Abstract
One common cancer chemotherapeutic strategy is to perturb cell division with anti-mitotic drugs. Paclitaxel, the classic microtubule-targeting anti-mitotic drug, so far still outperforms the newer, more spindle-specific anti-mitotics in the clinic, but the underlying cellular mechanism is poorly understood. In this study we identified post-slippage multinucleation, which triggered extensive DNA damage and apoptosis after drug-induced mitotic slippage, contributes to the extra cytotoxicity of paclitaxel in comparison to the spindle-targeting drug, Kinesin-5 inhibitor. Based on quantitative single-cell microscopy assays, we showed that attenuation of the degree of post-slippage multinucleation significantly reduced DNA damage and apoptosis in response to paclitaxel, and that post-slippage apoptosis was likely mediated by the p53-dependent DNA damage response pathway. Paclitaxel appeared to act as a double-edge sword, capable of killing proliferating cancer cells both during mitotic arrest and after mitotic slippage by inducing DNA damage. Our results thus suggest that to predict drug response to paclitaxel and anti-mitotics in general, 2 distinct sets of bio-markers, which regulate mitotic and post-slippage cytotoxicity, respectively, may need to be considered. Our findings provide important new insight not only for elucidating the cytotoxic mechanisms of paclitaxel, but also for understanding the variable efficacy of different anti-mitotic chemotherapeutics.
Keywords: anti-mitotic drug, mitotic slippage, apoptosis, multinucleation, DNA damage, mitotic arrest, paclitaxel, Kinesin-5 inhibitor
Introduction
Inhibiting cell proliferation by perturbing mitosis is a widely used therapeutic strategy for cancer treatment. Currently available anti-mitotic chemotherapeutics mainly include classic microtubule-targeting drugs, such as paclitaxel and vinca alkaloids, and the new spindle-specific drugs, such as inhibitors of Kinesin-5 (a.k.a., KSP, Eg5, KIF11), Polo kinase-1, and Aurora kinases. Anti-mitotic drugs work by activating prolonged mitotic arrest and subsequently triggering cancer cell death either during mitotic arrest or after mitotic slippage to an abnormal G1 state.1-5 Most of the available data, including our own, show that although cell death induced by anti-mitotics is mediated by the intrinsic apoptotic pathway in both cell cycle states, i.e., during mitotic arrest and after slippage, these 2 types of cell death involve very different mechanisms and molecular pathways.6-8 Death during mitotic arrest is mainly triggered by the loss of an anti-apoptotic protein, Mcl-1, due to an imbalance of synthesis and degradation,9-14 as transcription is silenced and translation is attenuated during mitosis.15-17 We further identified that the strong variation between cell lines in their sensitivity to apoptosis during mitotic arrest is determined by the variation in the expression level of another anti-apoptotic protein, Bcl-xL.9 Mcl-1 and Bcl-xL are thus the common, primary negative regulators of apoptosis during prolonged mitotic arrest induced by anti-mitotic drugs in general.
While the different anti-mitotic drugs induce a largely similar degree of cell death during mitotic arrest, their activities in triggering cytotoxicity after mitotic slippage appear to be highly variable. For instance, in cell culture, we found that Kinesin-5 inhibitors (K5Is) were much less pro-apoptotic than paclitaxel in activating cell death after mitotic slippage, although they induced similar duration of mitotic arrest and similar extent of death during mitotic arrest.5,9 Moreover, clinical data showed that so far paclitaxel is still more effective than the spindle-specific anti-mitotics,21 but the molecular origins of the extra cytotoxicity of paclitaxel are unclear. Compared with death during mitotic arrest, the molecular pathway activated by anti-mitotics, which links mitotic arrest, abnormal slippage out of the arrest, and death after slippage, is poorly understood. Previous studies showed that caspase activities, which were partially activated during prolonged mitotic arrest, would induce DNA fragmentation after mitotic slippage, subsequently leading to p53-mediated DNA damage response.18-20 However, such DNA damage did not necessarily activate post-slippage apoptosis.18 It also does not account for the significant variability in the extent of post-slippage cell death triggered by the different anti-mitotics. Identifying the mechanisms that govern post-slippage cell death is thus clearly needed so as to not only improve our understanding of the molecular basis underlying the variable efficacy of different anti-mitotic drugs, but also provide new insight for elucidating the large variation between cancer cell types in their response to anti-mitotics, leading to better prediction of the variable clinical outcomes.
Here we investigate the cellular and molecular mechanism by which paclitaxel activates more extensive post-slippage apoptosis than the spindle-specific anti-mitotic, K5I. In order to distinguish cells dying after slippage from those dying during mitotic arrest and examine molecular changes specific to the post-slippage population, we used time-lapse microscopy as the primary technique for measuring the extent and kinetics of apoptosis and other relevant cellular events at the single cell level. Our results revealed that formation of multiple small nuclei was a marked feature of cells that slip out of paclitaxel-induced mitotic arrest, while K5I typically resulted in one large nucleus after slippage. Moreover, the extent of DNA damage and apoptosis after mitotic slippage strongly depended on the degree of multinucleation. Together with data that showed post-slippage apoptosis was mediated by the p53-dependent DNA damage response pathway, our findings suggest that multinucleation induced by paclitaxel is a key trigger of post-slippage DNA damage and apoptosis, and likely renders the extra cytotoxicity of paclitaxel toward tumor cells as compared with the spindle-specific anti-mitotics.
Results
Multinucleation induced by paclitaxel engenders stronger post-slippage apoptosis than K5I
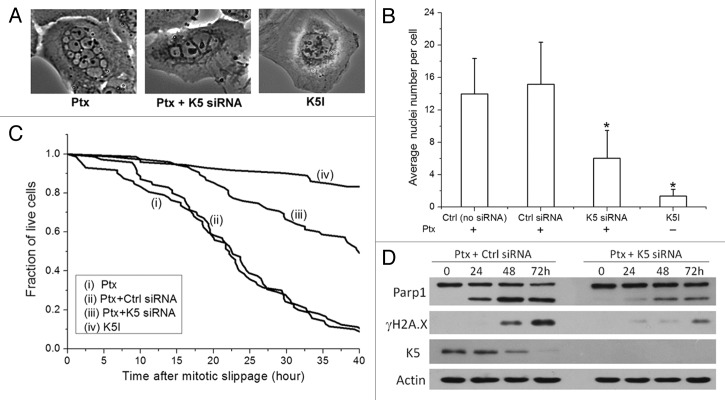
We previously profiled the anti-mitotic drug response of 11 mammalian cell lines, including cancer cell lines derived from the lung (A549, H460), colon (HCT116, HT29), breast (MCF7, MDAMB435S), ovary (OVCAR5), cervix (HeLa), prostate (PC3), and bone (U-2 OS) as well as a normal cell line, RPE.5 Our results showed that across all cell lines that were profiled, paclitaxel was consistently more cytotoxic than K5I and triggered substantially more cell death particularly after mitotic slippage (refer to Fig. S1 for the typical response dynamics to paclitaxel and K5I after mitotic slippage). The molecular pathway(s) that controls post-slippage apoptosis is evidently much more highly activated under paclitaxel. One striking post-slippage feature that we observed from the panel of mammalian cell lines was that cells that slip out of paclitaxel-induced mitotic arrest always have multiple small nuclei, while cells that slip out of K5I-induced arrest have mostly 1 and occasionally 2 large nuclei (Fig. 1A).
Figure 1. Characteristics of post-slippage multinucleation and apoptosis in response to anti-mitotic drugs. (A) Phase-contrast images of post-slippage U-2 OS cells treated with 150 nmol/L paclitaxel (Ptx), 150 nmol/L paclitaxel + K5 siRNA, and 1 μmol/L Kinesin-5 inhibitor (K5I), respectively. (B) Average number of post-slippage nuclei per cell (± SD) and (C) cumulative survival curves of U-2 OS cells under the indicated siRNA treatments in combination with 150 nmol/L paclitaxel or under treatment of K5I alone. Total number of cells analyzed for each condition ranges from 70–75. *P < 0.05 vs. Ctrl (no siRNA). Individual cells were monitored by time-lapse microscopy, and time from mitotic slippage to morphological death was measured and plotted as cumulative survival curves. (D) Knockdown of Kinesin-5 (K5) attenuated post-slippage apoptosis (indicated by Parp1 cleavage) and DNA damage (indicated by γH2A.X) induced by paclitaxel.
To determine whether the degree of multinucleation affects the extent of apoptosis after mitotic slippage, we attenuated multinucleation in paclitaxel-treated cells by knocking down Kinesin-5 (K5) using RNA interference (RNAi), as loss of K5 activity (e.g., by K5Is) promotes formation of a single nucleus in post-slippage cells. We chose to use K5 RNAi instead of the chemical inhibitor, K5I, to attenuate multinucleation, because in order to achieve the same attenuation effect as K5 RNAi, 5–10 μmol/L K5I, a concentration 10-fold higher than the saturating dosage for inducing mitotic arrest, i.e., 0.5–1 μmol/L,5 had to be used in combination with paclitaxel, and at such high K5I concentration, significant non-specific cytotoxic effects that were independent of mitotic arrest were observed. Hence, we opted to use K5 RNAi to attenuate post-slippage multinucleation in this study, as K5 RNAi alone induced less than 5% cell death after 36 h of siRNA transfection (the time when we added paclitaxel for experiment). We selected U-2 OS as the model cell line from the panel of 11 mammalian cell lines that we profiled before,5 as U-2 OS cells were found to be sensitive to post-slippage apoptosis and representative of the general drug response behaviors across the different cell lines.
Based on phase-contrast images from time-lapse microscopy (refer to “Materials and Methods” for image analysis method), we counted the number of post-slippage nuclei. Our results showed that the average number of nuclei per post-slippage cell was 6 under treatment of 150 nmol/L paclitaxel plus K5 knockdown as compared with 14 under paclitaxel alone (Fig. 1B). Cell death plotted as cumulative survival curves in Figure 1C demonstrated that both the kinetics and extent of post-slippage cell death were significantly attenuated in U-2 OS cells with K5 knockdown. Note that in Figure 1C we analyzed the time to death as the time from mitotic slippage to post-slippage cell death, as in this study we focused on understanding the mechanism of apoptosis activation specific to the post-slippage population. Under paclitaxel plus K5 knockdown, about 50% post-slippage cells died, and the average time from mitotic slippage to death was 27 h, in contrast to 91% post-slippage cell death and an average time to death of 20 h under paclitaxel alone. Western blot analysis of an apoptosis marker, Parp1, confirmed the single-cell imaging data, showing much higher percentage of Parp1 cleavage in cells treated with paclitaxel alone (Fig. 1D). We noted that the level of K5 decreased as cells died in response to paclitaxel. We suspect this may possibly be due to leakage of cytoplasmic K5 to the medium upon cell lysis (a signature event of cell death in culture), given that the decrease of K5 signal occurred when cells died. Overall our data strongly suggest that multinucleation plays a crucial role in rendering the strong post-slippage apoptosis in response to paclitaxel.
Post-slippage multinucleation and apoptosis depend on the duration of mitotic arrest
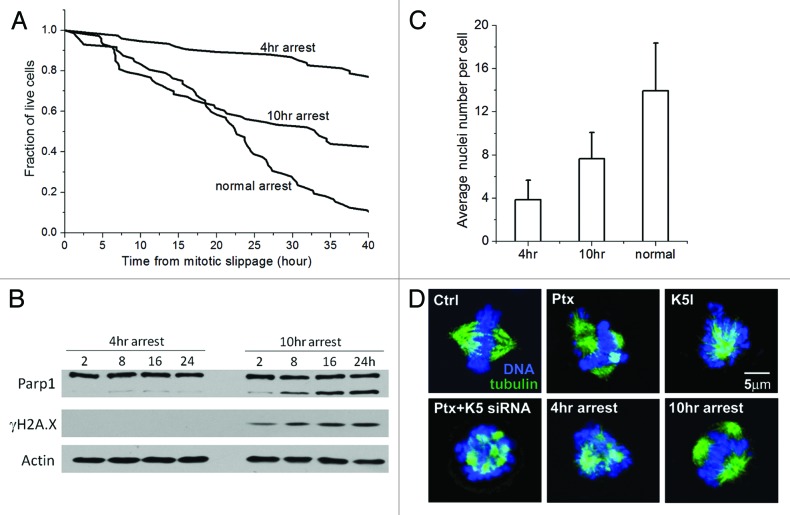
We next investigate whether the duration of mitotic arrest affects the extent of multinucleation and apoptosis after mitotic slippage. To obtain cells with distinct mitotic arrest time, we first synchronized U-2 OS cells in paclitaxel-induced mitotic arrest by mitotic shake-off and then induced slippage using an Aurora kinase inhibitor, VX-680.22,23 We found 2-h treatment of 150 nmol/L VX-680 was sufficient to induce >95% U-2 OS cells to slip out of mitotic arrest, and treatment of VX-680 for such a short period of time did not induce significant DNA damage or cell death (Fig. S2). Figure 2A shows cumulative survival curves of U-2 OS cells that were synchronized in mitotic arrest for 4 h (i.e., short arrest) and 10 h (i.e., long arrest) in comparison with cells that slip out of the normal paclitaxel-induced mitotic arrest (the average mitotic arrest time of unsynchronized U-2 OS cells is 13 h). It is evident that longer mitotic arrest increased and accelerated apoptosis after mitotic slippage. Only 27% of U-2 OS cells that underwent 4 h of mitotic arrest died 40 h after mitotic slippage, in contrast to 64% and 91% of cell death subsequent to 10-h and normal paclitaxel-induced arrest. This was further confirmed by western blot analysis of Parp1 cleavage (Fig. 2B). Moreover, the average number of nuclei per post-slippage cell was 4 after 4-h arrest and 8 after 10-h arrest in comparison to 14 after normal arrest, which again demonstrated the strong correlation between the extent of multinucleation and post-slippage apoptosis (Fig. 2C).
Figure 2. Post-slippage multinucleation and apoptosis depend on duration of mitotic arrest. (A) Cumulative survival curves of post-slippage U-2 OS cells that were synchronized in mitosis by 150 nmol/L paclitaxel for 4 h or 10 h in comparison with those that underwent normal arrest (no synchronization). Total number of cells analyzed for each condition ranges from 72–75. (B) Comparison of apoptosis induction (indicated by the percentage of Parp1 cleavage) at the indicated time points after slippage out of the 4-h and 10-h mitotic arrest induced by paclitaxel. (C) Average number of post-slippage nuclei (± SD) at the indicated conditions of mitotic arrest. (D) Confocal images of representative U-2 OS cells during mitotic arrest under the indicated treatments. Blue, DNA; green, microtubules.
The mechanism by which duration of mitotic arrest affects the degree of post-slippage multinucleation is unclear. Paclitaxel is known to generate multipolar spindle during mitotic arrest, with clusters of DNA attached to the multiple spindle asters.24,25 These well-separated multipolar structures can be clearly observed by confocal microscopy (Fig. 2D). In contrast, K5I induces the formation of only a single spindle pole. This raised the possibility that the post-slippage multinuclei induced by paclitaxel may originate from the multi-polar DNA clusters formed during mitotic arrest. Compared with cells that underwent normal and long mitotic arrest (i.e., 10 h), the spindle asters of U-2 OS cells that experienced short mitotic arrest (i.e., 4 h) aggregated closely and were not spatially well separated (Fig. 2D). Knockdown of K5 also inhibited separation of the multiple spindle asters under paclitaxel, which correlated with the attenuation of subsequent post-slippage multinucleation. These observations suggest that formation of stable and well-separated multipolar structures, around which the nuclear envelop may reform upon mitotic slippage, may be necessary for generating the multiple post-slippage nuclei. Since short mitotic arrest may not provide sufficient time for the spindle asters to separate, it resulted in less post-slippage nuclei than the long arrest. A previous study that imaged α-tubulin-GFP showed that the paclitaxel-induced spindle asters indeed underwent dynamic separation as mitotic arrest prolonged,25 providing evidence that support the above hypothesis.
Post-slippage apoptosis is mediated by p53-dependent DNA damage response
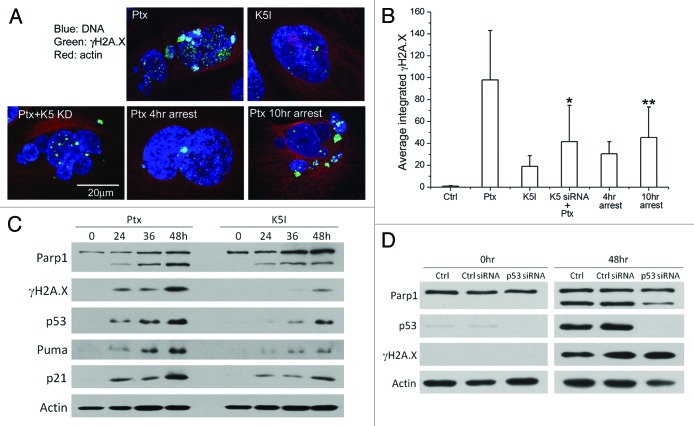
The multinucleated cells that slip out of paclitaxel-induced mitotic arrest were previously found to exhibit strong elevation of γH2A.X (a DNA damage marker) and p53.5,19 This indicated that multinucleation may promote post-slippage cell death by inducing DNA damage and subsequently activating the p53-mediated DNA damage response pathway that triggers apoptosis. To examine the correlation between multinucleation and DNA damage, we used immunofluorescence of γH2A.X to quantify the extent of DNA damage in anti-mitotics-treated cells. Figure 3A illustrates the confocal images of representative post-slippage U-2 OS cells under 5 treatment conditions, including 150 nmol/L paclitaxel, 1 μmol/L K5I, 150 nmol/L paclitaxel plus K5 knockdown, as well as induced slippage out of 4-h and 10-h mitotic arrest in 150 nmol/L paclitaxel. Paclitaxel evidently induced the highest level of γH2A.X (stained in green) in post-slippage cells. Quantification of the average integrated γH2A.X fluorescence showed a 5-fold higher damage signal in U-2 OS cells that slip out of paclitaxel-induced mitotic arrest than K5I-treated cells (Fig. 3B). Knockdown of K5 as well as short mitotic arrest (i.e., 4 h) both significantly reduced the level of DNA damage in post-slippage cells. Together with western blot results of ensemble γH2A.X level shown in Figures 1D, 2B, and 3C, our data demonstrated a strong correlation between the level of post-slippage DNA damage and the extent of multinucleation and apoptosis.
Figure 3. The extent of post-slippage multinucleation and apoptosis correlated with DNA damage level. (A) Confocal images of representative post-slippage U-2 OS cells under the indicated treatments. Ctrl, no treatment; Ptx, 150 nmol/L paclitaxel for 60 h; K5I, 1 μmol/L K5I for 60 h; Ptx+K5 siRNA, 150 nmol/L paclitaxel plus K5 knockdown by RNAi for 60 h; 4-h/10-h arrest, 40 h after induced slippage out of 4-h/10-h mitotic arrest in paclitaxel. Blue, DNA; green, γH2A.X; red, actin. (B) Average integrated fluorescence intensity of γH2A.X per cell (± SD) quantified from the confocal images. Twenty cells from 3 independent experiments were analyzed for each condition. *P < 0.05 vs. K5I; **P < 0.05 vs. 4-h arrest in paclitaxel. (C) Expression of selected DNA damage response genes in U-2 OS cells treated with paclitaxel (150 nmol/L) vs. K5I (1 μmol/L) by western blot analysis. (D) Knockdown of p53 attenuated apoptosis induced by paclitaxel.
One of the major signaling pathways that mediate DNA damage response is the p53 pathway, consisting of mainly the tumor suppressor protein p53 and its downstream transcriptional targets.26 In Figure 3C, we measured, by western blotting, the expression levels of p53 and 2 key p53 target genes, p21 and Puma, in paclitaxel- and K5I-treated cells. p53 as well as its target genes were all more highly induced under paclitaxel, correlating with the DNA damage level. Moreover, knocking down p53 by RNAi substantially attenuated post-slippage cell death in response to paclitaxel (Fig. 3D). These data thus suggest that post-slippage apoptosis induced by paclitaxel is p53-dependent and likely regulated by the p53-mediated DNA damage response pathway.
Both DNA damage and p53 induction occurred after mitotic slippage and in an asynchronous manner in individual nuclei
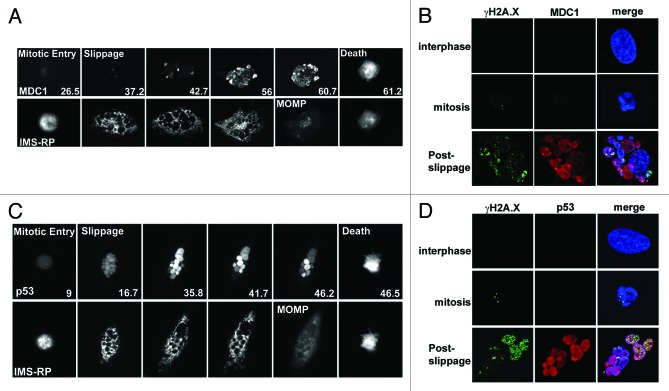
An important question regarding the observed DNA damage is how it occurs relative to mitotic arrest, slippage, and apoptosis activation. To simultaneously monitor the real-time dynamics of DNA damage induction and activation of post-slippage apoptosis, we imaged, by time-lapse microscopy, paclitaxel-treated U-2 OS cells that expressed 2 fluorescent reporters, including a green fluorescent reporter (MDC1-EGFP) of wild-type MDC1 (mediator of DNA damage checkpoint 1) fused with EGFP,27 and a red mitochondria reporter (IMS-RP) consisting of a monomeric red fluorescent protein targeted to the inter-membrane space of mitochondria by fusion to the leader peptide of SMAC.28 Upon DNA damage, MDC1 is phosphorylated by ATM (ataxia telangiectasia mutated) and then binds to γH2A.X, subsequently recruiting the downstream components involved in the damage response process.29 As shown in the upper panel of Figure 4A and Video S1, no increase in MDC1 signal was observable until U-2 OS cells slip out of the prolonged mitotic arrest. For the majority of cells that we analyzed, significant accumulation of MDC1 started to appear 4–6 h after mitotic slippage. Interestingly, individual nuclei belonging to the same post-slippage cell showed highly heterogeneous MDC1 signal and distinct kinetics of the signal increase, suggesting that DNA damage in the multiple small nuclei occurred in an asynchronous manner. Moreover, we observed mitochondrial outer membrane permeabilization (MOMP), the defining event of apoptosis activation, in the post-slippage cells based on a switch-like change from punctuate to diffused distribution in IMS–RP fluorescence (Fig. 4A, lower panel).28 Since post-slippage MOMP always occurred hours after the onset of MDC1 accumulation, our data pointed to induction of DNA damage being an upstream event of apoptosis, i.e., not a consequence of apoptotic damage. To confirm that the 2 DNA damage markers that we used, MDC1 and γH2A.X, indeed correlate, we performed confocal imaging of U-2 OS cells with co-staining of MDC1 and γH2A.X antibody. As shown in Figure 4B, fluorescent signaling of both MDC1 and γH2A.X only significantly increased after mitotic slippage, and the immunofluorescence of MDC1 largely correlated with that of γH2A.X, i.e., most nuclei with strong γH2A.X staining generally also exhibited high MDC1 accumulation. Overall the confocal data indicated that MDC1 and γH2A.X were likely to report similarly on DNA damage. We noticed that U-2 OS cells also showed a very small number of γH2A.X foci during mitotic arrest, but this γH2A.X signal was not accompanied by increase in MDC1 (Fig. 4B). It appeared that the γH2A.X signal arising in mitosis may not relate to conventional DNA damage, which agreed with findings from several previous studies.30-32
Figure 4. Induction dynamics of MDC1 and p53 relative to the activation of post-slippage apoptosis in U-2 OS cells treated with 150 nmol/L paclitaxel. (A) MDC1 dynamics relative to induction of post-slippage MOMP. Still images of representative U-2 OS cell were from fluorescence time-lapse movies. Upper panel, MDC1-EGFP; lower panel, IMS-RP. Elapsed time from paclitaxel addition is indicated at the bottom of the images (unit, hour). (B) Confocal images of γH2A.X (green), MDC1 (red), and DNA (blue) in interphase (pre-mitotic), mitotic, and post-slippage U-2 OS cells. (C) p53 dynamics relative to induction of post-slippage MOMP. Upper panel, p53-Venus; lower panel, IMS-RP. (D) Confocal images of γH2A.X (green), p53 (red), and DNA (blue) in interphase (pre-mitotic), mitotic, and post-slippage U-2 OS cells.
To determine the induction dynamics of p53 relative to post-slippage apoptosis, we used a clonal fluorescent U-2 OS reporter cell line, which stably expresses a p53-Venus construct as well as the MOMP reporter, IMS-RP.33 Dynamics of the p53–Venus construct had been confirmed to behave similarly to the endogenous wild-type p53 in response to DNA damage by previous studies.33,34 From the time-lapse movies, we observed strong induction of p53 in about 90% of the post-slippage cells treated with paclitaxel. Similar to MDC1, p53 level was found to significantly increase only after mitotic slippage, and MOMP was observed hours after p53 induction (Fig. 4C; Video S2). The different small nuclei present in the same cell again showed distinct kinetics and level of p53 activation, which correlated with the variable dynamics of DNA damage induction that we observed by the MDC1–EGFP measurement, indicating that the different nuclei may activate DNA damage response in an asynchronous manner. Some of the small nuclei showed a decrease in p53 fluorescence after the strong, initial post-slippage induction (Fig. 4C), which may be due to successful DNA damage repair. Confocal images of p53 and γH2A.X immunofluorescence confirmed the time-lapse data; i.e., increase in DNA damage and p53 level mainly occurred after mitotic slippage (Fig. 4D). The p53 immunofluorescence did not show a very large difference between the different nuclei as in the time-lapse data, although the nuclei exhibited significantly different γH2A.X level. We suspect this may be due to saturating signal from immunostaining and confocal imaging.
Discussion
Results from our study demonstrated that in addition to killing cancer cells during mitotic arrest, where different anti-mitotics exhibit largely similar efficacy, paclitaxel is much more potent in triggering cell death after mitotic slippage by engendering multiple small nuclei and, subsequently, strong DNA damage. Paclitaxel appears to exert dual cytotoxicity by 2 distinct mechanisms in the cell cycle state of mitosis and post-slippage interphase, capable of killing proliferating cancer cells by both triggering mitotic arrest and post-slippage DNA damage. For cancer cells that are resistant to apoptosis during mitotic arrest, such as those with a high expression level of Bcl-xL,9 paclitaxel is particularly desirable among the different anti-mitotics, as it is able to also induce extensive apoptosis after mitotic slippage by activating a strong DNA damage response. Moreover, given the dual cytotoxic activities of paclitaxel, our results suggest that to predict tumor cell response to paclitaxel, 2 distinct sets of biomarkers may need to be considered, one involving genes that regulate cell death during prolonged mitotic arrest (e.g., Mcl-1 and Bcl-xL) and the other involving genes that control DNA damage-dependent post-slippage death (e.g., p53 and possibly other components in the p53 pathway).
Recent work by Crasta et al. showed that DNA fragmentation and damage would arise as small micronuclei underwent defective and asynchronous replication.35 To test whether this is also the mechanism by which extensive DNA damage was induced in the multinucleated cells that slip out of paclitaxel-induced mitotic arrest, we inhibited DNA replication in the post-slippage cells using thymidine or mimosine.36 In order to generate post-slippage cells, we first synchronized cells in mitotic arrest by mitotic shake-off in paclitaxel and then divided the cells equally into treatment of paclitaxel alone or paclitaxel plus thymidine/mimosine. Neither thymidine nor mimosine attenuated DNA damage (as indicated by γH2A.X) or apoptosis in the post-slippage cells, as compared with paclitaxel treatment alone (Fig. S3A). In addition, bromodeoxyuridine (BrdU) staining of the post-slippage cells showed little BrdU incorporation into the chromosome, indicating that the post-slippage cells were likely arrested in the G1 state before DNA synthesis (Fig. S3B). It thus appeared unlikely that the post-slippage DNA damage observed in the paclitaxel-treated cells was originated from defected DNA replication of the multiple small nuclei.
Another possible mechanism by which DNA damage may be induced in the post-slippage cells is through partially activated caspase activities.18-20 Orth et al.18 showed that in response to K5I, partially activated caspases during prolong mitotic arrest may partly activate caspase-activated DNase (CAD), causing DNA damage and p53 induction after mitotic slippage. To examine whether this is the mechanism by which paxlitaxel activated post-slippage DNA damage and cell death, we compared the extent of post-slippage DNA damage and apoptosis with and without a pan-caspase inhibitor, zVAD-FMK. Quantification of the integrated γH2A.X fluorescence showed that treatment of zVAD-FMK significantly reduced the DNA damage level in response to paclitaxel by about 40%, indicating caspase activity is indeed involved. However, the reduced DNA damage level is still 3-fold higher than that induced by K5I treatment. So inhibition of caspase activity did not completely block the induction of post-slippage DNA damage under paclitaxel. More importantly, by imaging the MOMP reporter (IMS-RP), we found that 68% of the post-slippage cells treated with paclitaxel plus zVAD-FMK still underwent MOMP (Fig. S4A), although these cells did not die due to the lack of caspase activities to activate apoptotic events downstream of MOMP. Statistics of MOMP kinetics from the individual cells showed that the average time from mitotic slippage to MOMP was 18.7 h under paclitaxel plus zVAD-FMK, very similar to 19.2 h under paclitaxel alone (Fig. S4B). These preliminary data suggest that besides caspase activities, additional cellular components/pathways also contribute to the induction of DNA damage and apoptosis in the post-slippage multinucleated cells. Further, a more detailed mechanistic study is clearly needed to determine the molecular links between multinucleation, DNA damage, and apoptosis.
The origin of the post-slippage multinuclei was alluded to by our results but not completely resolved. We hypothesized that the DNA micro-clusters, which were attached to the multipolar mitotic spindles and formed during the prolonged mitotic arrest, may have given rise to the multiple post-slippage nuclei under paclitaxel. However, to determine whether they are indeed mechanistically related and identify the underlying molecular regulators, the cellular machinery that controls the slippage processes needs to be examined in detail, such as nuclear envelope reformation and chromosome unwinding. Results from such further study will also help to elucidate how duration of mitotic arrest impacts the extent of multinucleation and apoptosis in post-slippage cells.
Materials and Methods
Cell line and cell culture
U-2 OS cells were purchased from American Type Culture Collection (ATCC) and cultured under 37 °C and 5% CO2 in McCoy 5A supplemented with 10% fetal calf serum (FCS), 100 U/mL penicillin, and 100 μg/mL streptomycin. The fluorescent U-2 OS cell line that expresses the mitochondria reporter, IMS-RP (a generous gift from Dr Peter Sorger, Harvard Medical School), was generated by infecting U-2 OS cells with retrovirus encoding a construct consists of a monomeric red fluorescent protein targeted to the inter-membrane space of mitochondria by fusion to the leader peptide of SMAC.28 The fluorescent U-2 OS cell line that expresses the reporter of wild-type p53 fused with Venus was generated by lenti-viral infection of a p53-Venus construct (a generous gift from Dr Galit Lahav, Harvard Medical School).34
For mitotic shake-off experiment, we grew large volume of U-2 OS cells in 25 cm dishes to 90% confluency, then treated the cells with 150 nmol/L paclitaxel to induce mitotic arrest. After 4 h of drug treatment, the mitotic fraction of cells was collected by gently shaking and washing the dish to detach the mitotic cells from the bottom.
Chemicals
Paclitaxel was purchased from Sigma and used at 150 nmol/L (the saturating concentration for cell death induction). The potent Kinesin-5 inhibitor (EMD534085) was provided by Merck-Serono. Responses to EMD534085 have been shown to be similar to S-trityl-cysteine, a commercially available Kinesin-5 inhibitor. zVAD-FMK was purchased from Calbiochem and used at 100 μmol/L.
Time-lapse microscopy and analysis
Cells were plated in 35-mm imaging dish (μ-dish, ibidi) and cultured in phenol red-free CO2-independent medium (Invitrogen) supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cell images were acquired with the Nikon Ti-Eclipse-PFS inverted microscope enclosed in a humidified chamber maintained at 37 °C. Cells were imaged every 10 min using a motorized stage and a 20× objective.
Images were viewed and analyzed using the MetaMorph software (Molecular Dynamics). Based on the phase-contrast images, we scored by morphological tracking: interphase (by flat morphology), entry into mitosis (by cell rounding), mitotic slippage (by re-spreading without cytokinesis), and cell death (by blebbing followed by cell lysis) (Fig. S1). The time to death since mitotic slippage, which was plotted in Figures 1C and 2A, was calculated as the time of post-slippage death (scored by cell lysis) minus the time of mitotic slippage.
Gene knockdown by RNA interference (RNAi)
siRNA oligos were custom synthesized by Invitrogen. siRNA against Kinesin-5 (5′-AGGACAACUG CAGCUACUC-3′) was used at final concentration of 20 nmol/L for RNAi experiments. siRNA against p53 (5′-UGAACCAUUG UUCAAUAUCG UCCGG-3′) was used at final concentration of 5 nmol/L. Dharmacon On-Target plus siControl (D-001810-01) was used as non-targeting siRNA control. All siRNA transfections were performed using HiPerFect (Qiagen) according to manufacturers’ instructions. Experiments were conducted after 36 h (Kinesin-5 RNAi) or 24 h (p53 RNAi) of gene silencing.
Western blot analysis
Cell lysates were obtained using LDS sample buffer (NuPAGE, Invitrogen). Proteins were resolved on 8–15% Tris-glycine gels and transferred onto PVDF membranes. Blots were probed with commercial primary antibodies and chemiluminescent detection using ECL-plus (Amersham). Antibodies: PARP1 (#9542), tubulin (#2148), Kinesin-5 (#7625), p21 (#2947), and Puma (#4976) were purchased from Cell Signaling; p53 (#sc-126) from Santa Cruz; γH2A.X (#06–570) from Millipore. Anti-actin (#A5316) from Sigma was used as a loading control.
Immunofluorescence staining and analysis
Cells grown on glass coverslips were fixed in 3.7% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) for 15 min; permeabilized with 0.5% Triton-X100 for 30 min; washed 3 times in PBS; blocked in 5% BSA/0.1% Triton-X100/PBS for 1 h; incubated with primary antibody for 2 h; washed 3 times in PBS; incubated with fluorescently conjugated Alexa-Fluor secondary antibodies (Invitrogen) for 1 h; washed 3 times in PBS; stained with 1 μg/mL Hoechst (Invitrogen) for 2 min. All procedures were conducted at room temperature. The coverslips were then mounted on glass slides with ProLong Gold antifade reagent (Invitrogen). The stained cells were imaged using an Olympus FV1000 Laser Scanning Confocal microscope. Antibodies: tubulin (#2144) and p53 (#2527) were purchased from Cell Signaling; γH2A.X (#06-570) from Millipore; Actin (#A5316) from Sigma; and MDC1 (#ab11169) from abcam.
For quantification of the integrated γH2A.X fluorescence, nuclei of the cells were segmented using the DNA stain, and the total γH2A.X fluorescence intensity inside the nuclei was measured using the MetaMorph software.
Gene expression by transient transfection
To express MDC1-EGFP (a generous gift from Dr Jiri Lukas at the Institute of Cancer Biology, Denmark), U-2 OS cells were seeded in 6-well plate and transiently transfected with the plasmid using X-tremeGENE (Roche) according to manufacturer’s instruction. Time-lapse imaging experiment of U-2 OS cells expressing the MDC1-EGFP construct were conducted 36 h after the transfection.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr Jiri Lukas (Institute of Cancer Biology, Danish Cancer Society) for the MDC1-EGFP vector, Dr Perter Sorger (Systems Biology, Harvard Medical School) for the IMS-RP retroviral vector, and Dr Galit Lahav (Systems Biology, Harvard Medical School) for the p53-Venus lentiviral vector. This work was supported by the Hong Kong RGC (#261310) and HKBU (#FRG2/12-13/036) to J.S. EMD534085 was supplied by Merck-Serono. The authors declare no conflict of interest.
References
- 1.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 2.Weaver BA, Cleveland DW. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation, and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Orth JD, Tang Y, Shi J, Loy CT, Amendt C, Wilm C, Zenke FT, Mitchison TJ. Quantitative live imaging of cancer and normal cells treated with Kinesin-5 inhibitors indicates significant differences in phenotypic responses and cell fate. Mol Cancer Ther. 2008;7:3480–9. doi: 10.1158/1535-7163.MCT-08-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–22. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and Kinesin-5. Cancer Res. 2008;68:3269–76. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- 6.Tao W, South VJ, Zhang Y, Davide JP, Farrell L, Kohl NE, Sepp-Lorenzino L, Lobell RB. Induction of apoptosis by an inhibitor of the mitotic kinesin KSP requires both activation of the spindle assembly checkpoint and mitotic slippage. Cancer Cell. 2005;8:49–59. doi: 10.1016/j.ccr.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Wang LG, Liu XM, Kreis W, Budman DR. The effect of antimicrotubule agents on signal transduction pathways of apoptosis: a review. Cancer Chemother Pharmacol. 1999;44:355–61. doi: 10.1007/s002800050989. [DOI] [PubMed] [Google Scholar]
- 8.Park SJ, Wu CH, Gordon JD, Zhong X, Emami A, Safa AR. Taxol induces caspase-10-dependent apoptosis. J Biol Chem. 2004;279:51057–67. doi: 10.1074/jbc.M406543200. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Zhou Y, Huang HC, Mitchison TJ. Navitoclax (ABT-263) accelerates apoptosis during drug-induced mitotic arrest by antagonizing Bcl-xL. Cancer Res. 2011;71:4518–26. doi: 10.1158/0008-5472.CAN-10-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Pérez T, Ortiz-Ferrón G, López-Rivas A. Mitotic arrest and JNK-induced proteasomal degradation of FLIP and Mcl-1 are key events in the sensitization of breast tumor cells to TRAIL by antimicrotubule agents. Cell Death Differ. 2010;17:883–94. doi: 10.1038/cdd.2009.176. [DOI] [PubMed] [Google Scholar]
- 12.Harley ME, Allan LA, Sanderson HS, Clarke PR. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–20. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tunquist BJ, Woessner RD, Walker DH. Mcl-1 stability determines mitotic cell fate of human multiple myeloma tumor cells treated with the kinesin spindle protein inhibitor ARRY-520. Mol Cancer Ther. 2010;9:2046–56. doi: 10.1158/1535-7163.MCT-10-0033. [DOI] [PubMed] [Google Scholar]
- 14.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 15.Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/S0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- 16.Blagosklonny MV. Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle. 2007;6:70–4. doi: 10.4161/cc.6.1.3682. [DOI] [PubMed] [Google Scholar]
- 17.Sivan G, Elroy-Stein O. Regulation of mRNA Translation during cellular division. Cell Cycle. 2008;7:741–4. doi: 10.4161/cc.7.6.5596. [DOI] [PubMed] [Google Scholar]
- 18.Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell. 2012;23:567–76. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai SK, Wong CH, Lee YP, Li HY. Caspase-3-mediated degradation of condensin Cap-H regulates mitotic cell death. Cell Death Differ. 2011;18:996–1004. doi: 10.1038/cdd.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quignon F, Rozier L, Lachages AM, Bieth A, Simili M, Debatisse M. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 2007;26:165–72. doi: 10.1038/sj.onc.1209787. [DOI] [PubMed] [Google Scholar]
- 21.Jackson JR, Patrick DR, Dar MM, Huang PS. Targeted anti-mitotic therapies: can we improve on tubulin agents? Nat Rev Cancer. 2007;7:107–17. doi: 10.1038/nrc2049. [DOI] [PubMed] [Google Scholar]
- 22.Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–80. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 24.Bouchet BP, Bertholon J, Falette N, Audoynaud C, Lamblot C, Puisieux A, Galmarini CM. Paclitaxel resistance in untransformed human mammary epithelial cells is associated with an aneuploidy-prone phenotype. Br J Cancer. 2007;97:1218–24. doi: 10.1038/sj.bjc.6603936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornick JE, Bader JR, Tribble EK, Trimble K, Breunig JS, Halpin ES, Vaughan KT, Hinchcliffe EH. Live-cell analysis of mitotic spindle formation in taxol-treated cells. Cell Motil Cytoskeleton. 2008;65:595–613. doi: 10.1002/cm.20283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–23. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 27.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–11. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Nelson G, Buhmann M, von Zglinicki T. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle. 2009;8:3379–83. doi: 10.4161/cc.8.20.9857. [DOI] [PubMed] [Google Scholar]
- 31.Pankotai T, Hoffbeck AS, Boumendil C, Soutoglou E. DNA damage response in the absence of DNA lesions continued. Cell Cycle. 2009;8:4025–6. doi: 10.4161/cc.8.24.10564. [DOI] [PubMed] [Google Scholar]
- 32.Pospelova TV, Demidenko ZN, Bukreeva EI, Pospelov VA, Gudkov AV, Blagosklonny MV. Pseudo-DNA damage response in senescent cells. Cell Cycle. 2009;8:4112–8. doi: 10.4161/cc.8.24.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Chen J, Gan S, Guan H, Zhou Y, Ouyang Q, Shi J. DNA damage strength modulates a bimodal switch of p53 dynamics for cell-fate control. BMC Biol. 2013;11:73. doi: 10.1186/1741-7007-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackman J, O'Connor PM. Methods for synchronizing cells at specific stages of the cell cycle. Curr Protoc Cell Biol 2001; Chapter 8:Unit 8.3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.