Abstract
Background
Several molecular epidemiological studies have been conducted to examine the association between glutathione S-transferase M 1 (GSTM1) null genotype and lung cancer in Asians; however, the conclusions remained controversial. We therefore performed an extensive meta-analysis on 31 published case-control studies with a total of 5347 lung cancer cases and 6072 controls.
Material/Methods
PubMed and EMBASE were searched to identify case-control studies investigating the associations of GSTM1 null genotype with risk of lung cancer in Asians. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of association between lung cancer risk and polymorphism of GSTM1.
Results
GSTM1 null genotype was significantly associated with lung cancer risk (OR=1.43; 95% CI, 1.30–1.58). This result remained statistically significant when the adjusted ORs were combined (OR=1.38; 95% CI, 1.23–1.54). In the subgroup analysis by sex, there were significant associations in women and men. When stratifying for histology, this genotype showed increased adenocarcinoma risk and squamous cell carcinoma risk. In the subgroup analysis stratified by smoking status, lung cancer risk was increased in both smokers and non-smokers.
Conclusions
This study suggests that GSTM1 null genotype is a risk factor for lung cancer in Asians.
MeSH Keywords: Meta-Analysis, Lung Neoplasms - genetics, Polymorphism, Genetic - genetics
Background
Lung cancer is one of the leading causes of cancer-related deaths in the world. The mechanism of lung carcinogenesis is not understood. Although it is well known that smoking is the primary risk factor for lung cancer, lung cancer develops in less than 20% of people who smoke throughout their life. Moreover, lung cancer is a multi-cellular and multistage process involving a number of genetic changes in oncogenes, suggesting that genetic factors may play an important role in its development.
The glutathione S-transferases (GSTs) are a gene superfamily of phase II metabolic enzymes that detoxify free radicals, particularly in tobacco smoke, products of oxidative stress, and carcinogens such as benzopyrene and other polycyclic aromatic hydrocarbons [1]. GSTM1 has been mapped to the GST mu gene cluster on chromosome 1p13.3. One variant in GSTM1 has been identified: a deletion. The deletion (GSTM1 null variant) has been examined extensively in epidemiologic studies. Persons with a homozygous deletion of the GSTM1 locus have no enzymatic functional activity. Phenotype assays have confirmed this lack of function by demonstrating a strong concordance between phenotype and genotype [2]. Previous studies have suggested that individuals with null genotypes of GSTM1 may be unable to eliminate electrophilic carcinogens efficiently and have a high risk of lung cancer. However, the results from previous reported studies in Asians were inconclusive [3–33]. Therefore, we conducted a meta-analysis to explore the effect of GSTM1 null genotype on lung cancer risk in Asians.
Material and Methods
Selection of published studies
We searched the PubMed and EMBASE to identify published case-control studies investigating the associations of GSTM1 null genotype with risk of lung cancer in Asians. We used the following terms: ‘glutathione S-transferases’ or ‘GSTM1’ and ‘lung cancer’, without restriction on language. Additional studies were identified by a manual search of references of original studies or review articles. The inclusion criteria were: (1) original papers containing independent data; (2) studies should provide the sample size, odds ratios (ORs), and 95% confidence intervals (CIs), as well as the genetic distribution or the information needed to infer the results; and (3) case-control or cohort studies. The major exclusion criteria for studies were: (1) overlapping data; (2) insufficiently useful data; and (3) case-only studies or family-based studies; (4) reviews, abstracts, or commentaries; (5) not relevant to lung cancer or GSTM1; and (6) not conducted in Asians.
Data extraction
Two independent researchers extracted raw data according to the inclusion criteria. The following information was collected from each study using a data extraction form: the surname of the first author, year of publication, country of origin, sex of subjects, histology, smoking status, number of cases and controls, adjustment, and ORs and the corresponding 95% confidence intervals (CIs) of lung cancer risk.
Statistical analysis
For the GSTM1 gene, we estimated the risk effect of the null genotype on lung cancer compared with the non-null genotypes in the recessive model (null vs. heterozygous + wild type). The strength of the association between the GSTM1 gene and lung cancer risk was measured by ORs with 95% CIs.
The ORs with corresponding 95% CIs from individual studies were pooled using fixed or random effects models, according to the heterogeneity. When the P value for Cochran’s Q statistic was less than 0.1, and a significant heterogeneity existed across the included studies, the random effects model (DerSimonian and Laird method) was used for meta-analysis, or the fixed-effects model (Mantel-Haenszel method) was used. Sensitivity analysis was further performed by excluding a single study to assess the impact of an individual study on the pooled estimate. Subgroup analyses were stratified by sex, histology, and smoking status. Cumulative meta-analysis was also performed. Funnel plots and Egger’s regression test were used to assess the potential publication bias [34]. Data analysis was performed using STATA 12 (StataCorp LP, College Station, Texas, USA).
Results
Study characteristics
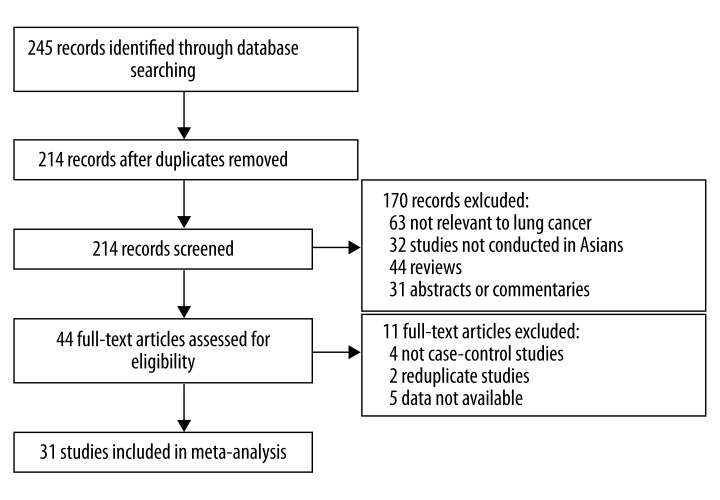
The flow chart shown in Figure 1 summarizes the study selection process. A total of 31 studies were retrieved based on the search criteria for lung cancer susceptibility related to the GSTM1 polymorphism [3–33]. The main study characteristics are summarized in Table 1. There are 5347 lung cancer cases and 6072 controls.
Figure 1.
Flow diagram of the literature search.
Table 1.
Characteristics of the case-control studies included in this meta-analysis
| First author | Year | Country | Sex | Histology | Smoking | Case | Control | Adjustment |
|---|---|---|---|---|---|---|---|---|
| Kihara | 1995 | Japan | Mixed | Mixed* | Smoker | 97 | 185 | No |
| Ge | 1995 | China | Mixed | Mixed | NA | 89 | 25 | No |
| Hong | 1998 | Korea | Mixed | Mixed* | NA | 85 | 63 | No |
| Gao | 1998 | China | Mixed | Mixed | Mixed | 70 | 46 | No |
| Gao | 1999 | China | Mixed | Mixed* | Mixed* | 59 | 132 | No |
| Kiyohara | 2000 | Japan | Mixed | Mixed | Mixed | 86 | 88 | Yes |
| Lan | 2000 | China | Mixed | Mixed | Mixed | 122 | 122 | Yes |
| London | 2000 | China | Men | Mixed | Mixed | 232 | 710 | Yes |
| Chen | 2001 | China | Mixed | Mixed | Mixed | 106 | 106 | No |
| Xue | 2001 | China | Mixed | Mixed | Mixed | 106 | 106 | No |
| Lu | 2002 | China | Mixed | Mixed | Mixed | 314 | 320 | No |
| Kiyohara | 2003 | Japan | Women | Mixed | Nonsmoker | 158 | 259 | Yes |
| Wang | 2003 | Japan | Mixed | AD | Mixed* | 112 | 119 | Yes |
| Chan-Yeung | 2004 | China | Mixed | Mixed* | Mixed | 130 | 117 | Yes |
| Chen | 2004 | China | Mixed | Mixed | Mixed | 97 | 197 | No |
| Yang | 2004 | China | Women | Mixed | Mixed | 200 | 144 | No |
| Gu | 2004 | China | Mixed | Mixed | Mixed* | 180 | 224 | No |
| Li | 2004 | China | Mixed | Mixed | Mixed* | 217 | 200 | No |
| Li | 2005 | China | Mixed | Mixed | Mixed* | 99 | 66 | Yes |
| Lee | 2006 | Korea | Mixed | Mixed* | Mixed | 171 | 196 | Yes |
| Chang | 2006 | China | Mixed | Mixed | Mixed* | 163 | 163 | No |
| Osawa | 2007 | Japan | Mixed | Mixed* | Mixed* | 113 | 121 | Yes |
| Yang | 2007 | Korea | Mixed* | Mixed* | Mixed | 318 | 353 | Yes |
| Jin | 2010 | China | Mixed | Mixed | Mixed* | 150 | 150 | Yes |
| Zheng | 2010 | China | Mixed | Mixed | Mixed | 265 | 307 | No |
| Zhu | 2010 | China | Women | Mixed | Nonsmoker | 160 | 160 | Yes |
| Kiyohara | 2012 | Japan | Mixed | Mixed | Mixed | 462 | 379 | Yes |
| Li | 2012 | China | Mixed | Mixed | Mixed* | 217 | 198 | Yes |
| Liu | 2012 | China | Mixed | Mixed* | Mixed* | 360 | 360 | No |
| Chen | 2012 | China | Mixed | Mixed | Mixed* | 200 | 200 | No |
| Wang | 2012 | China | Mixed | Mixed | Mixed | 209 | 256 | Yes |
More information can be extracted.
AD – adenocarcinoma.
Meta-analysis results
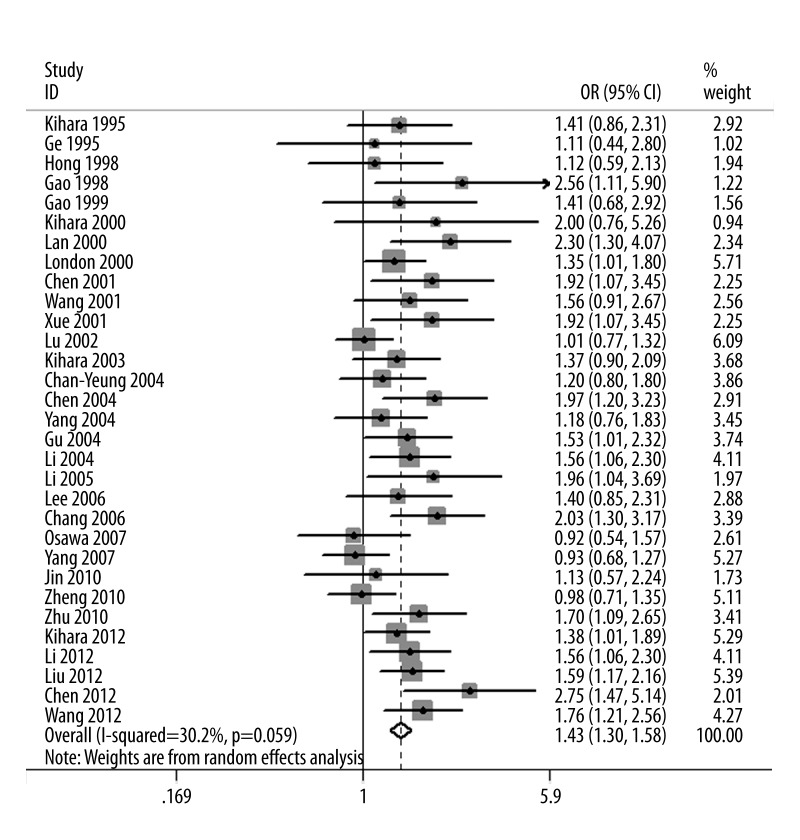
The evaluations of the association between GSTM1 polymorphism and lung cancer risk are summarized in Table 2. The null genotype of GSTM1 was associated with a significantly increased risk of lung cancer when compared with present genotype (OR=1.43; 95% CI, 1.30–1.58; Figure 2). Fifteen studies reported adjusted ORs. The combination of adjusted ORs for lung cancer was 1.38 (95% CI, 1.23–1.54).
Table 2.
Detailed results of meta-analysis.
| Test of association | Heterogeneity | |||
|---|---|---|---|---|
| OR (95% CI) | P Value | P Value | I2 (%) | |
| Overall | 1.43 (1.30–1.58) | <0.00001 | 0.06 | 30.0 |
| Men | 1.38 (1.06–1.78) | 0.02 | 0.78 | 0.0 |
| Women | 1.30 (1.03–1.64) | 0.03 | 0.24 | 28.0 |
| Adenocarcinoma | 1.27 (1.05–1.55) | 0.02 | 0.49 | 0.0 |
| SCC | 1.40 (1.10–1.78) | 0.006 | 0.48 | 0.0 |
| SCLC | 1.22 (0.81–1.83) | 0.35 | 0.18 | 38.0 |
| Non-smoker | 1.49 (1.25–1.79) | <0.00001 | 0.87 | 0.0 |
| Smoker | 1.78 (1.43–2.23) | <0.00001 | 0.37 | 8.0 |
SCC – squamous cell carcinoma; SCLC – small-cell lung cancer.
Figure 2.
Forest plot of lung cancer risk of GSTM1 polymorphism.
When stratified by sex, significantly elevated risks were observed in men (OR=1.38; 95% CI, 1.06–1.78) and women (OR=1.30; 95% CI, 1.03–1.64). In the subgroup analysis according to histology, significantly increased risks were observed in adenocarcinoma (OR=1.27; 95% CI, 1.05–1.55) and squamous cell carcinoma (OR=1.40; 95% CI, 1.10–1.78), but not in small-cell lung cancer (OR=1.22; 95% CI, 0.81–1.83). Subgroup analysis based on the smoking status showed that increased risks were found in non-smokers (OR=1.49; 95% CI, 1.25–1.79) and smokers (OR=1.78; 95% CI, 1.43–2.23).
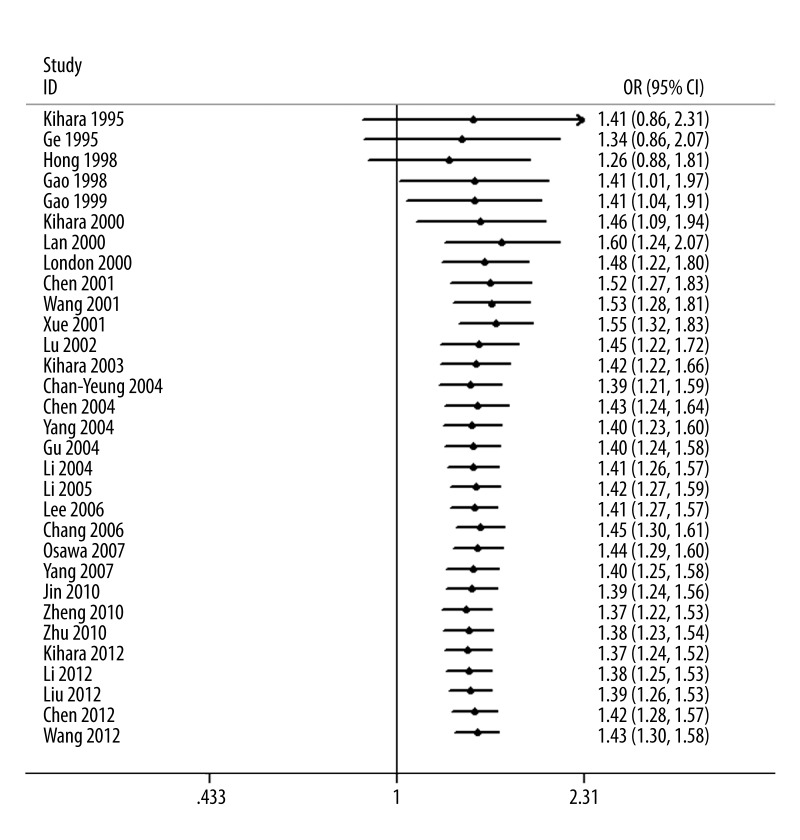
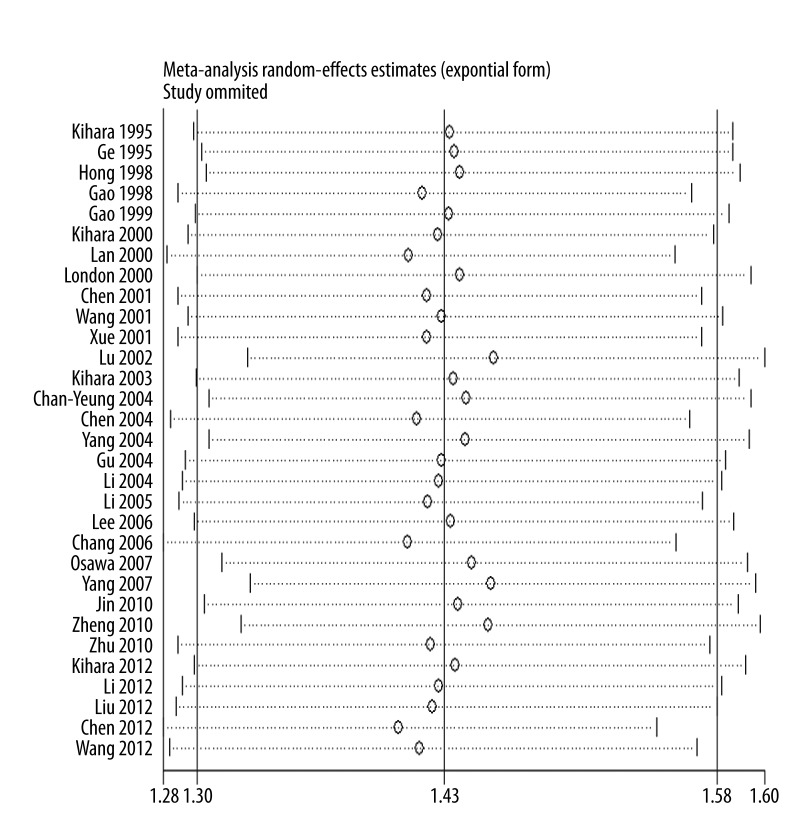
As shown in Figure 3, significant associations were evident with each addition of more data over time. The results showed that the pooled ORs tended to be stable. A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data set on the pooled ORs, and the corresponding pooled ORs were not materially altered (Figure 4).
Figure 3.
Cumulative meta-analysis of lung cancer risk of GSTM1 polymorphism.
Figure 4.
Sensitivity analysis of lung cancer risk of GSTM1 polymorphism.
Funnel plot and Egger’s test were used to assess the publication bias of the literature. Figure 5 shows the funnel plot for the assessment of publication bias. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (Figure 5). Egger’s test did not show evidence of publication bias (P=0.09).
Figure 5.
Funnel plot of association between GSTM1 polymorphism and lung cancer risk.
Discussion
The present meta-analysis, including 5347 lung cancer cases and 6072 controls from 31 case-control studies, explored the association of GSTM1 null genotype with lung cancer risk. We demonstrated that the null genotype of GSTM1 was associated with a significantly increased lung cancer risk in Asians. Furthermore, in the stratified analysis by sex, we found that both men and women with GSTM1 null genotype had increased lung cancer risk. However, it should be noted that the numbers of these studies were small. More studies are needed to assess the association between GSTM1 null genotype and lung cancer risk in males and females. Cigarette smoking is a pro-inflammatory stimulus and an important risk factor for lung cancer. Some studies explored the interaction between GSTM1 genotype and smoking habits. Our results showed significant associations between GSTM1 polymorphism and lung cancer risk among smokers and non-smokers. We also found that patients with GSTM1 null genotype had increased non–small-cell lung cancer (adenocarcinoma and squamous cell carcinoma) risk. However, we failed to find a significant relationship between GSTM1 null polymorphism and small-cell lung cancer risk. This result suggested that GSTM1 null polymorphism may play an important role in the development of non-small-cell lung cancer.
GSTs are biotransformation enzymes, and they are phase II enzymes with both catalytic activities and non-catalytic functions. Previous studies have shown that individuals with the GSTM1 null genotype have a decreased capacity to detoxify certain carcinogens. Thus, impaired GSTM1 function may lead to serious DNA damage and carcinogenesis. Thus, it is biologically plausible that the GSTM1 null genotype may increase risk of lung cancer.
Our study had some advantages. First, the methodological issues for meta-analysis such as one-way sensitivity analysis and cumulative meta-analysis were well investigated. Second, the main result remained statistically significant when the adjusted ORs were combined.
Results from one-way sensitivity analysis and cumulative meta-analysis suggested high stability and reliability of our results and significant heterogeneity was not observed in this meta-analysis. Moreover, funnel plots and Egger’s tests were used to find potential publication bias. The results indicated that there was no significant publication bias.
Some limitations in our meta-analysis should be mentioned. First, the numbers of published studies were not sufficient for a comprehensive analysis. Second, lack of the original data from the eligible studies limited the evaluation of the effects of the gene-gene and gene-environment interactions in the development of lung cancer. Third, only published studies were included in this meta-analysis; therefore, publication bias may have occurred even though the statistical test did not show it.
Conclusions
In conclusion, this meta-analysis suggests that an increased risk of lung cancer was associated with the null polymorphism of GSTM1 in Asians.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Li R, Boerwinkle E, Olshan AF, et al. Glutathione S-transferase genotype as a susceptibility factor in smoking-related coronary heart disease. Atherosclerosis. 2000;149:451–62. doi: 10.1016/s0021-9150(99)00483-9. [DOI] [PubMed] [Google Scholar]
- 2.Cotton SC, Sharp L, Little J, Brockton N. Glutathione S-transferase polymorphisms and colorectal cancer: a HuGE review. Am J Epidemiol. 2000;151:7–32. doi: 10.1093/oxfordjournals.aje.a010124. [DOI] [PubMed] [Google Scholar]
- 3.Kihara M, Kihara M, Noda K. Risk of smoking for squamous and small cell carcinomas of the lung modulated by combinations of CYP1A1 and GSTM1 gene polymorphisms in a Japanese population. Carcinogenesis. 1995;16:2331–36. doi: 10.1093/carcin/16.10.2331. [DOI] [PubMed] [Google Scholar]
- 4.Ge H, Lam WK, Lee J, et al. Analysis of L-myc and GSTM1 genotypes in Chinese non-small cell lung carcinoma patients. Lung Cancer. 1996;15:355–66. doi: 10.1016/0169-5002(95)00598-6. [DOI] [PubMed] [Google Scholar]
- 5.Hong YS, Chang JH, Kwon OJ, et al. Polymorphism of the CYP1A1 and glutathione-S-transferase gene in Korean lung cancer patients. Exp Mol Med. 1998;30:192–98. doi: 10.1038/emm.1998.28. [DOI] [PubMed] [Google Scholar]
- 6.Gao J, Ren C, Zhang Q. CYP2D6 and GSTM1 genetic polymorphism and lung cancer susceptibility. Zhonghua Zhong Liu Za Zhi. 1998;20:185–86. [PubMed] [Google Scholar]
- 7.Gao Y, Zhang Q. Polymorphisms of the GSTM1 and CYP2D6 genes associated with susceptibility to lung cancer in Chinese. Mutat Res. 1999;444:441–49. doi: 10.1016/s1383-5718(99)00092-3. [DOI] [PubMed] [Google Scholar]
- 8.Kiyohara C, Yamamura KI, Nakanishi Y, et al. Polymorphism in GSTM1, GSTT1, and GSTP1 and Susceptibility to Lung Cancer in a Japanese Population. Asian Pac J Cancer Prev. 2000;1:293–98. [PubMed] [Google Scholar]
- 9.Lan Q, He X, Costa DJ, et al. Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes, and lung cancer risk: a case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev. 2000;9:605–8. [PubMed] [Google Scholar]
- 10.London SJ, Yuan JM, Chung FL, et al. Isothiocyanates, glutathione S-transferase M1 and T1 polymorphisms, and lung-cancer risk: a prospective study of men in Shanghai, China. Lancet. 2000;356:724–29. doi: 10.1016/S0140-6736(00)02631-3. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Xue K, Xu L, et al. Polymorphisms of the CYP1A1 and GSTM1 genes in relation to individual susceptibility to lung carcinoma in Chinese population. Mutat Res. 2001;458:41–47. doi: 10.1016/s1383-5726(01)00011-5. [DOI] [PubMed] [Google Scholar]
- 12.Xue K, Xu L, Chen S, et al. Polymorphisms of the CYP1A1 and GSTM1 genes and their combined effects on individual susceptibility to lung cancer in a Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2001;18:125–27. [PubMed] [Google Scholar]
- 13.Lu W, Xing D, Qi J, et al. Genetic polymorphism in myeloperoxidase but not GSTM1 is associated with risk of lung squamous cell carcinoma in a Chinese population. Int J Cancer. 2002;102:275–79. doi: 10.1002/ijc.10712. [DOI] [PubMed] [Google Scholar]
- 14.Kiyohara C, Wakai K, Mikami H, et al. Risk modification by CYP1A1 and GSTM1 polymorphisms in the association of environmental tobacco smoke and lung cancer: a case-control study in Japanese nonsmoking women. Int J Cancer. 2003;107:139–44. doi: 10.1002/ijc.11355. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Deng Y, Cheng J, et al. GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Lett. 2003;201:185–93. doi: 10.1016/s0304-3835(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 16.Chan-Yeung M, Tan-Un KC, Ip MS, et al. Lung cancer susceptibility and polymorphisms of glutathione-S-transferase genes in Hong Kong. Lung Cancer. 2004;45:155–60. doi: 10.1016/j.lungcan.2004.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Chen HC, Cao YF, Hu WX, et al. Genetic polymorphisms of phase II metabolic enzymes and lung cancer susceptibility in a population of Central South China. Dis Markers. 2006;22:141–52. doi: 10.1155/2006/436497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XR, Wacholder S, Xu Z, et al. CYP1A1 and GSTM1 polymorphisms in relation to lung cancer risk in Chinese women. Cancer Lett. 2004;214:197–204. doi: 10.1016/j.canlet.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 19.Gu Y, Zhang S, Lai B, et al. Relationship between genetic polymorphism of metabolizing enzymes and lung cancer susceptibility. Zhongguo Fei Ai Za Zhi. 2004;7:112–17. doi: 10.3779/j.issn.1009-3419.2004.02.08. [DOI] [PubMed] [Google Scholar]
- 20.Li WY, Lai BT, Zhan XP. The relationship between genetic polymorphism of metabolizing enzymes and the genetic susceptibility to lung cancer. Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25:1042–45. [PubMed] [Google Scholar]
- 21.Li D, Zhou Q, Yuan T, et al. Study on the association between genetic polymorphism of CYP2E1, GSTM1 and susceptibility of lung cancer. Zhongguo Fei Ai Za Zhi. 2005;8:14–19. doi: 10.3779/j.issn.1009-3419.2005.01.03. [DOI] [PubMed] [Google Scholar]
- 22.Lee KM, Kang D, Lee SJ, et al. Interactive effect of genetic polymorphism of glutathione S-transferase M1 and smoking on squamous cell lung cancer risk in Korea. Oncol Rep. 2006;16:1035–39. [PubMed] [Google Scholar]
- 23.Chang F, Hu T, Wang G. Relationship between CYP1A1 and GSTM1 genetic polymorphisms and lung cancer susceptibility in population of Inner Mongolia. Zhongguo Fei Ai Za Zhi. 2006;9:413–17. doi: 10.3779/j.issn.1009-3419.2006.05.04. [DOI] [PubMed] [Google Scholar]
- 24.Osawa Y, Osawa KK, Miyaishi A, et al. NAT2 and CYP1A2 polymorphisms and lung cancer risk in relation to smoking status. Asian Pac J Cancer Prev. 2007;8:103–8. [PubMed] [Google Scholar]
- 25.Yang M, Choi Y, Hwangbo B, Lee JS. Combined effects of genetic polymorphisms in six selected genes on lung cancer susceptibility. Lung Cancer. 2007;57:135–42. doi: 10.1016/j.lungcan.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Jin Y, Xu H, Zhang C, et al. Combined effects of cigarette smoking, gene polymorphisms and methylations of tumor suppressor genes on non small cell lung cancer: a hospital-based case-control study in China. BMC Cancer. 2010;10:422. doi: 10.1186/1471-2407-10-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng D, Hua F, Mei C, et al. Association between GSTM1 genetic polymorphism and lung cancer risk by SYBR green I real-time PCR assay. Zhongguo Fei Ai Za Zhi. 2010;13:506–10. doi: 10.3779/j.issn.1009-3419.2010.05.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu XX, Hu CP, Gu QH. CYP1A1 polymorphisms, lack of glutathione S-transferase M1 (GSTM1), cooking oil fumes and lung cancer risk in non-smoking women. Zhonghua Jie He He Hu Xi Za Zhi. 2010;33:817–22. [PubMed] [Google Scholar]
- 29.Kiyohara C, Horiuchi T, Takayama K, Nakanishi Y. Genetic polymorphisms involved in carcinogen metabolism and DNA repair and lung cancer risk in a Japanese population. J Thorac Oncol. 2012;7:954–62. doi: 10.1097/JTO.0b013e31824de30f. [DOI] [PubMed] [Google Scholar]
- 30.Li W, Yue W, Zhang L, et al. Polymorphisms in GSTM1, CYP1A1, CYP2E1, and CYP2D6 are associated with susceptibility and chemotherapy response in non-small-cell lung cancer patients. Lung. 2012;190:91–98. doi: 10.1007/s00408-011-9338-8. [DOI] [PubMed] [Google Scholar]
- 31.Liu D, Wang F, Wang Q, et al. Association of glutathione S-transferase M1 polymorphisms and lung cancer risk in a Chinese population. Clin Chim Acta. 2012;414:188–90. doi: 10.1016/j.cca.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Chen CM, Jin YT, Xu HY, et al. Effects of CYP1A1 and GSTM1 gene polymorphisms and BPDE-DNA adducts on lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012;29:23–27. doi: 10.3760/cma.j.issn.1003-9406.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Wang N, Wu Y, Zhou X, Wu Y. Association between genetic polymorphism of metabolizing enzymes and DNA repairing enzymes and the susceptibility of lung cancer in Henan population. Wei Sheng Yan Jiu. 2012;41:251–56. [PubMed] [Google Scholar]
- 34.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]