Abstract
The enzyme Dicer is central to the production of small silencing RNAs such as microRNAs (miRNAs) and small interfering RNAs (siRNAs). Like other insects, Drosophila melanogaster uses different Dicers to make siRNAs and miRNAs: Dicer-1 produces miRNAs from pre-miRNAs, whereas Dicer-2 generates siRNAs from long double-stranded RNA (dsRNA). How do the 2 Dicers achieve their substrate specificity? Here, we review recent findings that inorganic phosphate restricts the substrate specificity of Dicer-2 to long dsRNA. Inorganic phosphate inhibits Dicer-2 from binding and cleaving pre-miRNAs, without affecting the processing of long dsRNA. Crystal structures of a fragment of human Dicer in complex with an RNA duplex identify a phosphate-binding pocket that recognizes both the 5′-monophosphate of a substrate RNA and inorganic phosphate. We propose that inorganic phosphate occupies the phosphate-binding pocket in the fly Dicer-2, blocking binding of pre-miRNA and restricting pre-miRNA processing to Dicer-1. Thus, a small molecule can alter the substrate specificity of a nucleic acid-processing enzyme.
Keywords: dicer, siRNA, miRNA, dsRNA, phosphate
Introduction
Drosha cleaves primary miRNA transcripts (pri-miRNAs) into precursor miRNAs (pre-miRNAs), and Dicer cleaves pre-miRNAs into miRNA duplexes (reviewed in ref. 1). Dicer also produces small interfering RNAs (siRNAs) from long dsRNA. Endogenous sources of long dsRNA include viral RNA genomes or replication intermediates, convergent mRNA transcription, transposon transcripts, and partially self-complementary hairpin RNAs. Long dsRNA can, of course, be introduced experimentally to trigger RNA interference (RNAi).2 After their production, miRNAs and siRNAs are loaded into members of the Argonaute family of proteins, for which they act as sequence-specific guides. miRNA–Argonaute complexes can repress mRNA translation and promote mRNA turnover, whereas siRNA–Argonaute complexes generally cleave their RNA targets.
In mammals, a single Dicer produces both miRNAs and siRNAs. In contrast, in Drosophila and other arthropods, Dicer-1 makes miRNAs (22–24 nt), while Dicer-2 produces siRNAs (~21 nt).3 What restricts each Dicer to its specific substrate? Here, we discuss the recent findings that inorganic phosphate (PO4), a small molecule found in all cells, restricts the substrate specificity of fly Dicer-2 to long dsRNA. We also discuss the recently reported crystal structures of a fragment of human Dicer, which contains a pocket that binds the 5′-monophosphate (PO4) of substrate RNA; inorganic phosphate can also bind this pocket. We propose that PO4 binds the phosphate-binding pocket of fly Dicer-2, blocking binding of inappropriate substrates such as pre-miRNAs.
Dicer Domain Architecture
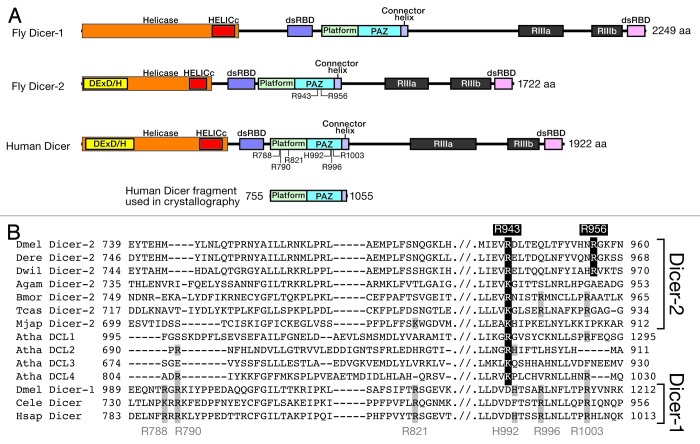
Drosophila Dicer-1 and Dicer-2 and human Dicer all share a common domain architecture (Fig. 1): an N-terminal “helicase” domain; a central, atypical dsRNA-binding domain (dsRBD, previously known as DUF283); a platform domain; a PAZ domain; a connector helix; 2 RNase III domains; and a C-terminal dsRBD. PAZ domains are found in both Dicer and Argonaute proteins and recognize the characteristic 2-nucleotide, 3′-overhang left by Drosha and Dicer cleavage.4,5 In addition, the PAZ domain of human Dicer recognizes the 5′ monophosphate of pre-miRNAs.6
Figure 1. Dicer domain architecture. (A) Domain structures of fly and human Dicers. DExD/H, DExD/DExH box helicase domain; HELICc, helicase conserved C-terminal domain; dsRBD, dsRNA-binding domain; PAZ, PAZ domain; RIIIa and RIIIb, Ribonuclease III domain. The fragment of human Dicer containing the platform domain, the PAZ domain, and the connector helix, whose crystal structure was determined in complex with dsRNA (Fig. 5)19 is also shown. (B) Alignment of the platform and PAZ domain sequences from Dicers from various arthropods, Arabidopsis, C. elegans, and humans. Dmel, Drosophila melanogaster; Dere, Drosophila erecta; Dwil, Drosophila willistoni; Agam, Anopheles gambiae; Bmor, Bombyx mori; Tcas, Tribolium castaneum; Mjap, Marsupenaeus japonicus; Atha, Arabidopsis thaliana; Cele, Caenorhabditis elegans; Hsap, Homo sapiens. In this review, we number human Dicer amino acid residues based on its 1922-aa full-length sequence, whereas in the Park et al. and Tian et al. papers6,19 and the PDB files therein, the amino acid residues are numbered based on 1,912 aa sequence, which is lacking the first 10 aa compared with the 1922 aa sequence.
Both Drosophila Dicer-1 and Dicer-2 contain an N-terminal “helicase” domain, but only the Dicer-2 helicase domain binds and hydrolyzes ATP.3,7-11 Dicer-2 is thought to use ATP to translocate along its dsRNA substrate, allowing it to produce siRNA duplexes processively from an end.12,13 In contrast, fly Dicer-1 as well as human Dicer need no ATP to produce miRNA/miRNA* duplexes from pre-miRNAs.8,14,15
Fly Dicer-2 produces shorter miRNAs with an altered seed sequence
Dicer-1 does not cleave long dsRNA, making it specific for pre-miRNAs.12,16 In contrast, purified, recombinant Dicer-2 efficiently cleaves pre-miRNAs, producing miRNA products that are 1–2 nt shorter than the authentic miRNA products produced by Dicer-1 (Fig. 2).12,16 For example, purified Dicer-1 cleaves pre-miR-8 mainly into 23-nt-long miR-8, which is the predominant isoform in vivo (Fig. 2).16 In contrast, Dicer-2 cleaves pre-miR-8 into 21- and 22-nt-long products. The target specificity of an miRNA is determined mainly by its seed sequence: positions 2–8 of the small RNA guide.17,18 Like ~60% of fly miRNAs, miR-8 resides on the 3′-arm of its pre-miRNA, so the shorter miR-8 isoforms produced by Dicer-2 have a seed sequence, AUACUGU or UACUGUC, that differs from that of the authentic miR-8 produced by Dicer-1, AAUACUG. Similarly, Dicer-2 cleaves pre-miR-79 into a 21-nt-long miR-79 isoform whose seed sequence, AAGCUAG, differs from that of the authentic 22-nt long miR-79 produced by Dicer-1 (AAAGCUA).16 The shorter miRNA products produced by Dicer-2 would bind and silence mRNAs different from those controlled by the canonical miRNAs and are therefore likely to have detrimental consequences.
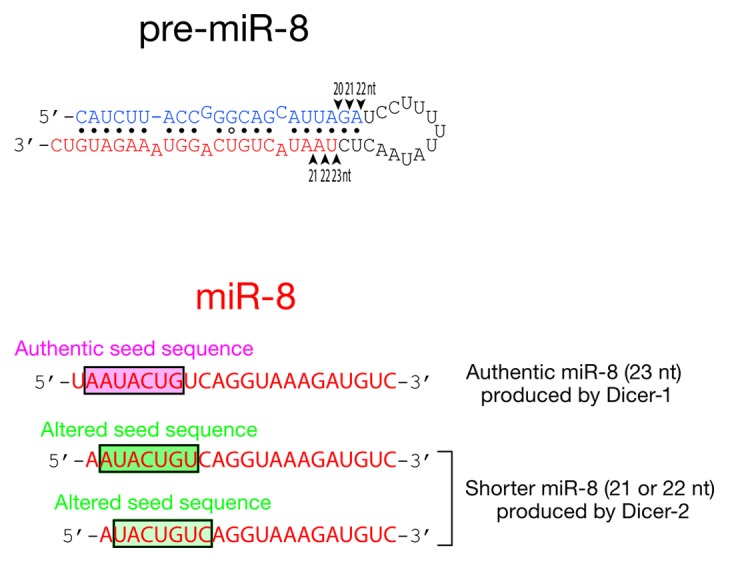
Figure 2. Purified fly Dicer-2 produces shorter RNA products from pre-miRNA than by purified fly Dicer-1. The shorter miR-8 isoforms (21 or 22 nt) produced by Dicer-2 have an altered seed sequence compared with the authentic miR-8 (23 nt) produced by Dicer-1.
Inorganic Phosphate Inhibits Dicer-2 from Cleaving Pre-miRNA
Although purified Dicer-2 can process pre-miRNAs, high-throughput sequencing analyses of the control and dicer-2 mutant flies show that pre-miRNAs are not processed by Dicer-2 in vivo,16 suggesting that some inhibitor exists in vivo to suppress pre-miRNA cleavage by Dicer-2.
Surprisingly, we found that inorganic phosphate, a small molecule found in all cells, restricts the substrate specificity of Dicer-2 to long dsRNA12,16 (Fig. 3). Dicer-2 cleavage of pre-miRNAs is suppressed by physiological concentrations (8–25 mM) of inorganic phosphate in a dose-dependent manner. Phosphate inhibition of Dicer-2 is specific: inorganic phosphate affects neither the cleavage by Dicer-2 of long dsRNA into siRNAs, nor the cleavage by Dicer-1 of pre-miRNA into miRNAs. Other anions do not inhibit the cleavage of pre-miRNA by Dicer-2.
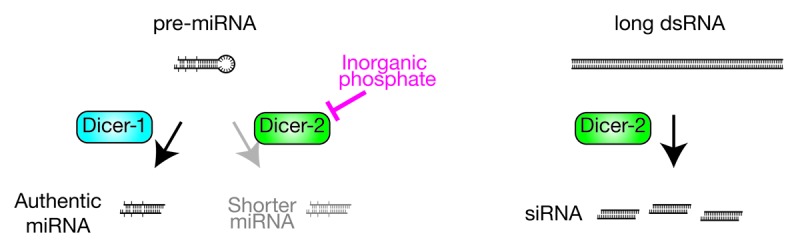
Figure 3. Inorganic phosphate inhibits fly Dicer-2 from cleaving pre-miRNA. Dicer-2 cleaves pre-miRNA in the absence of inorganic phosphate, and inorganic phosphate inhibits this pre-miRNA cleavage activity. Inorganic phosphate does not affect pre-miRNA cleavage by Dicer-1 or long dsRNA cleavage by Dicer-2.
Inorganic Phosphate Inhibits Dicer-2 from Cleaving Short RNA
What features of pre-miRNA and long dsRNA underlie the specific inhibition by inorganic phosphate of pre-miRNA cleavage by fly Dicer-2? To test if (1) mismatches in the stem, (2) a terminal loop, and/or (3) short length is the determinant, various substrate RNAs were designed and tested.16 The results revealed that short length is the determinant: inorganic phosphate inhibits Dicer-2 cleavage of short dsRNA (<38 bp) but not long dsRNA (>38 bp; Fig. 4). Binding and kinetics assays revealed that inorganic phosphate inhibits binding to Dicer-2 of short dsRNA and pre-miRNA, but not long dsRNA.
Figure 4. Fly Dicer-2 uses distinct mechanisms to cleave short and long dsRNAs. Summary of the in vitro RNA cleavage assay results using recombinant fly Dicer-2.16 Cleavage of short dsRNA (<38 bp) by Dicer-2 is inhibited by inorganic phosphate, requires an end with a 5′-monophosphate, a 2-nucleotide 3′-overhang, and 2 arginine residues (Arg943 and Arg956) in the PAZ domain, but does not require ATP. Cleavage of long dsRNA (>38 bp) by Dicer-2 is not inhibited by inorganic phosphate, does not require a specific terminal structure or the arginine residues, but requires ATP.
Fly Dicer-2 Uses Distinct Mechanisms to Cleave Short and Long dsRNAs
These results imply that Dicer-2 cleavage of short and long dsRNA are mechanistically different. In fact, cleavage of short dsRNA by Dicer-2 requires an end with a 5′-monophosphate and a 2-nucleotide 3′-overhang, but not ATP, whereas cleavage of long dsRNA requires no specific terminal structure, but does require ATP (Fig. 4).16
Furthermore, two (2) evolutionarily conserved arginine residues (Arg943 and Arg956) in the fly Dicer-2 PAZ domain are required for short dsRNA cleavage, but dispensable for long dsRNA cleavage. Arg943 is highly conserved in siRNA producing Dicer-2 proteins among arthropods; Arg956 is conserved in Dicer-2 enzymes among Drosophila species (Fig. 1). Neither residue is present in miRNA-producing enzymes such as fly Dicer-1, C. elegans Dicer, or human Dicer. We propose that the 2 arginines comprise a phosphate-binding pocket that recognizes the 5′-monophosphate of pre-miRNAs, and that inorganic phosphate occupies this pocket in vivo, blocking pre-miRNA binding.
Phosphate-Binding Pocket Identified in the Human Dicer Crystal Structures
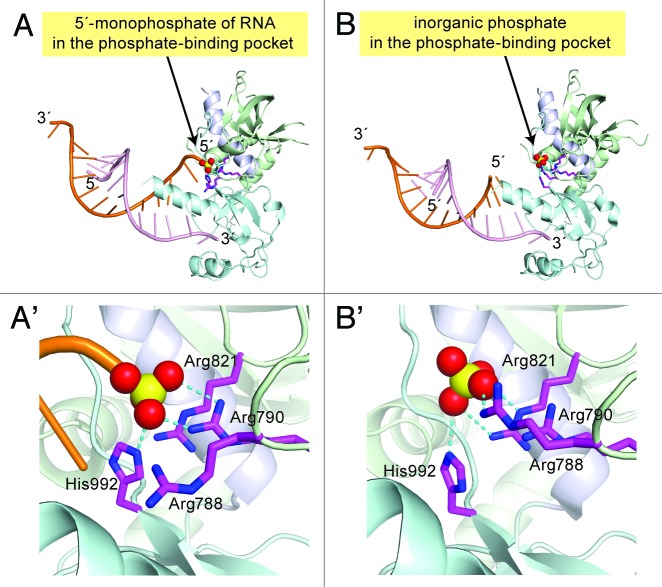
In 2011, the Kim group reported that human Dicer has a phosphate-binding pocket in its PAZ domain that recognizes the 5′-monophosphate of substrate RNAs.6 More recently, the Patel group reported several crystal structures of human Dicer fragment (“platform-PAZ-connector helix”, amino acid residues 765–1065 in complex with an RNA duplex (Figs. 1A and 5).19
Figure 5. Crystal structures of a human Dicer fragment complexed with dsRNA. (A) A human Dicer fragment bound with 10 bp dsRNA (light pink and orange) with 5′-monophosphorylated frayed ends (PDB ID: 4NH5). The platform domain, the PAZ domain, and the connector helix are colored light green, light cyan, and white, respectively (Fig. 1A). The dsRNA 5′-monophosphate (shown as a ball and stick model in yellow and red) is bound in the phosphate-binding pocket, where it is recognized by the side chains of Arg788, Arg790, Arg821, and His992 (shown as a ball and stick model in magenta). Direct hydrogen bonds between the phosphate and the basic side chains are shown as cyan dotted lines. A closer view of the phosphate-binding pocket is shown in (A'). (B) A human Dicer fragment bound with 10 bp dsRNA with 5′ hydroxyl and 1 nt overhang and 3′ 2-nucleotide overhang (PDB ID: 4NGG). The inorganic phosphate (shown as a ball and stick model in yellow and red) is bound in the phosphate-binding pocket and recognized by the side chains of Arg788, Arg790, Arg821, and His992. The 5′ end of dsRNA is away from the phosphate-binding pocket (the terminal 5′ OH is disordered). The closer view of the phosphate-binding pocket is shown in (B').
Interestingly, the structures reveal a phosphate-binding pocket. In one structure, the 5′-monophosphate of the bound dsRNA is recognized in a phosphate-binding pocket composed of the side chains of Arg788, Arg790, Arg821, His992, Arg996, Arg1003 (Fig. 5A and A').19 The first 3 residues are located in the platform domain, while the latter 3 reside in the PAZ domain. The side chains of Arg790 and His992 contact the non-bridging oxygen atoms of the 5′-monophosphate. In contrast, in another structure, the 5′ end (5′-hydroxyl) of dsRNA is not bound in the phosphate-binding pocket, but is instead exposed to the solvent (Fig. 5B and B').19 Interestingly, in this structure, inorganic phosphate is bound to the phosphate-binding pocket with the side chains of Arg788, Arg821, and His992 contacting the oxygen atoms of the phosphate.
A Phosphate-Binding Pocket in Fly Dicer-2?
By analogy to the human Dicer phosphate-binding pocket, we propose that fly Dicer-2 has a phosphate-binding pocket in its PAZ domain. The lack of sequence conservation between Dicer-2 and the related enzymes Dicer-1 and human Dicer suggests that the architecture of the Dicer-2 pocket differs from that in human Dicer (Fig. 1B). We envision that the phosphate-binding pocket of the Dicer-2 PAZ domain can bind the 5′-monophosphate of a pre-miRNA substrate, but that in vivo inorganic phosphate occupies the pocket, blocking pre-miRNA binding (Fig. 6). For long dsRNA, the end structure of the RNA substrate would not bind the PAZ domain. Alternatively, the bound inorganic phosphate might be displaced from the binding pocket by the 5′-monophosphate of long dsRNA because of high-affinity interactions between a long dsRNA substrate and other Dicer-2 domains, such as the helicase and dsRNA-binding domains.
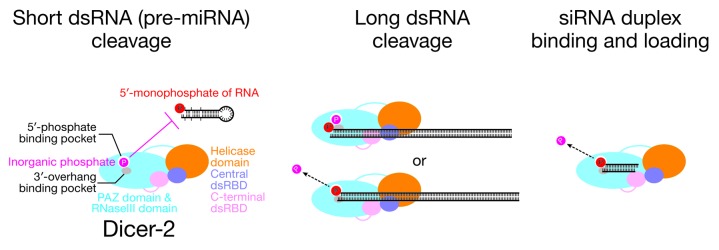
Figure 6. Models for the inhibition by inorganic phosphate of fly Dicer-2 processing of short, but not long, dsRNA and the role of the phosphate-binding pocket in siRNA loading. The PAZ domain of fly Dicer-2 has binding pockets for the 5′-monophosphate and the 2-nucleotide 3′-overhang of a pre-miRNA substrate. An inorganic phosphate molecule occupies the binding pocket in place of the 5′-monophosphate of the pre-miRNA, blocking its binding. Long dsRNA is recognized mainly by the helicase domain and/or the dsRBD domains and does not need a specific terminal structure to be efficiently cleaved. Long dsRNA is not blocked by the bound inorganic phosphate, since the strong interaction with the helicase domain or the dsRBD domains allows it either to bind tightly enough to Dicer-2 without additional interactions with the PAZ domain or to displace the inorganic phosphate from the binding pocket. The phosphate-binding pocket binds 5′-monophosphate of siRNA duplex in the loading step. The bound inorganic phosphate is displaced by the siRNA.
Function of the Phosphate-Binding Pocket In Fly Dicer-2
In human Dicer, the phosphate-binding pocket is required for accurate and efficient cleavage of pre-miRNA.6,19 In fly Dicer-2, the phosphate-binding pocket is dispensable for efficient and accurate cleavage of its natural substrate, long dsRNA;16 the pocket is only needed to cleave short dsRNA and pre-miRNA, an activity normally inhibited by inorganic phosphate. Why then has Dicer-2 retained the phosphate-binding pocket?
We propose that the phosphate-binding pocket of fly Dicer-2 participates in loading siRNA duplexes into Argonaute2 (Fig. 6). In addition to cleaving long dsRNA into siRNA duplexes, Dicer-2 collaborates with the dsRNA-binding protein R2D2 to sense the thermodynamic asymmetry of each siRNA duplex and to load the duplex into Argonaute2 in the correct orientation.20,21 During siRNA loading, Dicer-2 directly binds siRNA duplex.21 Considering that siRNA duplexes are short dsRNAs, we propose that binding of a siRNA duplex requires recognition of the 5′-monophosphate of the siRNA duplex by the phosphate-binding pocket of the Dicer-2 PAZ domain. We imagine that a bound inorganic phosphate may be displaced by the siRNA duplex and not inhibit loading (Fig. 6). Displacement of inorganic phosphate by the siRNA 5′-monophosphate may be facilitated by the association of Dicer-2 with R2D2. Notably, R2D2, like inorganic phosphate, also inhibits pre-miRNA processing by Dicer-2.12
Inorganic Phosphate Enhances DCL3 and Inhibits DCL4 in Plants
The plant Arabidopsis thaliana produces 4 Dicer enzymes (DICER-LIKE 1 through 4, DCL1–4).22,23 DCL1 acts as both Drosha and Dicer, sequentially cleaving pri-miRNA and pre-miRNA to produce miRNAs. Plant siRNAs are made from long dsRNA by DCL2, DCL3, and DCL4. The 24-nt siRNAs produced by DCL3 suppress transposons via RNA-directed DNA methylation, a form of transcriptional gene silencing, whereas the 21-nt siRNAs generated by DCL4 defend plants against viruses and regulate mRNA expression by post-transcriptional mechanisms.
Interestingly, the Fukuhara group found that in Arabidopsis seedling lysate, physiological concentrations of inorganic phosphate enhance DCL3 but inhibit DCL4 cleavage of a 50-bp dsRNA.24 The authors propose that inorganic phosphate may regulate substrate specificity in plants, just as in flies. DCL3 and DCL4 do not have basic residues corresponding to those in human Dicer (Arg788, Arg790, Arg821, His992), and in general, the degree of alignment in this region of the plant and human enzymes is poor (Fig. 1B). Intriguingly, DCL3 and DCL4, as well as DCL1 and DCL2, do have a basic residue in the corresponding position to Arg943 of fly Dicer-2. It is tempting to speculate that Lys913 of DCL3 and Lys1016 of DCL4 might be part of a phosphate-binding pocket.
Perspectives
The studies discussed here help explain how a small molecule can alter the substrate specificity of a nucleic acid-processing enzyme. Challenges for the future include testing the model that inorganic phosphate restricts the substrate specificity of Dicer-2 in vivo, perhaps by determining the structure of the fly Dicer-2 PAZ domain and using mutants with an altered affinity for phosphate. A structure should help define the proposed phosphate-binding pocket and uncover how the domain recognizes the 5′-monophosphate of RNA and inorganic phosphate.
The findings that a small molecule—inorganic phosphate—can bind human Dicer and alter the activities of fly Dicer-2 and plant DCL3 and DCL4 suggests that small-molecule drugs could be found that alter the activity or specificity of human Dicer. Such drugs might inhibit or enhance the production of miRNAs, such as those that promote or inhibiting tumorigenesis;25,26 promote destruction of toxic Alu RNA, causing macular degeneration;27,28 enhance anti-viral processing activity;29,30 or help restore Dicer function in neurodegenerative diseases.31
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by funds provided by the Department of Biological Chemistry at the Johns Hopkins School of Medicine.
References
- 1.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/S0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 4.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–9. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 5.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–7. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 6.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–5. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 8.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–9. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–22. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–74. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 11.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–32. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 12.Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42:172–84. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–99. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011;18:1153–8. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 15.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–43. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukunaga R, Colpan C, Han BW, Zamore PD. Inorganic phosphate blocks binding of pre-miRNA to Dicer-2 via its PAZ domain. EMBO J. 2014;33:371–84. doi: 10.1002/embj.201387176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–46. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Tian Y, Simanshu DK, Ma JB, Park JE, Heo I, Kim VN, Patel DJ. A phosphate-binding pocket within the platform-PAZ-connector helix cassette of human Dicer. Mol Cell. 2014;53:606–16. doi: 10.1016/j.molcel.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–5. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 21.Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–80. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- 22.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–26. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagano H, Fukudome A, Hiraguri A, Moriyama H, Fukuhara T. Distinct substrate specificities of Arabidopsis DCL3 and DCL4. Nucleic Acids Res. 2014;42:1845–56. doi: 10.1093/nar/gkt1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 26.Srikantan S, Gorospe M, Abdelmohsen K. Senescence-associated microRNAs linked to tumorigenesis. Cell Cycle. 2011;10:3211–2. doi: 10.4161/cc.10.19.17050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–59. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–4. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–8. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I, et al. miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci U S A. 2010;107:13111–6. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]