Cell-to-cell heterogeneity in gene expression is intrinsic to human cancers. Recent work has shown that heterogeneity provides a set of molecular states allowing for flexible cell fate decisions within a group of cancer cells.1,2 For example, tumor-propagating cells adopt a proliferative state to grow the overall population, whereas tumor-initiating cells can survive in harsh conditions and establish a new colony elsewhere in the body. For breast cancer, cells with tumorigenic potential are often identified by the expression of marker molecules, including CD44, CD133, α-6 integrin/CD49f, β-1 integrin/CD29, or the absence of CD24. Most markers described so far are cell-surface proteins that normally bind to adhesion ligands. Adhesion signaling enables a cell to respond dynamically to changes in its surroundings, and variability in adhesion-receptor expression implies differential single-cell adhesion. However, the functional importance of adhesion heterogeneity has not been addressed in the context of solid tumors.
We have developed a random-sampling approach, called stochastic profiling, that uncovers single-cell regulatory programs in adherent cells.3 By applying stochastic profiling to a basal-like human breast–epithelial clone cultured in basement membrane,4 we recently identified a pair of transcriptional regulatory states defined by transforming growth factor β receptor 3 (TGFBR3) and jun D proto-oncogene (JUND).5 JUND and TGFBR3 states are anticorrelated at the level of single cells and coexist at reciprocal frequencies in extracellular matrix (ECM)-attached cells (Fig. 1). By using various knockdown, overexpression, and addback approaches, we showed that proper regulation of these two states is critical for normal morphogenesis of 3D acini. Importantly, these states also appeared to become reengaged in premalignant breast lesions. We determined that the observed heterogeneity arises when the signaling–transcription circuit is excited by an environmental stimulus, causing cells to oscillate between states transiently and asynchronously. Surprisingly, we found that this regulatory circuit becomes disrupted upon ECM detachment.
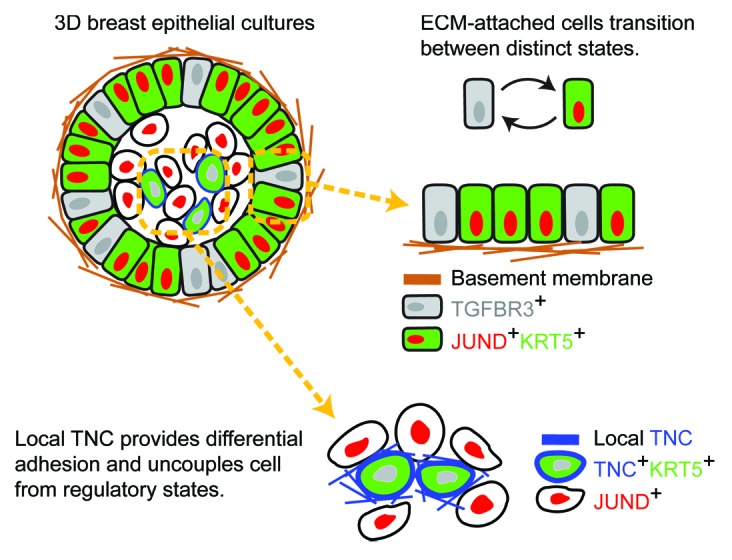
Figure 1. Adhesion ligands rewire transcriptional regulatory circuits to cause heterogeneity. ECM-attached cells in organotypic culture (upper left) mimic ECM-rich microenvironments, where cells undergo transient oscillations between states (upper right). Inner ECM-detached cells receive the juxtacrine TNC signal and are uncoupled from normal regulatory states (lower right).
Loss of ECM adhesion results in programmed cell death through a process called anoikis. However, before anoikis was evident in detached breast epithelial cells, we identified a small proportion of cells that dissolved their plasma and nuclear membranes and left behind keratinized skeletons of intermediate filament proteins. Keratinized cells display high levels of the diagnostic cytokeratin KRT5, whose heterogeneous expression is correlated with poor prognosis. Interestingly, we found that the breast cancer-associated ECM component, tenascin C (TNC), was also strongly upregulated in KRT5-positive, keratinized cells.5 TNC interacts with adjacent cells and allows them to survive in a JUND-positive state (Fig. 1). TNC adhesion thus uncouples the JUND–KRT5 regulatory states that are ordinarily tightly correlated when cells are adhered to basement membrane. We performed mouse xenograft experiments to show that TNC represents a crucial survival signal for cancer cells in ECM-poor environments. Our results suggest that local TNC provides differential adhesion that may drive functional differences in molecular states and cell-fate decisions.
Our finding opens up the possibility that cell adhesion might be an important input for regulatory circuits that act as multistate switches. Several regulatory circuits are crucial to the reversible transitions between cell states and govern normal development and tissue homeostasis.4,6 Further delineation of the ECM dependence of these circuits could enable us to understand how to set up and maintain specific molecular states in a given ECM context. Homogenization of single-cell states would be advantageous to drive more uniform therapeutic responses to anti-cancer drugs.
An interesting question for future studies is how widespread the extracellular triggers are that rewire the normal circuits to cause non-genetic cell-to-cell heterogeneity. Are other cancer-associated ECM molecules like TNC that may promote cancer cell survival by providing adhesive flexibility? Systematic analysis of non-genetic heterogeneity3,7 should provide important insight into the basis of adaptive cell states, which arise as a function of the local tissue context.
Wang CC, et al. Nat Cell Biol. 2014;16:345–56. doi: 10.1038/ncb2930.
References
- 1.Gupta PB, et al. Cell. 2011;146:633–44. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 2.Cleary AS, et al. Nature. 2014;508:113–7. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janes KA, et al. Nat Methods. 2010;7:311–7. doi: 10.1038/nmeth.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang CC, et al. Wiley Interdiscip Rev Syst Biol Med. 2012;4:51–78. doi: 10.1002/wsbm.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang CC, et al. Nat Cell Biol. 2014;16:345–56. doi: 10.1038/ncb2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janes KA. Cell Cycle. 2011;10:3461–5. doi: 10.4161/cc.10.20.18029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajikar SS, et al. Proc Natl Acad Sci U S A. 2014;111:E626–35. doi: 10.1073/pnas.1311647111. [DOI] [PMC free article] [PubMed] [Google Scholar]


