been investigated: i) synthesis of cyclic peptides,90 or peptide multimerization to enhance avidity with particular cell adhesion receptors,91 ii) using a more selective peptide sequence that is not based on RGD but contains other key sequences or iii) associating two different bioactive peptides derived from the same ECM protein92 or from different ones93 (Table 2). Thus, collagen-mimetic peptides,94,95 laminin-derived peptides85,86,96 and fibronectin-derived peptides or fragments83,92 are increasingly used for their higher selectivity.
Garcia et al.97 engineered polymer brushes of oligo(ethylene glycol) methacrylate on PDMS, which resisted biofouling and prevented cell adhesion. These polymer brushes were functionalized to display bioadhesive peptides, which were either tethered uniformly or constrained to micropatterned domains using standard peptide chemistry approaches. Benoit and Anseth92 showed that associating an RDG-containing peptide to another fibronectin-derived epitope like PHSRN not only made it possible to enhance the bioactivity of the functionalized surface compared to RGD only, but also to specifically target a particular integrin receptor α5β1. Each domain independently contributed little to binding, but when combined, they synergistically bound to α5β1 to provide stable adhesions.92,98
We are now progressively entering a new era, where peptides with higher specificity, high biological activity as well as targeting other receptors than integrins are being designed (Table 2). Indeed, it is now acknowledged that besides integrin receptors, other families of receptors including syndecans99 and growth factor receptors play key roles in early cellular events. Recent developments also include grafting the peptide sequence of growth factors, mostly bone morphogenetic protein 2 (BMP-2) derived peptides.100,101
3.1.2.2 Grafting ECM proteins to synthetic surfaces
Synthetic polymers such as PA and PDMS are often biofunctionalized by grafting proteins. For PA gels, three major methods, which are reviewed in,71 are commonly used. The first relies on carbodiimide coupling of proteins to poly(acrylic acid), which has to be inserted into the PA gel during gel formation. Another method uses molecules that have bi-functionality, one end of the molecules mediating the incorporation into polyacrylamide whereas the other end is reactive toward primary amines. Here again, we find acrylate and N-hydroxysuccinimide in the form of acrylic acid N-hydroxysuccinimide ester (NHS-acrylate), which is incorporated into a one-step polymerization reaction102 during the acrylamide polymerization reaction.
Recent developments include the fabrication of a synthetic interfacial hydrogel culture system, termed variable moduli Interpenetrating Polymer Networks (vmIPNs).103 The principle is to build at the first step a polyacrylamide gel by varying the concentration of acrylamide and bisacrylamide monomers to synthesize PA gels from 10 Pa to 10 kPa and of low swelling ratio (~2). Then, the IPN is created by polymerizing a second layer of amino-PEG (4 nm thick) within the top few nanometres of the first acrylamide layer for subsequent grafting of adhesion peptides. Such materials were then used to investigate the adhesion, proliferation and differentiation of adult neural stem-cells. Under mixed differentiation conditions with serum, softer gels were found to favor neurons, whereas harder gels promoted glial cultures.
PDMS, if untreated, exhibits high hydrophobicity and extremely low cell attachment.23 Different methods have thus been developed to biochemically modify the surface of PDMS for cell adhesion. Recently, Hinz et al. systematically compared the immobilization of cell-adhesive molecules to PDMS using electrostatic (simple protein adsorption and layer-by-layer deposition) and covalent surface coating procedures.104 They developed a functionalization protocol that is based on: (1) PDMS oxidation by oxygen plasma treatment, (2) binding of 3-aminopropyltriethoxysilane (APTES) to the oxidized surface and (3) covalent cross-linking of ECM proteins to the silane using glutaraldehyde. They found that the covalent linkage of adhesive molecules was superior to non-covalent methods in providing a coating that resisted to major deformations and that fully transmitted this stretch to cultured cells.
3.1.2.3 Grafting growth factors
There are only a few examples of covalent immobilization of entire growth factors on materials whose mechanical properties can also be modulated. One of the most studied “tethered” growth factor is epidermal growth factor (EGF).105 EGF plays an important physiological role in the maintenance of oro-esophageal and gastric tissue integrity. It was initially tethered to poly(methyl methacrylate)-graft-poly-(ethylene oxide) (PMMA-g-PEO) amphiphilic comb copolymers by activation with 4-nitrophenyl chloroformate (NPC) to target the N-terminal amine of murine EGF.105,106 In the latest work by this group,106 a biotinylated recombinant protein containing the 53 amino acid human EGF domain was linked to a biotonylated peptide hydrogel by neutravidin. This EGF-containing recombinant protein also contained a protease-resistant 20 amino acid hydrophilic spacer arm to provide optimal bioactivity. Another strategy consists in synthesizing photoreactive EGF via the reaction of primary amine groups in the growth factor with the N-hydroxysuccinimide functionality of Sulfo-SANPAH.107 In a subsequent step, EGF is covalently tethered to polystyrene by means of UV irradiation.
Nerve growth factor (NGF) has been grafted to 2-hydroxyethy methacrylate (HEMA) gels using ethylene dimethacrylate as crosslinker, ammonium persulfate as initiatior and tetramethy ethylenediamine (TEMED) as accelerator.108 By modifying these p(HEMA)-NGF gels with pAA, neuronal PC12 cells adhered and responded to the immobilized NGF by extending neuritis in a manner similar to that which is observed with soluble NGF. PC12 cell neurites were even observed to be thicker when cultured on immobilized NGF than when cultured in the presence of soluble NGF.
Very recently, vascular endothelial growth factor (VEGF) has been coupled to a PEG-hydrogel through photo-polymerization via laser scanning lithography.109 Endothelial cell cultures in these gels underwent accelerated tubulogenesis forming endothelium tubes that possess lumens only in the presence of tethered VEGF.
3.2. Biochemical functionalization by physical adsorption
The complex environment surrounding the cells in vivo is composed of ECM components (fibrillar proteins, proteoglycans, adhesion molecules) and soluble biomacromolecules such as cytokines, growth factors and other signaling molecules. Many of these biomolecules interact by non convalent interactions, including electrostatic, Van der Waals, hydrogen bonds and hydrophobic interactions, but also by ligand/receptor interactions. In vivo, these biomolecules are often presented by the ECM proteins or glycosaminoglycans in a “matrix-bound” manner. Thus, biomaterial scientists are also trying to associate different components of the in vivo cellular microenvironment to reproduce it in a simple way and to create biologically active materials. Three principal types of molecules or their fragments can be physisorbed on 2D biomaterials or entrapped in 3D biomaterials: ECM polysaccharides (glycosaminoglycans), ECM proteins (fibronectin, collagen, laminins) and growth factors (including EGF, VEGF, BMP-2 and fibroblast growth factor FGF2). In this part, we focus on non covalent interactions between proteins and polymers.
3.2.1 Adsorption of ECM proteins and of glycosaminoglycans
Due to the natural interactions between ECM proteins and natural polyelectrolytes, ECM proteins are often simply adsorbed on PEM films. Several parameters, including the amount of adsorbed protein (often in the range of several ng cm−2), the strength of the interaction (affinity) as well as protein conformation will depend on the physical and chemical parameters of the multilayer film: type of functional groups (sulfate, carboxylic, ammonium…), pH and ionic strength used during film buildup, and type of ending layer
Fibronectin (FN) is often used as an adhesive protein, due to its interaction with different types of integrin receptors. Wittmer et al.,110 investigated the LbL formation of films composed of PLL and dextran sulfate (DS), the adsorption of FN on to these films, and the subsequent spreading behavior of human umbilical vein endothelial cells. Overall, the FN-coated PLL monolayer and the FN-coated PLL-terminated multilayer were the best performing films in promoting cell spreading. They concluded that the presence of FN is an important factor (more than film charge or layer number) in controlling the interaction between cells and multilayer films. Semenov et al.111 also adsorbed or chemically coupled FN on to (PLL/HA) cross-linked films and demonstrated that film cross-linking strongly influenced FN surface distribution, leading to denser presentation of adhesion sites for cells.
Chen et al.28 modified synthetic (PAH/PAA) PEM films with type I collagen and the proteoglycan decorin. They showed that this did not alter substrate stiffness, but enhanced the retention of spheroids on surfaces and stabilized hepatic functions (such as albumin and urea secretion). Very interestingly, decorin was found to exhibit unique compliance-mediated effects on hepatic functions, down-regulating the hepatocyte phenotype when presented on highly compliant substrata, while up-regulating hepatocyte functions when presented on increasingly stiffer substrata. Collagen adsorption was also found to be important for the attachment and function of adult rat hepatocytes on cross-linked (PLL/ALG) and (PLL/PGA) multilayer films.58 Collagen112 and fibronectin can even be used as building blocks for layer-by-layer film buildup.
Besides ECM proteins, GAGs are more and more often used as main component of new biomimetic coatings and nano-particles, which were reviewed recently.54,113 They are often simply adsorbed and interact by non specific and/or specific interactions with other positively charged biomolecules.68
3.2.2 Adsorption of growth factors on thin films or matrices
Due to their utmost importance in signaling processes, GF are now often inserted into 2D and 3D biomaterials to achieve a specific function: to regulate cell proliferation for FGF2,114 or promote the formation of new vessels for VEGF,109 induce bone regeneration for BMP-2 or chondrogenesis for TGF-β1. There are several high affinity partners of growth factors among biological materials. Fibrin, a non globular fibrous protein, is involved in a large number of biological processes (blood clotting by polymerization of fibrin, signal transduction, platelet activation) and is a very interesting candidate for growth factor immobilization. Thus, natural interactions of fibrin with FGF, BMP-2 and EGF have been used to present these growth factors in their immobilized form115–117 (Fig. 5). Desorption experiments using radiolabeled proteins demonstrated that the patterns were retained in vitro with less than a 30% loss of growth factor over 9 days,116 which confirms the high affinity of growth factors for fibrin.
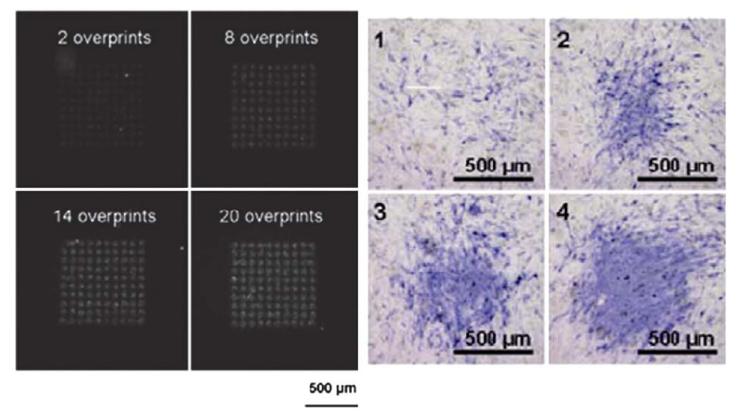
Fig. 5.
Bioprinting of BMP-2 growth factor, which is here fluorescently labeled, on a fibrin film. Left image: BMP-2 was printed at various surface concentrations by varying the number of overprints, leading to increased fluorescence on the surface. The four images correspond to, respectively: 2, 8, 14 and 20 overprints. Right images: corresponding expression of the bone marker alkaline phosphatase (in blue), which is expressed during differentiation of muscle derived stem cells in bone (from ref. 117, copyright AlphaMed Press).
The 12th–14th type three repeats of fibronectin (FN III12-14) also appear to be a natural affinity fragment for several GFs.118 In a recent study, Hubbell et al.118 showed by surface plasmon resonance that this FN fragment binds to GFs from different families, including most of the GFs from the platelet-derived growth factor (PDGF, VEGF and FGF families and some GFs from the TGF-β and neurotrophin families). Affinities were high in the nanomolar range, without inhibiting GF activity. These authors subsequently employed a fibrin-bound variant of FN III12-14 as 3D biomaterials and showed that it was highly effective as a GF delivery system. For instance, in matrices functionalized with FN III12-14, PDGF-BB-induced sprouting of human smooth muscle cell spheroids was greatly enhanced.
Other natural high affinity binding domains are those derived from heparin. Recently, Chow et al.119 created a self-assembled bioactive hierarchical membrane functionalized with a heparin-binding peptide amphiphile (HBPA). HBPA contains a consensus sequence to bind and display heparin loops on the surface of nanofibers in order to localize and activate potent angiogenic growth factors through their respective heparin-binding domains. Both VEGF and FGF2 are known to have heparin-binding domains along with potent angiogenic activity. Release of these factors was effectively lower in the presence of heparin in the membrane.
Polyelectrolyte multilayer films can also be employed to sequester growth factors and present them to cells in a “matrix-bound” manner. VEGF could be adsorbed on to (PAH/PSS)4 films and was shown to exhibit a specific bioactivity toward endothelial cells.120 (PLL/HA) crosslinked films or (PLL/HA-heparin) films can be used as a reservoir for BMP-2 delivery and controlled differentiation of myoblasts to osteoblasts.121,122 Very interestingly, these films are made of natural components, are biodegradable, and their mechanical properties can also be modulated.
4. Concluding remarks and outlook
Different types of synthetic and natural materials in various forms (thin films or gels) have been and are currently being developed to reproduce in vitro the in vivo cell microenvironment. Here, we have particularly focused on materials mechanical and biochemical properties. Biochemical functionalization has up to now mostly focused on improving cell adhesion by presenting cell adhesive ligands. However, many ligands are not highly specific and do not target a particular receptor type. Recent developments have begun to focus: i) on targeting a given type of integrin or non-integrin receptors (such as syndecans) and ii) to present not only adhesive signals but also signals triggered by growth factors (FGF, BMP, VEGF…), which affect cell proliferation and cell differentiation. There is no doubt that this direction will be further developed and studied in order to understand how different signals can act in synergy. The control over mechanical and biochemical properties will enable and foster further studies aimed at understanding possible synergies between these signals. Cross-talks between the different signaling pathways may be unveiled in a near future.
Natural materials that have some adhesive sites intrinsically and that can bind growth factors with a high affinity will be particularly interesting candidates as compared to their synthetic counterparts. Indeed, if multiple functionalities have to be added to synthetic materials, the strategy of coupling may become even more tedious and time-consuming. Ease of implementation is an important criterion for biomaterials scientists, as well as for biophysicists and cell biologists. Such experimental constraints should be kept in mind when designing materials, as only simple, easy-to-handle and easy-to-characterize materials would be used by cell biologists.
Creating anisotropic properties to mimic the natural environment is also a current challenge.123 Gradients in both mechanical properties and growth factors will thus be developed and used to understand how cells respond to these cues, from adhesion and proliferation to differentiation. We have seen that UV light is already widely used for the synthesis of biomaterials. Light-initiated cross-linking steps and gradients will probably be a valuable tool for basic cell-material interaction studies or advanced tissue engineering applications. It is also predicted that new methodological developments emerging from soft lithography and microfluidics will be combined to further develop these 2D and 3D biomaterials.124 Importantly, these technologies can be applied to a wide range of polymeric biomaterials currently in use. This will make it possible to incorporate spatial control which is crucial for developing complex microenvironments.125 Ultimately, control over biochemical and mechanical properties in a spatially-controlled manner will be achieved to investigate the respective role of each parameter as well as to produce innovative biomaterials.126
Acknowledgements
CP is a Junior Member of the “Institut Universitaire de France” whose support is gratefully acknowledged. CP wishes to thank the European Commission for support via an ERC Starting grant 2010 (GA 259370). VG thanks the Rhône-Alpes region for a fellowship via the cluster MACODEV.
Biography

Varvara Gribova received a Master’s degree in Cellular and Molecular Biology from the University of Pierre and Marie Curie in Paris in 2010. She is currently a graduate student at the Grenoble Institute of Technology and CNRS. Her project is focused on the design of biomimetic polyelectrolyte multilayer films for muscle and bone tissue engineering.

Thomas Crouzier completed a Master’s thesis in tissue engineering at Oklahoma University in 2006. He then received his PhD in Biological Engineering from the University of Montpellier in 2010 under the supervision of Prof. Picart. He is currently a post-doctoral fellow in K. Ribbeck’s laboratory at MIT.

Catherine Picart is Full Professor at the Grenoble Institute of Technology in Biomedical Engineering. She is group leader at the LMGP laboratory in MINATEC “Innovation pole in Nano and Micro-Technologies”. She received a doctoral degree in Biomedical Engineering from University Joseph Fourier, Grenoble in 1997 and completed her Habilitation in Biophysics and Biomaterials in 2002 at the University of Strasbourg. Her current research interests include self-assembly of biopolymers, protein/lipid interactions and design of layer-by-layer films for tissue engineering and regenerative medicine.
References
- 1.Langer R, Vacanti JP. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Newham P, Humphries MJ. Mol. Med. Today. 1996;2:304–313. doi: 10.1016/1357-4310(96)10021-6. [DOI] [PubMed] [Google Scholar]
- 3.Pelham RJ, Wang YL. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 5.Fan VH, Tamama K, Au A, Littrell R, Richardson LB, Wright JW, Wells A, Griffith LG. Stem Cells. 2007;25:1241–1251. doi: 10.1634/stemcells.2006-0320. [DOI] [PubMed] [Google Scholar]
- 6.Bongrand P, Capo C, Depieds R. Prog. Surf. Sci. 1982;12:217–285. [Google Scholar]
- 7.Rehfeldt F, Engler AJ, Eckhardt A, Ahmed F, Discher DE. Adv. Drug Delivery Rev. 2007;59:1329–1339. doi: 10.1016/j.addr.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luu O, David R, Ninomiya H, Winklbauer R. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4000–4005. doi: 10.1073/pnas.1010331108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrowsky TM, Panorchan P, Konstantopoulos K, Wirtz D. Biophysical Tools for Biologists, Vol 2: In Vivo Techniques. Vol. 89. Elsevier Academic Press Inc; San Diego: 2008. Live-Cell Single-Molecule Force Spectroscopy; p. 411. [DOI] [PubMed] [Google Scholar]
- 10.Janmey PA, Weitz DA. Trends Biochem. Sci. 2004;29:364–370. doi: 10.1016/j.tibs.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Shoichet MS. Macromolecules. 2010;43:581–591. [Google Scholar]
- 12.Nemir S, West JL. Ann. Biomed. Eng. 2010;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 13.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 14.Engler AJ, Rehfeldt F, Sen S, Discher DE. Methods Cell Biol. 2007;83:521–545. doi: 10.1016/S0091-679X(07)83022-6. [DOI] [PubMed] [Google Scholar]
- 15.Engler AJ, Griffin MA, Sen S, Bonnemann CG, Sweeney HL, Discher DE. J. Cell Biol. 2004;166:877–887. doi: 10.1083/jcb.200405004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solon J, Levental I, Sengupta K, Georges PC, Janmey PA. Biophys. J. 2007;93:4453–4461. doi: 10.1529/biophysj.106.101386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isenberg BC, Dimilla PA, Walker M, Kim S, Wong JY. Biophys. J. 2009;97:1313–1322. doi: 10.1016/j.bpj.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beningo KA, Hamao K, Dembo M, Wang YL, Hosoya H. Arch. Biochem. Biophys. 2006;456:224–231. doi: 10.1016/j.abb.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beningo KA, Dembo M, Wang YL. Proc. Natl. Acad. Sci. U. S. A. 2004;101:18024–18029. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudou T, Ohayon J, Picart C, Pettigrew RI, Tracqui P. Biorheology. 2009;46:191–205. doi: 10.3233/BIR-2009-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhart-King CA, Dembo M, Hammer DA. Biophys. J. 2005;89:676–689. doi: 10.1529/biophysj.104.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou SY, Cheng CM, LeDuc PR. Biomaterials. 2009;30:3136–3142. doi: 10.1016/j.biomaterials.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 23.Brown XQ, Ookawa K, Wong JY. Biomaterials. 2005;26:3123–3129. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Tzvetkova-Chevolleau T, Stephanou A, Fuard D, Ohayon J, Schiavone P, Tracqui P. Biomaterials. 2008;29:1541–1551. doi: 10.1016/j.biomaterials.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Decher G, Lehr B, Lowack K, Lvov Y, Schmitt J. Biosens. Bioelectron. 1994;9:677–684. [Google Scholar]
- 26.Thompson MT, Berg MC, Tobias IS, Rubner MF, Van Vliet KJ. Biomaterials. 2005;26:6836–6845. doi: 10.1016/j.biomaterials.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Thompson MT, Berg MC, Tobias IS, Lichter JA, Rubner MF, Van Vliet KJ. Biomacromolecules. 2006;7:1990–1995. doi: 10.1021/bm060146b. [DOI] [PubMed] [Google Scholar]
- 28.Chen AA, Khetani SR, Lee S, Bhatia SN, Van Vliet KJ. Biomaterials. 2009;30:1113–1120. doi: 10.1016/j.biomaterials.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Francius G, Hemmerle J, Ball V, Lavalle P, Picart C, Voegel J, Schaaf P, Senger B. J. Phys. Chem. C. 2007;111:8299–8306. [Google Scholar]
- 30.Kocgozlu L, Lavalle P, Koenig G, Senger B, Haikel Y, Schaaf P, Voegel JC, Tenenbaum H, Vautier D. J. Cell Sci. 2010;123:29–39. doi: 10.1242/jcs.053520. [DOI] [PubMed] [Google Scholar]
- 31.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713–722. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 32.Raeber GP, Lutolf MP, Hubbell JA. Biophys. J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Adv. Mater. 2003;15:888. [Google Scholar]
- 34.Yamamura N, Sudo R, Ikeda M, Tanishita K. Tissue Eng. 2007;13:1443–1453. doi: 10.1089/ten.2006.0333. [DOI] [PubMed] [Google Scholar]
- 35.Miron-Mendoza M, Seemann J, Grinnell F. Biomaterials. 2010;31:6425–6435. doi: 10.1016/j.biomaterials.2010.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDaniel DP, Shaw GA, Elliott JT, Bhadriraju K, Meuse C, Chung KH, Plant AL. Biophys. J. 2007;92:1759–1769. doi: 10.1529/biophysj.106.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halloran DO, Grad S, Stoddart M, Dockery P, Alini M, Pandit AS. Biomaterials. 2008;29:438–447. doi: 10.1016/j.biomaterials.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Wong M. Alginate in tissue engineering. Biopolymer Methods in Tissue Engineering. 2004. pp. 77–86. [DOI] [PubMed]
- 39.Augst AD, Kong HJ, Mooney DJ. Macromol. Biosci. 2006;6:623–633. doi: 10.1002/mabi.200600069. [DOI] [PubMed] [Google Scholar]
- 40.Kong HJ, Wong E, Mooney DJ. Macromolecules. 2003;36:4582–4588. [Google Scholar]
- 41.Lee KY, Rowley JA, Eiselt P, Moy EM, Bouhadir KH, Mooney DJ. Macromolecules. 2000;33:4291–4294. [Google Scholar]
- 42.Rowley JA, Mooney DJ, Biomed J. Mater. Res. 2002;60:217–223. doi: 10.1002/jbm.1287. [DOI] [PubMed] [Google Scholar]
- 43.Genes NG, Rowley JA, Mooney DJ, Bonassar LJ. Arch. Biochem. Biophys. 2004;422:161–167. doi: 10.1016/j.abb.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 44.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat. Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jouon N, Rinaudo M, Milas M, Desbrieres J. Carbohydr. Polym. 1995;26:69–73. [Google Scholar]
- 46.Zimmerman E, Geiger B, Addadi L. Biophys. J. 2002;82:1848–1857. doi: 10.1016/S0006-3495(02)75535-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ladam G, Vonna L, Sackmann E. J. Phys. Chem. B. 2003;107:8965–8971. [Google Scholar]
- 48.Shu XZ, Liu Y, Luo Y, Roberts MC, Prestwich GD. Biomacromolecules. 2002;3:1304–1311. doi: 10.1021/bm025603c. [DOI] [PubMed] [Google Scholar]
- 49.Young JL, Engler AJ. Biomaterials. 2011;32:1002–1009. doi: 10.1016/j.biomaterials.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balgude AP, Yu X, Szymanski A, Bellamkonda RV. Biomaterials. 2001;22:1077–1084. doi: 10.1016/s0142-9612(00)00350-1. [DOI] [PubMed] [Google Scholar]
- 51.Leipzig ND, Shoichet MS. Biomaterials. 2009;30:6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Boudou T, Crouzier T, Ren K, Blin G, Picart C. Adv. Mater. 2010;22:441–467. doi: 10.1002/adma.200901327. [DOI] [PubMed] [Google Scholar]
- 53.Picart C, Mutterer J, Richert L, Luo Y, Prestwich GD, Schaaf P, Voegel J-C, Lavalle P. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12531–12535. doi: 10.1073/pnas.202486099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crouzier T, Boudou T, Picart C. Curr. Opin. Colloid Interface Sci. 2010;15:417–426. [Google Scholar]
- 55.Richert L, Boulmedais F, Lavalle P, Mutterer J, Ferreux E, Decher G, Schaaf P, Voegel J-C, Picart C. Biomacromolecules. 2004;5:284–294. doi: 10.1021/bm0342281. [DOI] [PubMed] [Google Scholar]
- 56.Francius G, Hemmerle J, Ohayon J, Schaaf P, Voegel J-C, Picart C, Senger B. Microsc. Res. Tech. 2006;69:84–92. doi: 10.1002/jemt.20275. [DOI] [PubMed] [Google Scholar]
- 57.Boudou T, Crouzier T, Auzely-Velty R, Glinel K, Picart C. Langmuir. 2009;25:13809–13819. doi: 10.1021/la9018663. [DOI] [PubMed] [Google Scholar]
- 58.Wittmer CR, Phelps JA, Lepus CM, Saltzman WM, Harding MJ, Van Tassel PR. Biomaterials. 2008;29:4082–4090. doi: 10.1016/j.biomaterials.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ren K, Crouzier T, Roy C, Picart C. Adv. Funct. Mater. 2008;18:1378–1389. doi: 10.1002/adfm.200701297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phelps JA, Morisse S, Hindie M, Degat MC, Pauthe E, Van Tassel PR. Langmuir. 2011;27:1123–1130. doi: 10.1021/la104156c. [DOI] [PubMed] [Google Scholar]
- 61.Hillberg AL, Holmes CA, Tabrizian M. Biomaterials. 2009;30:4463–4470. doi: 10.1016/j.biomaterials.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 62.Pozos Vazquez C, Boudou T, Dulong V, Nicolas C, Picart C, Glinel K. Langmuir. 2009;25:3556–3563. doi: 10.1021/la803577t. [DOI] [PubMed] [Google Scholar]
- 63.Peyton SR, Kim PD, Ghajar CM, Seliktar D, Putnam AJ. Biomaterials. 2008;29:2597–2607. doi: 10.1016/j.biomaterials.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim PD, Peyton SR, VanStrien AJ, Putnam AJ. Biomaterials. 2009;30:3854–3864. doi: 10.1016/j.biomaterials.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 65.Brigham MD, Bick A, Lo E, Bendali A, Burdick JA, Khademhosseini A. Tissue Eng. A. 2009;15:1645–1653. doi: 10.1089/ten.tea.2008.0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Q, Feng Q, Hu K, Cui F. J. Mater. Sci.: Mater. Med. 2008;19:629–634. doi: 10.1007/s10856-007-3180-9. [DOI] [PubMed] [Google Scholar]
- 67.Croll TI, O’Connor AJ, Stevens GW, Cooper-White JJ. Biomacromolecules. 2006;7:1610–1622. doi: 10.1021/bm060044l. [DOI] [PubMed] [Google Scholar]
- 68.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. Garland Publishing Inc; New York: 1994. [Google Scholar]
- 69.Macri L, Silverstein D, Clark RA. Adv. Drug Delivery Rev. 2007;59:1366–1381. doi: 10.1016/j.addr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 70.Hynes RO. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kandow CE, Georges PC, Janmey PA, Beningo KA. Methods Cell Biol. 2007;83:29–46. doi: 10.1016/S0091-679X(07)83002-0. [DOI] [PubMed] [Google Scholar]
- 72.Grabarek Z, Gergely J. Anal. Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 73.Kuo JW, Swann DA, Prestwich GD. Bioconjugate Chem. 1991;2:232–241. doi: 10.1021/bc00010a007. [DOI] [PubMed] [Google Scholar]
- 74.Boontheekul T, Hill EE, Kong HJ, Mooney DJ. Tissue Eng. 2007;13:1431–1442. doi: 10.1089/ten.2006.0356. [DOI] [PubMed] [Google Scholar]
- 75.Doran MR, Frith JE, Prowse ABJ, Fitzpatrick J, Wolvetang EJ, Munro TP, Gray PP, Cooper-White JJ. Biomaterials. 2010;31:5137–5142. doi: 10.1016/j.biomaterials.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 76.Picart C, Elkaim R, Richert L, Audoin F, Da Silva Cardoso M, Schaaf P, Voegel J-C, Frisch B. Adv. Funct. Mater. 2005;15:83–94. [Google Scholar]
- 77.Mann BK, Schmedlen RH, West JL. Biomaterials. 2001;22:439–444. doi: 10.1016/s0142-9612(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 78.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 79.Hern DL, Hubbell JA. J. Biomed. Mater. Res. 1998;39:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 80.Khetan S, Burdick JA. Biomaterials. 2010;31:8228–8234. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 81.Almany L, Seliktar D. Biomaterials. 2005;26:2467–2477. doi: 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]
- 82.Ruoslahti E. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 83.Petrie TA, Raynor JE, Reyes CD, Burns KL, Collard DM, Garcia AJ. Biomaterials. 2008;29:2849–2857. doi: 10.1016/j.biomaterials.2008.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lutolf MP, Weber FE, Schmoekel HG, Schense JC, Kohler T, Muller R, Hubbell JA. Nat. Biotechnol. 2003;21:513–518. doi: 10.1038/nbt818. [DOI] [PubMed] [Google Scholar]
- 85.Hozumi K, Yamagata N, Otagiri D, Fujimori C, Kikkawa Y, Kadoya Y, Nomizu M. Biomaterials. 2009;30:1596–1603. doi: 10.1016/j.biomaterials.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 86.Urushibata S, Hozumi K, Ishikawa M, Katagiri F, Kikkawa Y, Nomizu M. Arch. Biochem. Biophys. 2010;497:43–54. doi: 10.1016/j.abb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 87.Doran MR, Frith JE, Prowse AB, Fitzpatrick J, Wolvetang EJ, Munro TP, Gray PP, Cooper-White JJ. Biomaterials. 2010;31:5137–5142. doi: 10.1016/j.biomaterials.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 88.Raeber GP, Lutolf MP, Hubbell JA. Acta Biomater. 2007;3:615–629. doi: 10.1016/j.actbio.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 89.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 90.Hsiong SX, Boontheekul T, Huebsch N, Mooney DJ. Tissue Eng. A. 2009;15:263–272. doi: 10.1089/ten.tea.2007.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suzuki N, Nakatsuka H, Mochizuki M, Nishi N, Kadoya Y, Utani A, Oishi S, Fujii N, Kleinman HK, Nomizu M. J. Biol. Chem. 2003;278:45697–45705. doi: 10.1074/jbc.M304667200. [DOI] [PubMed] [Google Scholar]
- 92.Benoit DS, Anseth KS. Biomaterials. 2005;26:5209–5220. doi: 10.1016/j.biomaterials.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 93.Rezania A, Healy KE. Biotechnol. Prog. 1999;15:19–32. doi: 10.1021/bp980083b. [DOI] [PubMed] [Google Scholar]
- 94.Reyes CD, Petrie TA, Burns KL, Schwartz Z, Garcia AJ. Biomaterials. 2007;28:3228–3235. doi: 10.1016/j.biomaterials.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weber LM, Hayda KN, Haskins K, Anseth KS. Biomaterials. 2007;28:3004–3011. doi: 10.1016/j.biomaterials.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 96.Werner S, Huck O, Frisch B, Vautier D, Elkaim R, Voegel JC, Brunel G, Tenenbaum H. Biomaterials. 2009;30:2291–2301. doi: 10.1016/j.biomaterials.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Wu YZ, Coyer SR, Ma HW, Garcia AJ. Acta Biomater. 2010;6:2898–2902. doi: 10.1016/j.actbio.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garcia AJ, Schwarzbauer JE, Boettiger D. Biochemistry. 2002;41:9063–9069. doi: 10.1021/bi025752f. [DOI] [PubMed] [Google Scholar]
- 99.Bellin RM, Kubicek JD, Frigault MJ, Kamien AJ, Steward RL, Jr, Barnes HM, Digiacomo MB, Duncan LJ, Edgerly CK, Morse EM, Park CY, Fredberg JJ, Cheng CM, LeDuc PR. Proc. Natl. Acad. Sci. U. S. A. 2009;106:22102–22107. doi: 10.1073/pnas.0902639106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He XZ, Ma JY, Jabbari E. Langmuir. 2008;24:12508–12516. doi: 10.1021/la802447v. [DOI] [PubMed] [Google Scholar]
- 101.Zouani OF, Chollet C, Guillotin B, Durrieu MC. Biomaterials. 2010;31:8245–8253. doi: 10.1016/j.biomaterials.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 102.Wang YL, Pelham RJ., Jr Methods Enzymol. 1998;298:489–496. doi: 10.1016/s0076-6879(98)98041-7. [DOI] [PubMed] [Google Scholar]
- 103.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophys. J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wipff PJ, Majd H, Acharya C, Buscemi L, Meister JJ, Hinz B. Biomaterials. 2009;30:1781–1789. doi: 10.1016/j.biomaterials.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 105.Kuhl PR, Griffith-Cima LG. Nat. Med. 1996;2:1022–1027. doi: 10.1038/nm0996-1022. [DOI] [PubMed] [Google Scholar]
- 106.Mehta G, Williams CM, Alvarez L, Lesniewski M, Kamm RD, Griffith LG. Biomaterials. 2010;31:4657–4671. doi: 10.1016/j.biomaterials.2010.01.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puccinelli TJ, Bertics PJ, Masters KS. Acta Biomater. 2010;6:3415–3425. doi: 10.1016/j.actbio.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kapur TA, Shoichet MS. J. Biomed. Mater. Res. 2004;68:235–243. doi: 10.1002/jbm.a.10168. [DOI] [PubMed] [Google Scholar]
- 109.Leslie-Barbick JE, Shen C, Chen CS, West JL. Tissue Eng. A. 2011;17:221–229. doi: 10.1089/ten.tea.2010.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wittmer CR, Phelps JA, Saltzman WM, Van Tassel PR. Biomaterials. 2007;28:851–860. doi: 10.1016/j.biomaterials.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Semenov OV, Malek A, Bittermann AG, Voros J, Zisch A. Tissue Eng. A. 2009;15:2977–2990. doi: 10.1089/ten.TEA.2008.0602. [DOI] [PubMed] [Google Scholar]
- 112.Zhang J, Senger B, Vautier D, Picart C, Schaaf P, Voegel J-C, Lavalle P. Biomaterials. 2005;26:3353–3361. doi: 10.1016/j.biomaterials.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 113.Boddohi S, Kipper MJ. Adv. Mater. 2010;22:2998–3016. doi: 10.1002/adma.200903790. [DOI] [PubMed] [Google Scholar]
- 114.Sahni A, Sporn LA, Francis CW. J. Biol. Chem. 1999;274:14936–14941. doi: 10.1074/jbc.274.21.14936. [DOI] [PubMed] [Google Scholar]
- 115.Campbell PG, Miller ED, Fisher GW, Walker LM, Weiss LE. Biomaterials. 2005;26:6762–6770. doi: 10.1016/j.biomaterials.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 116.Miller ED, Fisher GW, Weiss LE, Walker LM, Campbell PG. Biomaterials. 2006;27:2213–2221. doi: 10.1016/j.biomaterials.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 117.Phillippi JA, Miller E, Weiss L, Huard J, Waggoner A, Campbell P. Stem Cells. 2008;26:127–134. doi: 10.1634/stemcells.2007-0520. [DOI] [PubMed] [Google Scholar]
- 118.Martino MM, Hubbell JA. FASEB J. 2010;24:4711–4721. doi: 10.1096/fj.09-151282. [DOI] [PubMed] [Google Scholar]
- 119.Chow LW, Bitton R, Webber MJ, Carvajal D, Shull KR, Sharma AK, Stupp SI. Biomaterials. 2011;32:1574–1582. doi: 10.1016/j.biomaterials.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Muller S, Koenig G, Charpiot A, Debry C, Voegel J, Lavalle P, Vautier D. Adv. Funct. Mater. 2008;18:1767–1775. [Google Scholar]
- 121.Crouzier T, Ren K, Nicolas C, Roy C, Picart C. Small. 2009;5:598–608. doi: 10.1002/smll.200800804. [DOI] [PubMed] [Google Scholar]
- 122.Crouzier T, Szarpak A, Boudou T, Auzely-Velty R, Picart C. Small. 2010;6:651–662. doi: 10.1002/smll.200901728. [DOI] [PubMed] [Google Scholar]
- 123.Kutejova E, Briscoe J, Kicheva A. Curr. Opin. Genet. Dev. 2009;19:315–322. doi: 10.1016/j.gde.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 124.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marklein RA, Burdick JA. Soft Matter. 2010;6:136–143. [Google Scholar]
- 126.Huebsch N, Mooney DJ. Nature. 2009;462:426–432. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]