Abstract
Heterochromatin protein 1α (HP1α), a bona fide factor of silent chromatin, is required for establishing as well as maintaining the higher-order chromatin structure in eukaryotes. HP1α is decorated with several post-translational modifications, and many of these are critical for its cellular functions. HP1α is heavily phosphorylated; however, its physiological relevance had remained to be completely understood. We have recently demonstrated that human HP1α is a mitotic target for NDR kinase, and the phosphorylation at the hinge region of HP1α at the G2/M phase of the cell cycle is crucial for mitotic progression and Sgo1 loading at mitotic centromeres (Chakraborty et al., 2014). We now demonstrate that the dephosphorylation of HP1α within its hinge domain occurs during mitosis, specifically soon after prometaphase. In the absence of the hinge-specific HP1α phosphorylation, either as a consequence of depleting NDR1 or in cells expressing a non-phosphorylatable HP1α mutant, the cells arrest in prometaphase with several mitotic defects. In this study we show that NDR1-depleted cells expressing hinge-specific phosphomimetic HP1α mutant rescues the prometaphase arrest but displays defects in mitotic exit, suggesting that the dephosphorylation of HP1α is required for the completion of cytokinesis. Taken together, our results reveal that the phosphorylation–dephosphorylation cycle of HP1α orchestrates accurate progression of cells through mitosis.
Keywords: HP1α, NDR kinases, cell cycle, dephosphorylation, mitosis, phosphorylation
Introduction
The heterochromatinization of almost all the eukaryotic genome is facilitated by the protein HP1, a heterochromatin-associated protein originally discovered from the silenced region in Drosophila polytene chromosome.1 All the isoforms of HP1 (α, β, and γ) proteins have a conserved architecture consisting of an N-terminal chromodomain (CD), a C-terminal chromoshadow domain (CSD), and a flexible hinge region that links together the chromodomain and the chromoshadow domain.2-4 Similar to histones, HP1 isoforms are also excellent targets for post-translational modifications including acetylation, phosphorylation, methylation, sumoylation, and formylation,5-7 and often these modifications represent a response to inter- and intracellular signals. Phosphorylation of HP1/Swi6 in fission yeast promotes its chromatin binding and therefore is required for efficient heterochromatin organization.8 In Drosophila, HP1 phosphorylation is developmentally regulated and also required for heterochromatin formation.9,10 In mammals, recent proteomic analyses have identified multiple phosphorylation sites on HP1 in vivo.6,11 Phosphorylation at the N-terminal serine residues (S11–14) in human and mouse cells have been implicated in enhancing the binding affinity of HP1α for the H3K9 trimethyl chromatin mark.12 On the other hand, casein-kinase-II (CKII)-mediated phosphorylation of HP1β at Thr-51 occurs during DNA damage response, and this modification facilitates the release of HP1-β from the chromatin.13 The HP1γ isoform phosphorylated at Ser-83 demonstrates impaired silencing activity and is associated with transcriptional elongation.14 Abundant phosphorylation at Ser-93, -95, and -176 as well as formylation at S-176 of mammalian HP1γ have also been reported in human cells, but their exact roles remain to be determined.15 In mouse cells, initial targeting of HP1α to pericentric heterochromatin is promoted by the SUMOylation of HP1α at the hinge domain.16 In addition to its bona fide role in heterochromatin assembly and gene silencing, HP1 regulates proper chromosome segregation and accurate sister chromatid cohesion.17-19
The decision to enter mitosis brings a systemic and extensive physiological and structural reorganization in animal cells which is dependent on the activation of various mitotic kinases.20 Consequently, mitosis-specific phosphorylation events occur on a large number of substrates, including HP1α.21-26 In a recent study, we have identified a mitotic specific phosphorylation of mammalian HP1α at its hinge region. HP1α interacts with NDR (Nuclear-Dbf2-related) kinase at its chromoshadow domain and is phosphorylated at its hinge domain, predominantly during G2/M phase, and localizes to the kinetochores during early mitosis. Upon NDR kinase depletion, cells arrest at prometaphase with defects in chromosome alignment and a defect in Sgo1 binding to mitotic centromeres. Moreover, phospho-dead hinge mutants of HP1α phenocopy the mitotic defects that are observed upon NDR1 depletion. These results suggest that NDR1 kinase-mediated phosphorylation of HP1α is crucial for mitotic entry and proper chromosome alignment.27
NDR (nuclear-Dbf2-related) kinases are a highly conserved subfamily of serine/threonine kinases that control crucial cellular processes including mitotic exit, cytokinesis, cell growth, and proliferation and differentiation.28 In budding yeast, the NDR kinase ortholog, Dbf2p is required for mitosis exit network (MEN), whereas the Sid2p in fission yeast regulates septation initiation network (SIN).29-32 In humans, NDR kinases have been implicated in centrosome duplication,33 G1/S transition by regulating the stability of the cell cycle protein p21,34 and accurate chromosome alignment during mitosis.35 However, how NDR kinases regulate mitosis in mammalian cells had remained to be understood. We have recently shown that mammalian NDR kinases phosphorylate HP1α during G2/M phase, and that this phosphorylation is crucial for mitotic progression.27
In this “Extra Views” article we have further dissected the role of hinge-phosphorylated HP1α during the progression through mitosis. Our results suggest that dephosphorylation of hinge-phosphorylated HP1α begins at prometaphase, continues progressively through mitosis, and culminates by the end of mitosis, and this is necessary for the successful mitotic exit in mammalian cells. We suggest that multiple mechanisms regulate MEN in mammalian cells, and that the NDR kinase and HP1α dephosphorylation are key regulators of MEN.
Results
Dynamic phosphorylation and dephosphorylation states within the hinge domain of HP1α governs mitotic progression
We recently demonstrated that HP1α is phosphorylated within the hinge domain (serine-95) during G2/M phase of the cell cycle.27 This phosphorylation is catalyzed by NDR1 kinase. Further, depletion of NDR1 kinase shows significantly reduced phosphorylation at S-95, concomitant with cells arresting in prometaphase with chromosome alignment defects. Further, the binding of Sgo1, a protein crucial for sister chromatid cohesion, at the centromeres is compromised. Based on these results, we suggest that the S-95 phosphorylation of HP1α mediated by NDR1 helps Sgo1 localization at mitotic centromeres.27
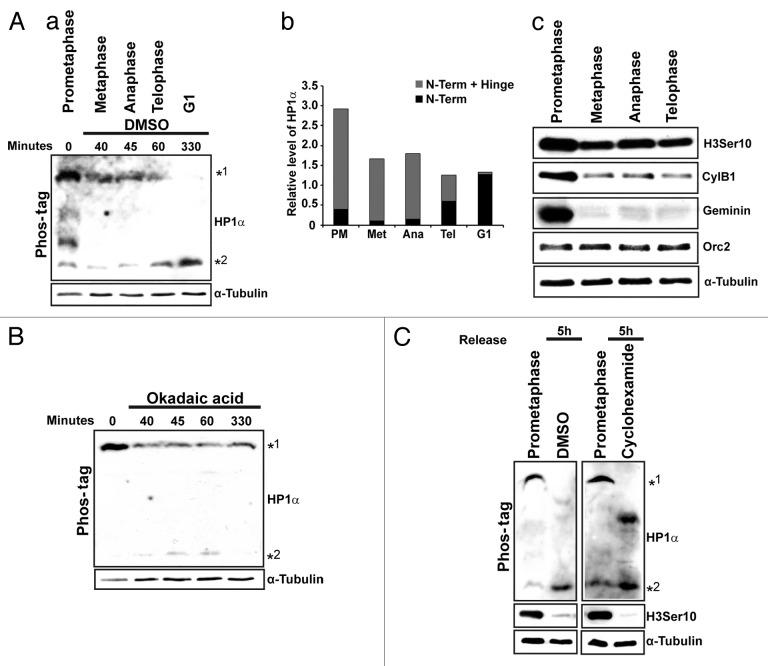
We next examined the status of the hinge phosphorylation of HP1α during various sub-stages of mitosis including prometaphase, metaphase, anaphase, and telophase. Cells were synchronized using nocodazole. The release from nocodazole arrest was monitored under the microscope; cells enriched at various mitotic substages and G1, were collected at specific time points, and the lysates were subject to immunoblot and phos-tag analysis (Fig. 1A; a and c). We observed that during prometaphase the hinge-specific phosphorylation of HP1α was the predominant one (Fig. 1A; panel a, slower migrating band *1). As the cells progressed into metaphase and anaphase, there was a clear decrease in the slower migrating form. By telophase the decrease in the slower migrating form (*1) concomitant with an increase in the faster migrating N-terminal phosphorylation form of HP1α (*2) was observed (Fig. 1A; a and b). Finally, G1 cells contain only the N-terminal phosphorylated form of HP1α (Fig. 1A; a and b). To address if the appearance of the chromo-specific phosphorylated form at the end of mitosis was a result of specific dephosphorylation at the hinge region of the already existing HP1α or due to the synthesis of new HP1α, we treated nocodazole-arrest-followed-by-release cells with Okadaic acid, a PP1 and PP2A phosphatase inhibitor or with cycloheximide to prevent new protein synthesis. The release was monitored under the microscope as well as by flow cytometry. Okadaic acid-treated cells continue to accumulate in mitosis with only a minor population of cells reaching G1 even 5 h post-release and continued to stay as doublets. The majority of these mitotic-released cells showed the presence of hinge-specific phosphorylation of HP1α even after 5 h post-release, and these continued to show reduced levels of the N-terminal phosphorylated form of HP1α (Fig. 1B). Both control (DMSO) and cycloheximide-treated mitotic cells showed the presence of the faster migrating form of HP1α (N-terminal phosphorylation) after 5 h of release from prometaphase, indicating that new protein synthesis is not required for the appearance of the N-terminal alone phosphorylated form of HP1α in post-mitotic cells (Fig. 1C). Our results suggest that the S-95 in the hinge region of HP1α is dephosphorylated during the later stages of mitosis. Occasionally, we observe a form of HP1α, especially in cycloheximide-treated mitotic cells, that has mobility in between the 2 phosphorylated forms (Fig. 1C). We suspect that this form may represent the hinge-only phosphorylated form.
Figure 1. Dephosphorylation of HP1α is required for mitotic exit. (A; a) Phos-tag PAGE analysis of lysates from different sub-stages of mitosis. Immunoblot analysis was performed using HP1α antibody. (b) Quantitation of the relative levels of N-terminal+ hinge and N-terminal phosphorylation of HP1α during different sub-stages of mitosis based on the immunoblot in (Aa). Note the accumulation of slower migrating (*1) form during prometaphase. Only the faster migrating form is evident during G1 (*2). (c) Immunoblot analysis of lysates collected from different mitotic substages. (B). Okadaic acid treatment of nocodazole arrest followed by release. Phos-tag PAGE analysis of these samples and immunoblot analysis with HP1α antibody. Note the retention of the slower migrating form of HP1α. (C). Phos-tag analysis of lysates (that were nocodazole arrested followed by release) treated with DMSO or cycloheximide.
It is therefore interesting to note that the phosphorylation of HP1α that facilitates Sgo1 recruitment to centromeres occurs only during a narrow time window when Sgo1 is needed at centromeres. Sgo1 binding to centromeres is significantly reduced during anaphase and completely lost during telophase.36 Concomitantly there is a loss of phosphorylation of HP1α as cells exit mitosis, and this could, in turn, result in the release of Sgo1 from the centromeres.
Dephosphorylation of HP1α orchestrates the successful completion of mitosis
To further evaluate the relevance of the phosphorylation/dephosphorylation of HP1α during mitosis, we carried out depletion of NDR1 in cells stably expressing YFP-HP1α-WT, YFP-HP1α-S95A-a phospho-dead mutant, and YFP-HP1α-S95E-a phospho-mimetic mutant. We have previously demonstrated that depletion of NDR1 kinase in cells expressing YFP-HP1α-WT shows accumulation of cells in prometaphase27 (Fig. 2A and B). Depletion of NDR1 kinase in YFP-HP1α-S95A mutant showed more dramatic increase in the population of prometaphase cells (68 ± 3% vs. 32 ± 3% in control; Fig. 2B). Further, depletion of NDR1 kinase in YFP-HP1α-S95E expressing cells, contrary to our expectations of rescuing the prometaphase arrest, showed even further increase in 4C DNA content (Fig. 2A). Detailed examination of the mitotic cells revealed that the cells were in telophase (52 ± 2% in telophase compared with 14 ± 0.5% in control; Fig. 2B and D). Immunoblot analysis showed accumulation of geminin in NDR-depleted YFP-HP1α-WT and YFP-HP1α-S95A-expressing cells consistent with a mitotic arrest (Fig. 2C). Interestingly, NDR depletion in YFP-HP1α-S95E showed a reduction in geminin levels consistent with cells being at the end of mitosis (Fig. 2C). These results suggested that hinge phosphorylation of HP1α facilitated by NDR kinases is required for mitotic progression, whereas its dephosphorylation is required for mitotic exit and completion of cytokinesis (Fig. 3). It is important to note that S95E shows this phenotype only in cells lacking NDR kinases, suggesting that multiple mechanisms are operable that govern the exit out of mitosis.
Figure 2. Dephosphorylation of HP1α is required for mitotic exit. (A) Depletion of NDR1 kinase using siRNA in YFP-HP1α-WT, YFP-HP1α-S95A, and YFP-HP1α-S95E cells. Note the flow profile shows increased G2/M accumulation in NDR1-si treated cells and a further increase in the YFP-HP1α-S95E background. (B) Distribution of cells at various sub-stages of mitosis following NDR1 siRNA treatment in YFP-HP1α-WT, YFP-HP1α-S95A, and YFP-HP1α-S95E cells. Note a significant increase in the telophase population in YFP-HP1α-S95E cells treated with NDR1 siRNA. Error bars represent s.d. of 3 independent experiments. Statistical significance was determined by Student’s t test. Mean ± s.d., *P < 0.05, **P < 0.01 and ***P < 0.001. (C) Immunoblot analysis of YFP-HP1α-WT, YFP-HP1α-S95A, and YFP-HP1α-S95E stable cells depleted of NDR1. (D). Phenotypic analysis of YFP-HP1α-S95E stable cells depleted of NDR1. Scale bar 10 µm.
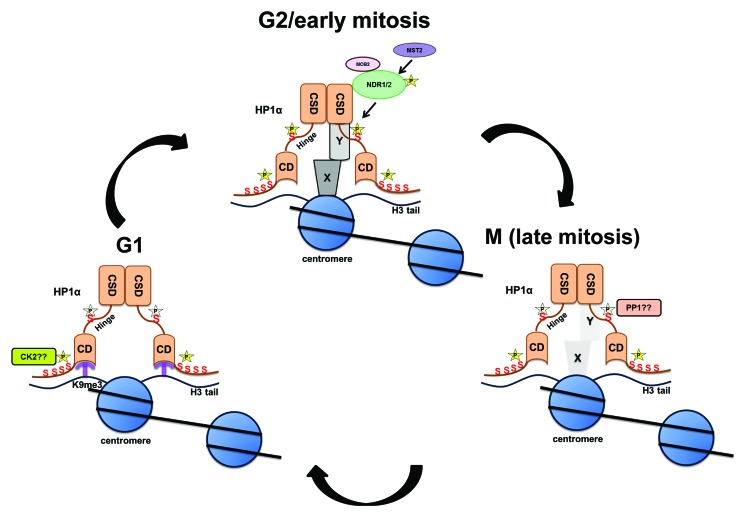
Figure 3. Model depicting the dynamics of HP1α phosphorylation during G1, G2/early mitosis, and in later parts of mitosis. Phosphorylation of the N terminus of HP1α facilitates its association to the chromatin mark H3trimethylK9. NDR kinase associates with the chromo-shadow domain of HP1α and phosphorylates the hinge domain. Hinge-phosphorylated form of HP1α localizes to centromeres, associates with centromeric proteins (X/Y denote unknown proteins at centromere), and governs chromosome alignment (schematic adapted and modified from Chakraborty et al., 2014). Starting at metaphase, HP1α is dephosphorylated (presumably by PP1/PP2A), and this releases this form of HP1α and associated partners (bound specifically to hinge-phosphorylated HP1α) from the centromere.
Discussion
The accumulation and destruction of cyclins and the simultaneous activation and inactivation of CDKs ensures that the eukaryotic cell cycle progresses accurately and unidirectionally. The progression of mitosis, which ensures the faithful segregation of newly synthesized DNA into 2 daughter nuclei depends on many factors, including the phosphorylation and degradation of several proteins. The most important mitotic kinase that determines the fate of interphase to mitotic transition is the cyclin-dependent kinase 1 (CDK1). Recent studies have also highlighted additional kinases, including Aurora kinase, Polo-like kinase, NIMA family kinases, mitotic checkpoint, and mitotic exit kinases.20 The activation of mitotic kinases is always associated with the phosphorylation of a wide range of mitotic substrates; the significance of such phosphorylation is often unclear.21-26 Once the chromosomes achieve the proper bipolar attachment to the spindle and the spindle assembly checkpoint is eased, the cell cycle moves toward the mitotic exit, which is accompanied by the proteolysis of key mitotic factors and dephosphorylation of a broad range of mitotic substrates. The regulation of mitotic exit relies on a fine-tuned balance between mitotic kinase inactivation and counteracting protein phosphatase activation.37 In budding yeast, the process of mitotic exit is quite well understood, where the phosphatase Cdc14 takes care of both CDK1 inactivation and the dephosphorylation of its substrates.38 The Cdc14 early anaphase release (FEAR) and MEN regulatory networks are required for the activation of Cdc14 in budding yeast.39 Unlike budding yeast, the mitotic exit network in animal cells is independent of CDC14 and relies on the regulatory network of the protein phosphatase PP1 and PP2A families.37
Studies from fission yeast,40,41 Drosophila42 as well as in mammalian cells,43,44 have established the requirement of HP1 for accurate chromosome segregation. During mitosis, Aurora B kinase-mediated phosphorylation at serine 10 of H3 (H3S10) disrupts the HP1 chromodomain (CD) interaction to the H3K9me3.3,45 Consequently most of the HP1α is released from chromatin except a small fraction that is left at the centromeres.46 In human cells, HP1α recruitment at mitotic centromeres is facilitated by INCENP through the CSD–PXVXL/I interaction.47,48 This centromeric pool of HP1α has previously been proposed to target Sgo1 at the mitotic centromere.49 A recent study has suggested that this mechanism is dispensable in sister chromatid cohesion in human cells.48 However, in fission yeast, the loading of the cohesins to the centromere and the establishment of the centromeric histone H3 variant CENP-ACnp1 rely on HP1/Swi6, which is crucial to regulate sister centromere cohesion and proper chromosome segregation.40,50-52 We have recently demonstrated that mammalian HP1α is a mitotic substrate for NDR (nuclear Dbf2-related) kinase and undergoes a hinge specific phosphorylation as the cells progress from G2 to mitosis.27 Constitutive phosphorylation within the N terminus12,27 of HP1α strengthens its binding to chromatin.12 We have observed that depletion of NDR1 causes the reduction of hinge-specific phosphorylation of HP1α and impaired Sgo1 loading to mitotic centromere.27 Further, NDR-depleted cells arrest in prometaphase with mitotic defects. This is consistent with a previous report that NDR1 kinase activity is maximal during mitosis, and that the Furry-mediated activation of NDR1 is crucial for chromosome alignment.35
Our results demonstrate that as the cells progress through mitosis, a hinge-specific dephosphorylation event occurs, which is required for mitotic exit. Post-mitotic cells treated with a PP1 and PP2A phosphatase inhibitor retained the hinge-specific phosphorylation, suggesting that PP1 may be the major contributor that dephosphorylates HP1α during mitosis.53 Expression of a hinge-specific phospho-mimetic mutant in the absence of NDR1 kinase resulted in the accumulation of cells in telophase. This is similar to what is observed upon depletion of LATS1 (another NDR kinase family member) in human cells. Loss of LATS1 results in cells accumulating in telophase, suggesting that LATS1 coordinates mitotic exit.54 Our results imply that dephosphorylation of HP1α during later stages of mitosis is an essential event that coordinates the mitotic exit network (MEN) program in mammalian cells. In yeast, the NDR kinase orthologs are required for the MEN and SIN (septation initiation network).55-57 While the MEN pathway coordinates accurate chromosome segregation and completion of mitosis, the SIN pathway regulates formation of the septum. Dbf2 orthologs and its upstream co-activators that constitute the Hippo pathway coordinate key cellular processes, including cell growth and proliferation.58-60 On the other hand, in fission yeast, NDR ortholog Orb6p has been implicated in cell polarity and morphogenesis, whereas Sid2p regulates cell division.61,62 In Drosophila, members of the NDR family have been shown to play key roles in cell proliferation and morphogenesis.63 Finally, in humans, NDR1, NDR2, LATS1 (large tumor suppressor-1), and LATS2 together constitute the NDR family of kinases.28 NDR kinases have been shown to be required for G1/S transition, centrosome duplication, and for mitotic chromosome alignment.34 Human LATS1 has been assigned as a kinase associated with mitotic exit network.54 Multiple mechanisms regulate MEN in mammalian cells; we propose that the dephosphorylation of HP1α is critical for this process.
Phosphorylation/dephosphorylation events during mitosis ensure the unidirectionality of the cell cycle.64 While phosphorylation of various mitotic substrates is required for reorganization of the mitotic spindle, the dephosphorylation of these substrates ensures accurate completion of mitosis. We demonstrate that the NDR-mediated phosphorylation of HP1α within its hinge domain is essential for mitotic progression, and that the subsequent dephosphorylation of HP1α is needed for mitotic exit. Failure to dephosphorylate HP1α during mitosis results in cells accumulating in telophase followed by cell death. Based on our results, we propose that the phosphorylation and dephosphorylation of HP1α at the hinge region governed by the NDR family of kinases and PP1/PP2A phosphatases, respectively, controls accurate mitotic progression (Fig. 3).
Materials and Methods
Cell culture, transfection, and generation of stable cell lines and antibodies
Human U2OS cells used in this study were grown in Dulbecco modified Eagle medium (DMEM) containing high glucose supplemented with 10% fetal bovine serum (FBS; Hyclone). Lipofectamine 2000 (Invitrogen) was used to transfect cells as per the manufacturer’s protocol. For generation of stable YFP-HP1α-WT, YFP-HP1α-mS95A, and YFP-HP1α-mS95E cell lines, corresponding plasmid constructs were transfected in U2OS cells followed by selecting and maintaining them in the cell culture media containing G418.
The antibodies used in this study were as follows: anti-HP1α (Chemicon), anti-α-tubulin (Sigma), and anti-geminin (Santa Cruz).
Cell synchronization
U2OS cells were synchronized at prometaphase by treating them with 50 ng/ml nocodazole for 12–16 h. Nocodazole-arrested cells were released in the fresh medium by washing them in PBS. The release from nocodazole arrest was monitored under the microscope; cells enriched at various mitotic substages and G1 were collected. Synchronized samples were evaluated by flow cytometry and immunoblot analysis.
Depletion of human NDR1/2
Small interfering RNA (siRNAs) targeting human NDR1 (IDT, USA) against 3′ UTR (sense: 5′ CCAAUAUGUC AUAGUAAAGU CUCCT3′, anti-sense: 3′GUGGUUAUAC AGUAUCAUUU CAGAGGA5′) were delivered into cells twice at a gap of 24 h, in the presence of Lipofectamine RNAimax (Invitrogen) at a final concentration of 10 µM. siRNA against control luciferase were described elsewhere.65,66
Flow cytometry and phos-tag analysis
For flow cytometry, cells were fixed in chilled ethanol overnight after resuspending in PBS + 1% NGS. After 2 rounds of washing, cells were resuspended in PBS + 1% NGS with 120 µg/ml propidium iodide (PI) and 10 µg/ml RNase A followed by 30 min incubation at 37 °C. DNA content was measured by flow cytometry.
To detect the phosphorylation based on band shift, phos-tag SDS-PAGE was employed according to the manufacturer’s protocol (AAL-107; NARD Institute). Protein samples treated with CIP (NEB) was used as dephosphorylated control.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs Pierre Chambon, Mirek Dundr, Brian Hemmings, Yasushi Hiraoka, J Nakayama, Kannanganattu Prasanth, Hongtao Yu, D Spector, and B Stillman for providing reagents and suggestions. We would like to thank Drs K Prasanth and S Ceman for critical reading of the paper. This work was supported by NSF career (1243372) and NIH (1RO1GM099669) awards to S.G.P.
References
- 1.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–72. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–73. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 4.Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164–74. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissenberg JC, Elgin SC. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103–10. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeRoy G, Weston JT, Zee BM, Young NL, Plazas-Mayorca MD, Garcia BA. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–42. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maison C, Bailly D, Roche D, Montes de Oca R, Probst AV, Vassias I, Dingli F, Lombard B, Loew D, Quivy JP, et al. SUMOylation promotes de novo targeting of HP1α to pericentric heterochromatin. Nat Genet. 2011;43:220–7. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 8.Shimada A, Murakami Y. Dynamic regulation of heterochromatin function via phosphorylation of HP1-family proteins. Epigenetics. 2010;5:30–3. doi: 10.4161/epi.5.1.10605. [DOI] [PubMed] [Google Scholar]
- 9.Eissenberg JC, Ge YW, Hartnett T. Increased phosphorylation of HP1, a heterochromatin-associated protein of Drosophila, is correlated with heterochromatin assembly. J Biol Chem. 1994;269:21315–21. [PubMed] [Google Scholar]
- 10.Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12:1671–85. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–34. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 12.Hiragami-Hamada K, Shinmyozu K, Hamada D, Tatsu Y, Uegaki K, Fujiwara S, Nakayama J. N-terminal phosphorylation of HP1alpha promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 14.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–15. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 15.LeRoy G, Weston JT, Zee BM, Young NL, Plazas-Mayorca MD, Garcia BA. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–42. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 17.Ainsztein AM, Kandels-Lewis SE, Mackay AM, Earnshaw WC. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Biol. 1998;143:1763–74. doi: 10.1083/jcb.143.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obuse C, Iwasaki O, Kiyomitsu T, Goshima G, Toyoda Y, Yanagida M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat Cell Biol. 2004;6:1135–41. doi: 10.1038/ncb1187. [DOI] [PubMed] [Google Scholar]
- 20.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 21.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengeveld RC, Hertz NT, Vromans MJ, Zhang C, Burlingame AL, Shokat KM, Lens SM. Development of a chemical genetic approach for human aurora B kinase identifies novel substrates of the chromosomal passenger complex. Mol Cell Proteomics. 2012;11:47–59. doi: 10.1074/mcp.M111.013912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Körner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;8:4553–63. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- 24.Nousiainen M, Silljé HH, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci U S A. 2006;103:5391–6. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 26.Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, Schmidt A, Silljé HH, Körner R, Nigg EA. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics. 2011;10:004457. doi: 10.1074/mcp.M110.004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty A, Prasanth KV, Prasanth SG. Dynamic phosphorylation of HP1α regulates mitotic progression in human cells. Nat Commun. 2014;5:3445. doi: 10.1038/ncomms4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–64. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 29.Grallert A, Connolly Y, Smith DL, Simanis V, Hagan IM. The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nat Cell Biol. 2012;14:738–45. doi: 10.1038/ncb2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol. 2001;11:784–8. doi: 10.1016/S0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 31.Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–13. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visintin R, Amon A. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol Biol Cell. 2001;12:2961–74. doi: 10.1091/mbc.12.10.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–34. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Human NDR kinases control G(1)/S cell cycle transition by directly regulating p21 stability. Mol Cell Biol. 2011;31:1382–95. doi: 10.1128/MCB.01216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiba S, Ikeda M, Katsunuma K, Ohashi K, Mizuno K. MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr Biol. 2009;19:675–81. doi: 10.1016/j.cub.2009.02.054. [DOI] [PubMed] [Google Scholar]
- 36.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12:469–82. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- 38.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–32. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 39.Geymonat M, Jensen S, Johnston LH. Mitotic exit: the Cdc14 double cross. Curr Biol. 2002;12:R482–4. doi: 10.1016/S0960-9822(02)00963-6. [DOI] [PubMed] [Google Scholar]
- 40.Ekwall K, Javerzat JP, Lorentz A, Schmidt H, Cranston G, Allshire R. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science. 1995;269:1429–31. doi: 10.1126/science.7660126. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama J, Klar AJ, Grewal SI. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell. 2000;101:307–17. doi: 10.1016/S0092-8674(00)80840-5. [DOI] [PubMed] [Google Scholar]
- 42.Kellum R, Alberts BM. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J Cell Sci. 1995;108:1419–31. doi: 10.1242/jcs.108.4.1419. [DOI] [PubMed] [Google Scholar]
- 43.Quivy JP, Gérard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–9. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- 44.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 46.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–80. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 47.Hayakawa T, Haraguchi T, Masumoto H, Hiraoka Y. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J Cell Sci. 2003;116:3327–38. doi: 10.1242/jcs.00635. [DOI] [PubMed] [Google Scholar]
- 48.Kang J, Chaudhary J, Dong H, Kim S, Brautigam CA, Yu H. Mitotic centromeric targeting of HP1 and its binding to Sgo1 are dispensable for sister-chromatid cohesion in human cells. Mol Biol Cell. 2011;22:1181–90. doi: 10.1091/mbc.E11-01-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–5. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 50.Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–42. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- 51.Folco HD, Pidoux AL, Urano T, Allshire RC. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–7. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 53.Wu JQ, Guo JY, Tang W, Yang CS, Freel CD, Chen C, Nairn AC, Kornbluth S. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol. 2009;11:644–51. doi: 10.1038/ncb1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. Human LATS1 is a mitotic exit network kinase. Cancer Res. 2005;65:6568–75. doi: 10.1158/0008-5472.CAN-05-0862. [DOI] [PubMed] [Google Scholar]
- 55.Grallert A, Connolly Y, Smith DL, Simanis V, Hagan IM. The S. pombe cytokinesis NDR kinase Sid2 activates Fin1 NIMA kinase to control mitotic commitment through Pom1/Wee1. Nat Cell Biol. 2012;14:738–45. doi: 10.1038/ncb2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol. 2001;11:784–8. doi: 10.1016/S0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- 57.Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–13. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–73. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 59.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–97. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 60.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–92. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–90. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verde F, Wiley DJ, Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci U S A. 1998;95:7526–31. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geng W, He B, Wang M, Adler PN. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics. 2000;156:1817–28. doi: 10.1093/genetics/156.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Merrick KA, Fisher RP. A virtual cycle: theory and experiment converge on the exit from mitosis. F1000 Biol Rep. 2010;2 doi: 10.3410/B2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 66.Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci U S A. 2010;107:15093–8. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]