Abstract
Long non-coding RNAs (lncRNAs) have been found to perform various functions in a wide variety of important biological processes. To make easier interpretation of lncRNA functionality and conduct deep mining on these transcribed sequences, it is convenient to classify lncRNAs into different groups. Here, we summarize classification methods of lncRNAs according to their four major features, namely, genomic location and context, effect exerted on DNA sequences, mechanism of functioning and their targeting mechanism. In combination with the presently available function annotations, we explore potential relationships between different classification categories, and generalize and compare biological features of different lncRNAs within each category. Finally, we present our view on potential further studies. We believe that the classifications of lncRNAs as indicated above are of fundamental importance for lncRNA studies, helpful for further investigation of specific lncRNAs, for formulation of new hypothesis based on different features of lncRNA and for exploration of the underlying lncRNA functional mechanisms.
Keywords: long non-coding RNA, lncRNA, lncRNA classification, RNA transcripts
Introduction
In contrast to a small proportion of the mammalian genome (e.g., human, mouse) that are transcribed into mRNAs, the vast majority of the genome is transcribed into what was previously regarded as “dark matter”—non-coding RNAs (ncRNAs) that do not encode information about proteins.1-5 Among these ncRNAs, long ncRNAs (lncRNAs) represent the most prevalent and functionally diverse class.6-8 There is no definition of lncRNA that is based on biological argumentation and widely accepted in the community. The most commonly used definition is based on the threshold of 200 nucleotides (nt) of the RNA length.6,7,9 It conventionally divides ncRNAs into lncRNAs that have more than 200 nt in length and the remaining ones that are considered “small” RNAs. Small ncRNAs include many different RNAs, such as microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), piwiRNAs (piRNAs) (Taft et al., 2010). Undoubtedly, the definition of lncRNA merely based on length is arbitrary. One attempt to distinguish lncRNAs from small ncRNAs, based more on the biological argumentation is proposed by Amaral et al. defining lncRNAs as those ncRNAs that function either as primary or spliced transcripts, independent of extant known classes of small ncRNAs.10 Therefore, there are some lncRNAs that do not exceed the arbitrary threshold in length (such as BC1and snaR, which are less than or close to 200 nt but included in lncRNAdb10).
LncRNAs are observed in a large diversity of species, including animals,11,12 plants,13 yeast,14 prokaryotes15 and even viruses.16 However, lncRNAs are poorly conserved among different species when compared with the well-studied RNAs (such as mRNAs, miRNAs, snoRNAs),4,17,18 invoking uncertainty about whether a given lncRNA is functional at all due to poor interspecies conservation, or it conveys functional species-specific characteristics. In addition, lncRNAs are usually low expressed,4,19,20 making them to look like more as transcriptional noise. Despite this, a lot of evidence has accumulated showing that lncRNAs play a significant role in a wide variety of important biological processes,21,22 including transcription,23,24 splicing,25,26 translation,27,28 protein localization,29,30 cellular structure integrity,31,32 imprinting,33-35 cell cycle36,37 and apoptosis,16,38 stem cell pluripotency39 and reprogramming40 and heat shock response.41,42 It has been suggested that lncRNAs may regulate cancer progression43 and development of many other human diseases.44 Moreover, a considerable number of lncRNAs are 3′ polyadenylated, 5′ capped, multi-exonic2,4,20 and exhibit transcriptional activation activity similar to that of mRNAs.4,45,46 As a consequence, all these functions and biological features of lncRNAs make them interesting and important research topic.
Recent advances in experimental and computational technologies make it feasible to conduct deep mining on more and more transcribed sequences.2,20,47-49 At present, there are 73,370 lncRNA entries from 1,239 organisms according to NONCODE v3.050 (a database of literature documented lncRNAs). Conversely, among all these lncRNAs, only a small proportion (less than 200 according to LncRNAdb,10 a database of lncRNA annotation) has been functionally annotated. To better understand their functional significance, it helps to classify lncRNAs into different groups that are useful for exploring their underlying mechanisms of actions, for formulating new hypotheses and for providing insights in differences of such major classes of lncRNAs. Here, we summarize classification methods of lncRNAs according to their different features as discussed in what follows, including their (1) genome location and context, (2) exerted effect on DNA sequences, (3) mechanism of functioning and (4) targeting mechanism. Finally, we provide our perspectives on potential further studies.
Genomic Location and Context
Intergenic lncRNAs and intronic lncRNAs
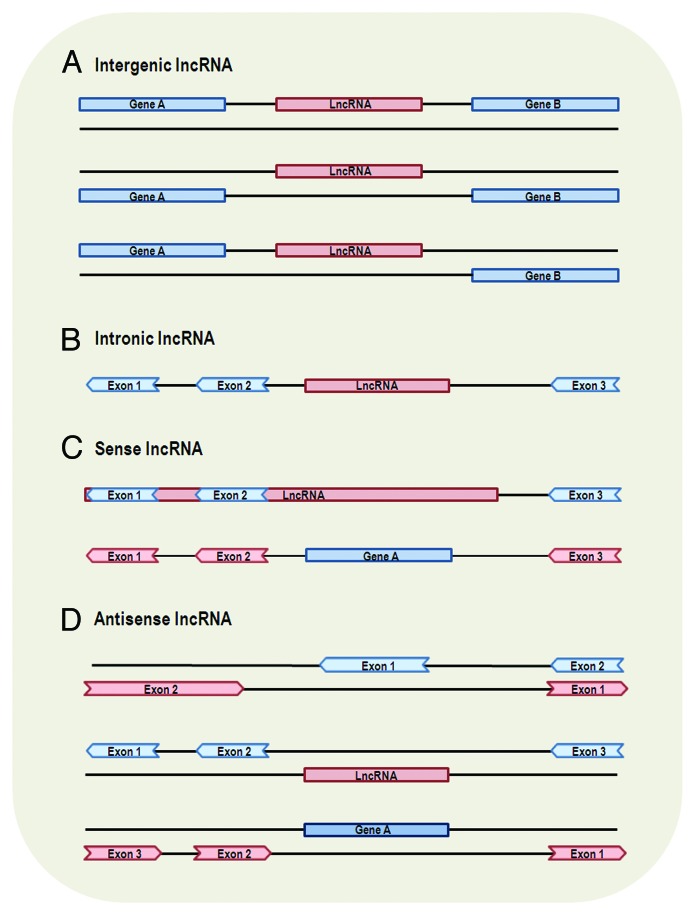
There is a large number of non-coding regions (accounting for 98–99% in the human genome) interspersed between coding regions.51,52 Since lncRNAs are located and transcribed from different genomic locations, those transcribed from intergenic regions are named intergenic lncRNAs (Fig. 1A) and, in contrast, those transcribed entirely from introns of protein-coding genes are named intronic lncRNAs (Fig. 1B).
Figure 1. Genomic location and context of lncRNAs. Protein-coding genes and their exons are represented by blue color, while lncRNAs and their exons are represented by red color. Panels are mainly based on lncRNA location annotation of GENCODE.4 (A) Intergenic lncRNA, transcribed intergenically from both strands. (B) Intronic lncRNA, transcribed entirely from introns of protein-coding genes. (C) Sense lncRNA, transcribed from the sense strand of protein-coding genes and contain exons from protein-coding genes, overlapping with part of protein-coding genes or covering the entire sequence of a protein-coding gene through an intron. (D) Antisense lncRNA, transcribed from the antisense strand of protein-coding genes, overlapping with exonic or intronic regions or covering the entire protein-coding sequence through an intron.
It is suggested that intergenic lncRNAs and intronic lncRNAs are most likely regulated through different transcription activation mechanisms53 and may have different poly(A) modifications and manifest activities in different cellular locations.48 However, only a small portion of intronic lncRNAs has been explored regarding their function. In contrast, there is a large number of long intergenic non-coding RNAs (lincRNAs) that function through different types of mechanisms: cis or trans transcriptional regulation (described below), translational control, splicing regulation, other post-transcriptional regulation, etc. (see Table 1). Also, lincRNAs have been extensively studied about their expression feature and conservation among species.
Table 1. Functional mechanisms and genomic locations of lncRNAs.
| Function mechanism | Gene symbol | |
|---|---|---|
| Transcriptional regulationa | ||
| Cis | Aird, Alpha 250/ Alpha 280d, ANRILd, Beta-globin transcriptsb, Beta-MHC antisense transcriptsd, CAR Intergenic 10d, CCND1 associated ncRNAsb, COLDAIRe, COOLAIRd, DHFR upstream transcriptsd, Emx2osd, Evf2d, fbp1+ promoter RNAsb, GAL10-ncRNAd, H19b, H19 antisensec, H19 upstream conserved 1 and 2b, H19 ICR ncRNAs, HOTAIRM1b, HOTTIPd, Hoxa11asd, ICR1c, Kcnq1ot1d, Khps1ad, L1PA16b, LINoCRb, MEG3b, Mistralb, Msx1asd, Nespasd, ncR-Uparb, PHO5 lncRNAd, PHO84 antisensed, pRNAb, PWR1d, RTLd, SRG1b,c, TEA ncRNAsc, TIR1axutd, TPO1axutd, Tsixb, Xistb | |
| Trans | 7SKb, B2 SINE RNAb, GAS5b, HOTAIRd, Jpxb, LXRBSVc, PR antisense transcriptsd, VL30 RNAs | |
| Unclear | Adapt33b, antiPeg11d, Gtl2-asd, HOXA3asd, HOXA6asd, linc1242b, linc1257b, linc1368b, linc1547b, linc1582b, linc1609b, linc1610b, lincRNA-p21b, lincRNA-RoR b, Malat1-asb, MEG9b, NDM29e, NEAT1b, PANDAb, PCAT-1 b, Rianb, SatIII transcriptsb, SNHG3c, SRAc, Tmevpg1b, TncRNAb, TUG1b | |
| Translational regulation | BC1b, BC200b, Gadd7, SNHG1b, SNHG6d, snaRb, Zeb2NATd | |
| Splicing regulation | MIATb, LUSTd, Malat1b, SAFd, VL30 RNAs, Zeb2NATd | |
| Other post-transcriptional regulation | 21Ab, 1/2-sbsRNA1 c, At4 b, BACE1ASd, CDR1 ASd, Dio3os b, E2F4 antisensed, Emx2os d, Gadd7, H19 b, HULCb, HSUR1 and HSUR2, IPS1b, KRASP1b, Linc-MD1 b, psvA antisense RNAd, PTENP1b, tie-1asd, WT1-ASd | |
| Other functional mechanisms | 7SLb, Beta 2.7 RNA, Centromeric α-satellite RNAb, ENOD40c, EBER1 and EBER2 RNAs, G22b, L1PA16b, hsr omega transcriptsb, meiRNA, Maternal RNA templatesc,d, Maternal somatic nucleus RNAsc,d, MER11Cb, NRONd, rncs-1b, roX1 and roX2b, sfRNA, TERRA, TERCb, VAI and VAII RNAs, VegT RNAc, Xlsirts, Y RNAsb |
Note: lncRNAs listed are collected from the database of lncRNAdb and published papers. aCis, lncRNAs that regulate expression of genes in close genomic proximity; Trans, lncRNAs that regulate expression of distant genes; Unclear, lncRNAs that regulate gene expression at transcriptional level, either in cis or trans. bLincRNAs. cSense lncRNAs. dAntisense lncRNAs. eIntronic lncRNAs.
It is found that lincRNAs are transcriptionally activated similarly to mRNAs,45,46,54,55 as they are more conserved than introns45,46,55 and antisense transcripts,45 more tissue-specifically expressed than protein-coding genes4,20,45 and more stable than intronic lncRNAs56. “K4-K36” domain (with histone H3K4 trimethylation at their 5′ end and histone H3K36 trimethylation in the body of the gene), an indicator of active transcription in protein-coding genes, is found to prevalently exist in transcriptionally active lincRNAs.45,46,54,55 Approximately 70% of lincRNAs with “K4-K36” domain show evidence of RNA transcription, which is similar to the proportion (~72%) of protein-coding genes.46 Most importantly, nearly 70% of the transcription active domains (K4-K36 domain) of lincRNAs in human are conserved in the orthologous region of mouse, which is comparable to the corresponding proportion (80%) of protein-coding genes.46 In addition, lincRNAs are found to be conserved across multiple vertebrate species.20 The above evidences (active transcription, a degree of domain conservation, tissue-specific expression, stability) strongly indicate the functional importance of lincRNAs. In reality, lincRNAs are found to perform important functions in many cellular processes, from embryonic stem cell pluripotency to cell proliferation and cancer progression.38,46,53,54
Sense and antisense lncRNAs
Sense lncRNAs are transcribed from the sense strand of protein-coding genes, containing exons from protein-coding genes. They may overlap with part of protein-coding genes, or cover the entire sequence of a protein-coding gene (Fig. 1C). Antisense lncRNAs, to the contrary, are transcribed from the antisense strand of protein-coding genes. According to GENCODE (a database of manually curated lncRNAs) annotation,4 antisense lncRNAs may appear in three scenarios: (1) transcripts from the antisense strand of protein-coding genes overlap an exon of a sense gene through lncRNAs’ exons, (2) transcripts from the intron of a sense gene do not have exon-exon overlap with this sense gene and (3) transcripts cover the entire sequence of a sense gene through an intron (Fig. 1D). Sense and antisense lncRNAs are proved to be genuine transcripts by strand-specific assay or sequencing,45,47 qRT-PCR validation45 and by sequencing 5′ and 3′ ends of full-length cDNA2 and, thus, they do not represent truncated CDSs or transcriptional noise. Most of the lncRNAs that come from protein-coding genes or the antisense of the protein-coding genes can be obtained using CAGE (cap-analysis gene expression) and oligo-dT guided reverse transcription, suggesting that they also possess mRNA-like features of 3′ polyadenylation and 5′ capping.2 Moreover, sense and antisense lncRNAs can also be multi-exonic.45,57
Many antisense lncRNAs function through different types of mechanisms (similar to lincRNAs) (Table 1). It has been found that as many as 87% coding transcripts have antisense partners in the mouse genome49 and ~32% of the human lncRNAs are antisense to coding genes,4 suggesting that antisense regulation is likely to be commonly utilized. However, in comparison with lincRNAs and antisense lncRNAs, sense lncRNAs have been less explored for their functions (Table 1). It is suggested4 that most lncRNAs tend not to have protein-coding potential. Intriguingly, some sense lncRNAs are special in the sense that they can function as both RNA and protein-coding gene. For instance, SRA (steroid receptor RNA activator) can translate into protein, and the RNA sequence can also act as a scaffold for several co-activator and repressor proteins to form complexes that regulate gene transcription;58 ENOD40 (early nodulin 40) can translate into proteins59 and is also needed for correct subcellular localization of RNP particles in legume plants.30 These findings have challenged our understanding of gene classification and, in the meantime, broadened the known roles of lncRNA. Such special relationship between the sense lncRNAs and protein-coding genes may provide novel insights into the evolution of gene function.
Present studies are mainly focused on lincRNAs and antisense lncRNAs (especially lincRNAs), though less is known about intronic lncRNAs and sense lncRNAs. Some biological features of lncRNA are quite prominent, such as the high conservation of lincRNAs among mammals.45,46,55 Also, another evidence, albeit not extensive, may indicate the special feature of the sense lncRNAs’ coding potential.58,59 Additionally, as antisense lncRNAs and sense lncRNAs are correlated differently with coding genes, they are most likely to exert different effects on gene locus or mRNAs. Therefore, genomic location and context can be used for classification of lncRNA, though a classification of lncRNA using exclusively genomic localization and context may not be fully adequate.
Effects Exerted on DNA Sequences
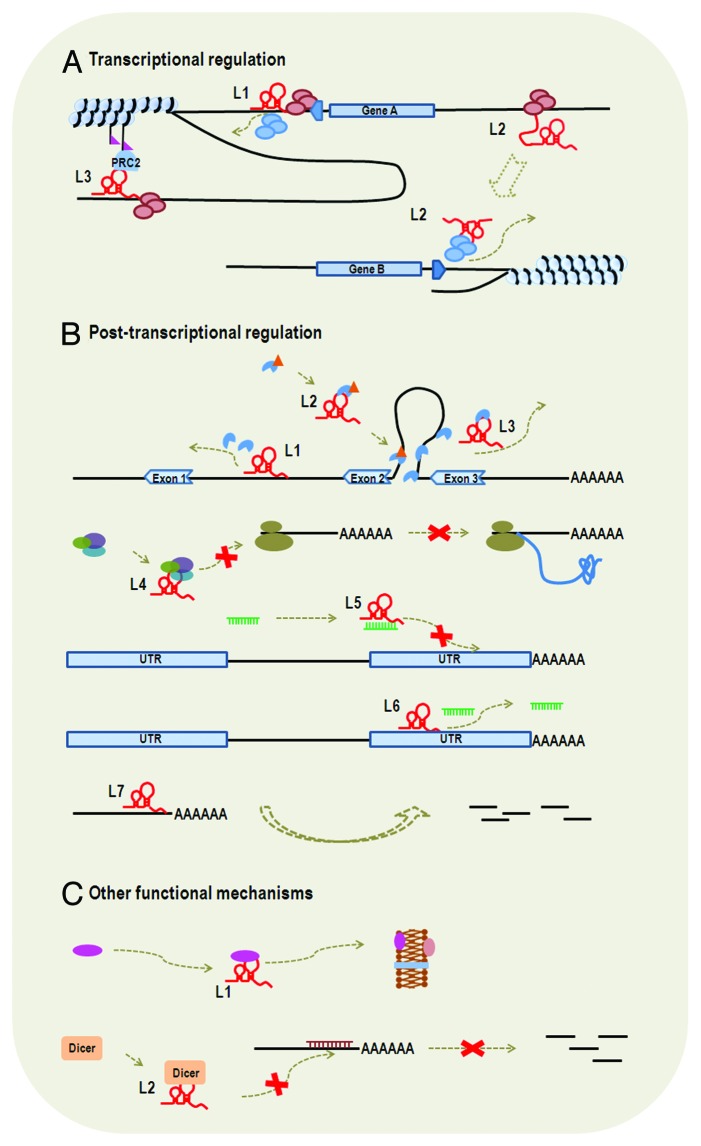
It has been found that lncRNAs are predominately localized in nucleus and chromatin,4 suggesting that lncRNAs may have a significant impact on DNA sequences. Also, a large proportion of lncRNAs are involved in transcriptional regulation (~42% of the 182 assessed entries according to lncRNAdb; Table 1). Therefore, it is meaningful to classify lncRNAs based on their effects exerted on DNA sequences: cis-lncRNAs (cis-acting lncRNAs) that regulate the expression of genes in close genomic proximity and trans-lncRNAs (trans-acting lncRNAs) that regulate the expression of distant genes (Fig. 2A).
Figure 2. Functional mechanisms of lncRNAs. LncRNA is represented by a letter “L” and a number appended. (A) Transcriptional regulation: We listed examples of cis-lncRNA (L1 and L3) and trans-lncRNA (L2). L1 is transcribed from the promoter region of gene A and its binding to promoter of gene A blocks the binding of transcription factors, thus affecting transcription initiation of gene A. L3 functions to modify chromatin protein in its vicinity through recruiting the complex of PRC2. L2 influences transcription of gene B from a distant region through interaction with transcription factor or RNA polymerase. Therefore, L1 and L2 also function through transcriptional interference, whereas L3 functions through chromatin modification. (B) Post-transcriptional regulation: L1, L2 and L3 all influence gene splicing. Specifically, L1 binds to intronic area to inhibit binding of splicing factor, L2 functions to modulate the pool of modified (such as phosphorylation) splicing factor and L3 binds to splicing factor to block spliceosomal complex formation. L4 interacts with translational factors to inhibit translation. L5 and L6 are two examples of ceRNAs, which interact directly or indirectly with miRNAs. L5 binds to miRNA and, thus, inhibits the binding of miRNA to the 3′ UTR of target mRNA. L6 binds to the 3′ UTR of target mRNA, which also blocks the binding of miRNA to the target gene. L7 serves as natural antisense inhibitor to promote degradation of mRNA. (C) Other functional mechanisms. L1 is involved in protein transportation and L2 binds to Dicer to influence RNA interference.
Cis-lncRNAs
It seems plausible that cis-lncRNAs function through transcriptional interference or chromatin modification, ranging from yeast [e.g., SRG1 (regulatory gene 1) RNAs60,61] to plants [e.g., COLDAIR (cold assisted intronic noncoding RNA) 62] to mammals [e.g., DHFR (dihydrofolate reductase) upstream transcripts,23,63 Xist (X inactive-specific transcript)64] (detailed below). Regarding the mechanism of transcriptional interference, lncRNAs may influence the transcription activity of target genes through promoter binding to block PIC (preinitiation complex) formation,23 or by interacting with transcription factors23,60,61 (Fig. 2A). Such cis-lncRNAs may be transcribed from genes’ promoter regions. For instance, DHFR upstream transcripts, ~0.8–7.3 kb lncRNAs from the promoter region of DHFR, can form stable triplex structures with the promoter of DHFR23,63 and interact with TFIIB to efficiently dissociate PIC.23 Likewise, SRG1 RNAs, ~0.4–1.9 kb lncRNAs from the promoter region of SER3 (Ser3p), are found to cover the promoter of SER3 coding gene to prevent transcription factor binding to the promoter and, thus, repress the expression of SER3.60,61 Also, the transcription of lncRNAs in promoter regions may induce chromatin remodeling and, thus, activate the downstream protein-coding genes’ transcription, such as fbp1 promoter RNAs.65
Cis-lncRNAs that function through chromatin modification often recruit chromatin modification complexes, e.g., PRC (polycomb repressive complex) or Rpd3S HDAC (Rpd3 small histone deacetylase complexes). The most studied chromatin modification complex is PRC and one well-known example is Xist (a 19 kb lncRNA in human), which binds to PRC2 to induce H3K27me3 modification and, thus, leads to transcriptional silencing of genes on the X chromosome.64 Similar examples can be found in MEG3 (maternally expressed 3) (a ~1.6 kb lncRNA)66 and COLDAIR (a ~1.1 kb lncRNA).62 Also, there are other chromatin modification complexes that are recruited by lncRNAs. GAL10-ncRNA (Gal10p-noncoding RNA) (a ~4 kb lncRNA) has been reported to recruit the Rpd3S HDAC complex, resulting in a decrease in some histone 3 acetylation to repress the expression of GAL1.14 Although the above-mentioned examples of PRC and Rpd3S HDAC are all implicated in negative regulation of gene expression, interaction of lncRNA with chromatin modification complexes is also involved in positive regulation. A case example is HOTTIP (HOXA transcript at the distal tip) (a ~3.8 kb lncRNA in human) that recruits a MLL chromatin modifying complex to maintain a domain of active chromatin over the 5′ end of HOXA (homeobox A cluster) gene cluster.67
Trans-lncRNAs
Although it may be easier for lncRNAs to influence genes in their immediate vicinity probably based on sequence complimentary to the locus from which they are transcribed, lncRNAs can also function in trans-acting mode to target distant gene loci. For instance, HOTAIR (HOX antisense intergenic RNA), a ~2.2 kb lncRNA that is transcribed from the HOXC (homeobox C cluster) gene locus in chromosome 12, can be transported by the Suz-Twelve protein to regulate the homologous target sites at HOXD (homeobox D cluster) gene locus in chromosome 224. Also, HOTAIR are found to bind to many other genomic loci that tend to possess specific DNA motifs and influence gene expression by recruiting chromatin modification complexes.68,69
Therefore, unlike cis-lncRNAs, trans-lncRNAs may function independently of sequence complementary to target gene locus. In addition to chromatin modification complexes,68,69 they may bind to transcription elongation factors70,71 or RNA polymerases41 to affect transcription. It is reported that 7SK RNA (a ~330 bp lncRNA) functions as a central scaffold to coordinate protein-protein interactions in 7SK snRNP (small nuclear ribonucleoproteins), which comprises transcription elongation factor—P-TEFb (positive transcription elongation factor b). This activity consequently leads to repression of transcription elongation at many gene loci.70,71 Another lncRNA, B2 SINE RNA, has been found to stably bind to polymerase II complex to block its activity during heat shock response.41
Mechanisms of Functioning
While only a small number of lncRNAs has been well documented, it is believed that lncRNAs are involved in a wide variety of cellular molecular functions. According to their mechanisms of functioning, lncRNAs roughly fall into three groups that affect transcriptional regulation, post-transcriptional regulation or other functions (Fig. 2).
Transcriptional regulation
As mentioned above, there is a large number of lncRNAs that regulate gene transcription through transcriptional interference (e.g., DHFR upstream transcripts, SRG1 RNAs, 7SK snRNA, B2 SINE RNA) and chromatin remodeling (e.g., fbp1 (fructose-1,6-bisphosphatase-1) promoter RNAs, Xist, MEG3, GAL10-ncRNA, HOTAIR, HOTTIP and COLDAIR). Therefore, lncRNAs responsible for transcription regulation can be sub-divided according to the mechanism of their functioning: (1) transcriptional interference and (2) chromatin remodeling (Fig. 2A). Besides, there are other related functional mechanisms, for example, the regulation effect: a set of lncRNAs transcribed from enhancers are termed eRNAs (enhancer RNAs) as they positively regulate genes’ transcription, such as ncRNA-a1 (activating long ncRNA 1),54 Evf-2 (embryonic ventral forebrain-2) RNA,72 Alpha-250/Alpha-280.73
Post-transcriptional regulation
There are two common post-transcriptional regulation mechanisms that lncRNAs get involved in, namely, splicing regulation and translational control (Fig. 2B). LncRNAs that influence mRNA splicing may function through binding to74 or modulating26 splicing factors, or directly hybridizing with mRNA sequences to block splicing.25,28 MIAT (myocardial infarction associated transcript), a ~9–10 kb lncRNA, contains strong intron branch point sequences (UACUAAC repeats) and is able to bind to SF1 (splicing factor 1) to inhibit splicing and spliceosomal complex formation.74 Malat1 (metastasis-associated lung adenocarcinoma transcript 1), a ~7 kb lncRNA, can bind to SR splicing factor [serine–arginine (SR)-rich splicing factor] and regulate its distribution in nuclear speckle domains.26 Also, it is suggested that Malat1 may modulate the pools of phosphorylated SR and, thus, influence alternative splicing of pre-mRNAs.26 Additionally, other splicing regulation mechanisms may exist. LUST (LUCA-15-specific transcript), a ~1.4–2.4 kb lncRNA, is the antisense transcript of RBM5 (RNA binding motif protein 5) and is hypothesized to regulate the expression of RBM5 splice variants through masking a sense-strand regulatory sequence.25
LncRNAs that participate in translational control may function through binding to translation factors27,75 or ribosome.76,77 There are two lncRNAs, BC1 (brain cytoplasmic RNA 1) and BC200 (200 nt brain cytoplasmic RNA), which can bind eIF4A (eukaryotic translation initiation factor 4A), PABP (poly(A)-binding protein) and other factors, to repress translation initiation by blocking assembly of the required complex.27,75 snaR (small NF90-associated RNAs), a cytoplasmic lncRNAs, can bind to ribosome, presumably influencing translation of mRNAs.76 Gadd7 (growth arrested DNA-damage inducible gene 7), which is associated with active translation, is hypothesized to bind to ribosome.77 It should be noted that in some cases translation and splicing are associated with each other. Zeb2 (zinc finger E-box-binding homeobox 2) translation requires retention of an intron. Zeb2NAT (Zeb2 natural antisense transcript) (a lncRNA that is more than 1.2 kb), which overlaps the 5′ splicing site of an intron, can inhibit splicing of the intron to allow translation of Zeb2.28
Aside from splicing regulation and translational control, there are other post-transcriptional regulation mechanisms utilized by lncRNAs. The findings of siRNA (small interfering RNA) mechanism78,79 and competing endogenous RNAs80-83 have opened up new aspects of post-transcriptional regulation and recent studies suggest that lncRNAs are also implicated in these processes, displaying direct siRNA mechanisms78,79 or interacting with miRNAs.80-83
LncRNAs may function as natural antisense inhibitors to promote degradation of mRNA78,79 (Fig. 2B). It has been found that 21A, a ~300 bp lncRNA, which shows high sequence homology to CENP-F (centromere protein F) intronic portions, can reduce CENP-F expression at both mRNA and protein level through antisense inhibitor.78 1/2-sbsRNA1 (half-STAU1-binding site RNA1), a ~0.7 kb lncRNA, has been found to bind to mRNAs’ 3′ UTR through Alu elements, and reduces mRNA abundance.79
Moreover, there are many lncRNAs that interact directly or indirectly with miRNAs to stabilize target mRNAs80-83 (Fig. 2B). These lncRNAs are called ceRNAs (competing endogenous RNAs).80-83 For instance, linc-MD1 (long intergenic ncRNA that is associated with muscle differentiation), a ~0.5 kb lncRNA, acts as sponge/target mimic of miR-133 and miR-135 to regulate the expression of two transcription factors: MAML1 (mastermind-like protein 1) and MEF2C (myocyte-specific enhancer factor 2C), which activate the expression of muscle-specific genes.80 Similar examples can be found in IPS1 (induced by phosphate starvation 1) RNA84 and HULC (highly up-reglated in liver cancer) RNA.85 Some pseudogenes can function as a sponge/target mimic for miRNAs to stabilize their homologous mRNAs, such as KRASP1 and PTENP1, which are pseudogenes of KRAS (V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog) and PTEN (phosphatase and tensin homolog), respectively.83,86 In addition, some antisense lncRNAs may bind to mRNA to mask the binding sites of miRNA and thus stabilize mRNA. BACE1AS (BACE1 antisense RNA), a ~2 kb lncRNA, has been reported to be activated in Alzheimer disease to form an RNA duplex with BACE1 (β-secretase 1) mRNA,87 which may mask the binding site for miR-485-5p and, thus, prevent translational repression of BACE1 mRNA by miRNA.88
Other mechanisms of lncRNA functioning
In addition to transcriptional regulation and post-transcriptional regulation, lncRNAs may function through other mechanisms (Fig. 2C), such as protein localization,29,30 telomere replication,89 RNA interference,90 beyond transcription and translation regulation, etc. Considering the limitation of the currently available knowledge, it is difficult to categorize these lncRNAs into some more “stable” groups and, thus, we roughly place them into lncRNAs with “other functional mechanisms.” For instance, meiRNA, a ~0.5 kb lncRNA, is required for nuclear localization of Mei2.29 ENOD40 RNA is required for correct subcellular localization of RNP particles in legume plants.30 TERC (telomerase RNA component), which is part of telomerase reverse transcriptase, acts as template to extend telomere during DNA replication in eukaryote.89 In ciliated protozoa, maternal RNA templates have been found to guide reproducible rearrangement during the transition from germline nucleus to somatic nucleus.91 In addition, lncRNAs may be involved in RNA interference by regulation of Dicer1. It is suggested that rncs-1 (RNA noncoding and starvation upregulated) reduces Dicer-generated siRNA and affects levels of Dicer-regulated genes.90
Targeting Mechanisms of lncRNAs
According to their mode of action, lncRNAs may also be classified based on their targeting mechanisms, mainly associated with the following categories92: (1) signal: show cell type-specific expression and respond to diverse stimuli, such as Xist,64 COLDAIR62; (2) decoy: bind and titrate away a protein target, but does not exert any additional functions, such as DHFR upstream transcripts,23 PANDA;93 (3) guide: bind proteins and then direct the localization of ribonucleoprotein complex to specific targets, such as Xist,64 HOTAIR;24,68 (4) scaffold: serve as central platforms to bring together multiple proteins to form ribonucleoprotein complexes, such as HOTAIR,24,68 7SL.94,95 Alternatively, lncRNAs can be grouped based on the types of interactions they make with their targets: RNA-RNA pairings, RNA-DNA hybrids, RNA structure mediated interactions and protein linkers.96
However, a single targeting archetype in referring to mode of action may not be sufficient to fully describe one lncRNA since one lncRNA may contain multiple archetypes. There are many lncRNAs that are induced by endogenous and exogenous signals to express, and they also can possess the binding sites of chromatin modification complexes (such as PRC) to repress or activate expression of a set of genes, e.g., Xist,64 Air,97 COLDAIR,62 HOTTIP,67 HOTAIR,24,68 lincRNA-p21.38 Therefore, these lncRNAs operate in a dual mode as both signal and guide. Some signal lncRNAs may also function as decoys that bind and titrate away a protein target, e.g., PANDA.93 More complexly, some lncRNAs have more than two archetypes, such as HOTAIR that functions according to three archetypes: as anatomic signal, guiding the chromatin-modifying complexes to the target gene, and as a scaffold for PRC2 and LSD1.24,68
Albeit, there are multiple groups of lncRNAs based on different classification methods as mentioned above, different groups appear to be linked closely to one another. LncRNAs that function via transcriptional interference mechanism may target gene loci through RNA-DNA hybrids and operate as the archetypes of decoy, e.g., DHFR upstream transcripts;23 lncRNAs that affect chromatin modification may target gene loci through RNA structure mediated interaction, which, thus, acts as the archetypes of signal, guide and scaffold, e.g., HOTAIR;24,68 ceRNAs that directly or indirectly regulate mRNA level target miRNAs or mRNAs through RNA-RNA pairing, may also operate as the archetype of decoy, for example, KRASP1 and PTENP1;86 lncRNAs that regulate splicing may function through RNA-RNA pairing to influence their targets and operate as the archetype of decoy (e.g., Malat126).
Perspectives
Here, we summarized classification methods of lncRNAs according to their four major characteristics (genomic location and context, effects on DNA sequences, functional mechanisms and targeting mechanisms). Based on function annotations presently available, we explored potential relationships between different classification categories and investigated biological features of different lncRNAs within each category. Although lncRNA could be described in terms of other features, such classifications of lncRNAs as summarized here are of fundamental importance for lncRNA studies, helpful for further investigation of specific groups of lncRNA, for generation of new hypothesis based on different lncRNA groups and for exploration of lncRNA underlying functional mechanisms.
Classification of lncRNAs
When studying lncRNAs, it is straightforward to investigate their functional features by classifying them into different groups. However, classification of lncRNAs is highly dependent on the current existing knowledge, thus requiring frequent validation of the classification system, exploring new classification systems and, when necessary, abandoning old ones. One possible example is the definition of lncRNA that is longer than 200 nt. We investigated the length of lncRNAs and compared their distribution between human and mouse (data from NONCODE V3.050). Clearly, lncRNAs can be further divided into different groups based on their length distribution (Fig. 3): small-lncRNA (200~950 nt), medium-lncRNA (950~4,800 nt), large-lncRNA (4,800 nt~). In human, the majority of lncRNAs are small-lncRNAs (58%), whereas the majority of lncRNAs in mouse are medium-lncRNAs (78%). In addition, human genome contains more small-lncRNAs and large-lncRNAs than mouse, but less medium-lncRNAs. However, the number of manually annotated lncRNAs by GENCODE released recently is less than half of NONCODE V3.0,4,50 suggesting the uncertainty of the comparative results. While the differences could be due to the annotation levels, if proved to reflect the genuine situation, such differences may imply distinct evolutionary process of lncRNAs between human and mouse and need further evolutionary analysis.
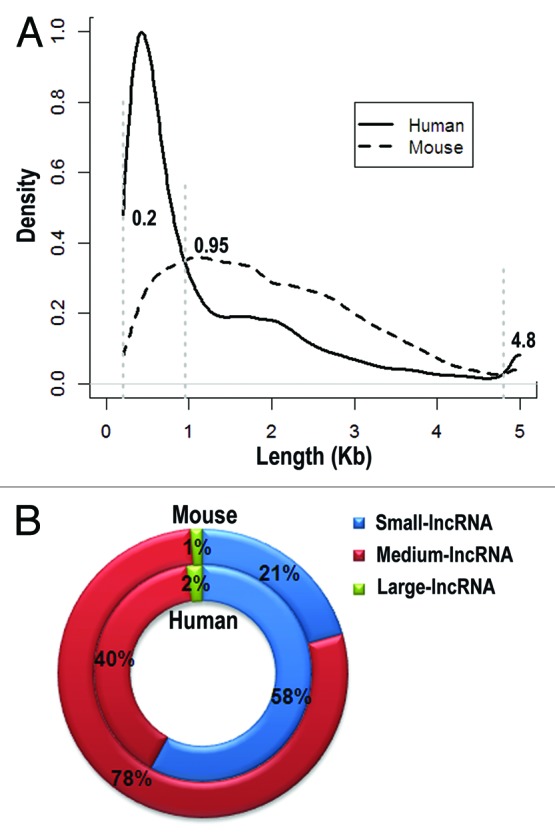
Figure 3. Length distribution of lncRNAs in human and mouse. LncRNAs are divided into three groups based on their length distribution: small-lncRNA (200~950 nt), medium-lncRNA (950~4,800 nt) and large-lncRNA (4,800 nt~). Density distributions of lncRNA length are shown in (A) and percentages of three lncRNA groups are depicted in (B).
Investigation of specific groups of lncRNAs
Classification of lncRNAs is useful for focusing on specific groups. Currently, lincRNAs that recruit chromatin modification complexes have attracted a lot of attention. It is reported that lincRNAs tend to bind to specific chromatin-modifying complexes,55 so that lincRNA-associated chromatin modification may represent a specific regulation mechanism. Additionally, lincRNAs recruiting the same chromatin-modifying complex may diverge greatly between different species.66 Therefore, lincRNAs may affect specific variety of target genes in different biological processes, such as PCAT-1, which regulates a variety of prostate cancer implicated genes through recruiting PRC2.53 These findings shed lights on the functional significance of the lincRNA group (that functions through recruiting PRC2) and stimulate further work on other chromatin-modifying complexes. Notably, there is a large number of antisense lncRNAs, which function through recruiting chromatin modification complexes to regulate transcription activity according to available evidences (Table 1). It has been suggested that most of the PRC2 binding transcripts are antisense and sense lncRNAs rather than lincRNAs.66 Therefore, it would be desirable for future studies to explore more chromatin complex binding features of the antisense and sense lncRNAs.
Regulation network of lncRNA and small ncRNA
The discovery of lncRNAs and their regulatory roles challenges the initially miRNA-centered regulatory networks, which may help comprehensively understand gene regulation by both small ncRNA and lncRNA and, accordingly, provide new insights into complex processes of gene regulation. As mentioned above, ceRNAs, a group of lncRNAs interacting with miRNAs, can impose an additional level on post-transcriptional regulation.80-88 In addition to those lncRNAs that act as sponge/target mimic of miRNAs,80-86 it should be noted that more than 50% of the sense and antisense transcript pairs may be composed of coding and antisense non-coding transcripts,49 and the formed RNA duplex may also influence the interaction between miRNAs and their target mRNAs.87,88 According to our previous integrative analysis of miRNAs and mRNAs in NSCLC (non-small cell lung carcinoma),98 we found that most of the target mRNAs do not vary significantly, in spite of the dramatic increase or decrease of their miRNAs, indicating that gene regulation network may be more sophisticated.
Considering their similar background in gene expression and function overlap between lncRNA and small ncRNA, it is attractive to investigate whether there is a certain evolutionary association between the two components. It is found that some lncRNAs contain miRNAs in their gene locus.99-101 H19 (gene comes from colon pH19), a ~2.3 kb lncRNA, contains mir-675 in its exon and also serves as a precursor of this miRNA in addition to its transcription regulation activity.99 Also, lncRNA LOC554202 contains mir-31 in its intron, and both miR-31 and the host lncRNA are found to be lowly expressed in triple-negative breast cancer.100 It has been found that five lncRNAs, namely, MEG3, MEG8 (maternally expressed 3), MEG9 (maternally expressed 3), antiPeg11 (antisense transcript to Peg11/Rtl1) and Rian (RNA imprinted and accumulated in nucleus), contain a lot of miRNAs and snoRNAs and function through transcriptional regulation; interestingly, these five lncRNAs and their endogenous small ncRNA may target the same gene,101 but it remains unclear whether those five lncRNAs can serve as precursors of their corresponding miRNAs. In addition, it is suggested that some lncRNAs in human may be preferentially post-processed into snoRNAs.4,102 Taken together, future studies focused on this aspect may bring new unexpected insights into the evolutionary relationships between small ncRNA and lncRNA.
Acknowledgments
We thank Hao Wu and Gang Wu for their valuable comments on this manuscript. This work was supported by grants from National Natural Science Foundation of China (Grant No. 31200978 to L.M.), King Abdullah University of Science and Technology research funds (to V.B.B.), the “100-Talent Program” of Chinese Academy of Sciences (Y1SLXb1365 to Z.Z.) and National Programs for High Technology Research and Development (863 Program; Grant No. 2012AA020409 to Z.Z.), the Ministry of Science and Technology of the People’s Republic of China.
Glossary
Abbreviations:
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- snoRNAs
small nucleolar RNAs
- piRNAs
piwiRNAs
- lincRNAs
long intergenic non-coding RNAs
- CAGE
cap-analysis gene expression
- SRA
steroid receptor RNA activator
- ENOD40
early nodulin 40
- SRG1
SER3 regulatory gene 1
- COLDAIR
cold assisted intronic noncoding RNA
- DHFR
dihydrofolate reductase
- Xist
X inactive-specific transcript
- SER3
Ser3p
- PRC
polycomb repressive complex
- Rpd3S HDAC
Rpd3 small histone deacetylase complexes
- MEG3/8/9
maternally expressed 3/8/9
- GAL10-ncRNA
Gal10p-noncoding RNA
- HOTTIP
HOXA transcript at the distal tip
- HOXA
homeobox A cluster
- HOTAIR
HOX antisense intergenic RNA
- HOXC
homeobox C cluster
- HOXD
homeobox D cluster
- snRNP
small nuclear ribonucleoproteins
- P-TEFb
positive transcription elongation factor b
- B2 SINE RNA
B2 short interspersed element RNA
- fbp1
fructose-1,6-bisphosphatase-1
- eRNAs
ncRNA-a1, activating long ncRNA 1
- Evf-2
embryonic ventral forebrain-2
- MIAT
myocardial infarction associated transcript
- SF1
splicing factor 1
- Malat1
metastasis-associated lung adenocarcinoma transcript 1
- SR splicing factor
serine–arginine (SR)-rich splicing factor
- LUST
LUCA-15-specific transcript
- RBM5
RNA binding motif protein 5
- BC1
brain cytoplasmic RNA 1
- BC200
200nt brain cytoplasmic RNA
- eIF4A
eukaryotic translation initiation factor 4A
- PABP
poly(A)-binding protein
- Gadd7
growth arrested DNA-damage inducible gene 7
- snaR
small NF90-associated RNAs
- Zeb2
zinc finger E-box-binding homeobox 2
- Zeb2NAT
Zeb2 natural antisense transcript
- siRNA
small interfering RNA
- CENP-F
centromere protein F
- 1/2-sbsRNA1
half-STAU1-binding site RNA1
- ceRNAs
competing endogenous RNAs
- linc-MD1
long intergenic ncRNA that is associated with muscle differentiation
- MAML1
mastermind-like protein 1
- MEF2C
myocyte-specific enhancer factor 2C
- IPS1
induced by phosphate starvation 1
- HULC
highly up-reglated in liver cancer
- KRAS
V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog
- PTEN
phosphatase and tensin homolog
- BACE1
beta-secretase 1
- BACE1AS
BACE1 antisense RNA
- Mei2
meiosis protein 2
- TERC
telomerase RNA component
- rncs-1
RNA noncoding and starvation up-regulated
- NSCLC
non-small cell lung carcinoma
- H19
gene comes from colon pH19
- Rian
RNA imprinted and accumulated in nucleus
- antiPeg11
antisense transcript to Peg11/Rtl1
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group Phase I & II Team Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JM, Edwards S, Shoemaker D, Schadt EE. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 2005;21:93–102. doi: 10.1016/j.tig.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr., et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–39. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 9.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 10.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39(Database issue):D146–51. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–42. doi: 10.1016/0092-8674(92)90520-M. [DOI] [PubMed] [Google Scholar]
- 12.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–75. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 14.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–95. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein HD, Zopf D, Freymann DM, Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci U S A. 1993;90:5229–33. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 2007;316:1345–8. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 17.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, et al. Mouse transcriptome: neutral evolution of 'non-coding' complementary DNAs. Nature. 2004;431:757. doi: 10.1038/nature03016. [DOI] [PubMed] [Google Scholar]
- 19.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin Cell Dev Biol. 2011;22:366–76. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 24.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rintala-Maki ND, Sutherland LC. Identification and characterisation of a novel antisense non-coding RNA from the RBM5 gene locus. Gene. 2009;445:7–16. doi: 10.1016/j.gene.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol. 2002;321:433–45. doi: 10.1016/S0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 28.Beltran M, Puig I, Peña C, García JM, Alvarez AB, Peña R, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–69. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe Y, Yamamoto MS. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell. 1994;78:487–98. doi: 10.1016/0092-8674(94)90426-X. [DOI] [PubMed] [Google Scholar]
- 30.Campalans A, Kondorosi A, Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–59. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–57. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- 32.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–59. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, et al. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–31. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 34.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 35.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–5. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 36.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A. 2006;103:5781–6. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–46. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 38.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–37. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–7. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–9. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 42.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–60. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 43.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–42. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–54. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 49.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. RIKEN Genome Exploration Research Group. Genome Science Group (Genome Network Project Core Group) FANTOM Consortium Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 50.Bu D, Yu K, Sun S, Xie C, Skogerbø G, Miao R, et al. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012;40(Database issue):D210–5. doi: 10.1093/nar/gkr1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 53.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. 2010;151:939–47. doi: 10.1210/en.2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leygue E. Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signal. 2007;5:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Girard G, Roussis A, Gultyaev AP, Pleij CW, Spaink HP. Structural motifs in the RNA encoded by the early nodulation gene enod40 of soybean. Nucleic Acids Res. 2003;31:5003–15. doi: 10.1093/nar/gkg721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–4. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 61.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 63.Blume SW, Meng Z, Shrestha K, Snyder RC, Emanuel PD. The 5′-untranslated RNA of the human dhfr minor transcript alters transcription pre-initiation complex assembly at the major (core) promoter. J Cell Biochem. 2003;88:165–80. doi: 10.1002/jcb.10326. [DOI] [PubMed] [Google Scholar]
- 64.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–4. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 66.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–5. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 71.Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature. 2001;414:317–22. doi: 10.1038/35104575. [DOI] [PubMed] [Google Scholar]
- 72.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–84. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tasheva ES, Roufa DJ. Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes Dev. 1995;9:304–16. doi: 10.1101/gad.9.3.304. [DOI] [PubMed] [Google Scholar]
- 74.Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells. 2011;16:479–90. doi: 10.1111/j.1365-2443.2011.01502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin D, Pestova TV, Hellen CU, Tiedge H. Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol. 2008;28:3008–19. doi: 10.1128/MCB.01800-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parrott AM, Tsai M, Batchu P, Ryan K, Ozer HL, Tian B, et al. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res. 2011;39:1485–500. doi: 10.1093/nar/gkq856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crawford DR, Schools GP, Salmon SL, Davies KJ. Hydrogen peroxide induces the expression of adapt15, a novel RNA associated with polysomes in hamster HA-1 cells. Arch Biochem Biophys. 1996;325:256–64. doi: 10.1006/abbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 78.Pagano A, Castelnuovo M, Tortelli F, Ferrari R, Dieci G, Cancedda R. New small nuclear RNA gene-like transcriptional units as sources of regulatory transcripts. PLoS Genet. 2007;3:e1. doi: 10.1371/journal.pgen.0030001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–57. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 85.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–83. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, et al. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 90.Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci U S A. 2008;105:12897–902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nowacki M, Landweber LF. Epigenetic inheritance in ciliates. Curr Opin Microbiol. 2009;12:638–43. doi: 10.1016/j.mib.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zwieb C, van Nues RW, Rosenblad MA, Brown JD, Samuelsson T. A nomenclature for all signal recognition particle RNAs. RNA. 2005;11:7–13. doi: 10.1261/rna.7203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–8. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 96.Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010;7:582–5. doi: 10.4161/rna.7.5.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 98.Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng F, et al. An integrated analysis of miRNA and mRNA expressions in non-small cell lung cancers. PLoS One. 2011;6:e26502. doi: 10.1371/journal.pone.0026502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–6. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer. 2012;11:5. doi: 10.1186/1476-4598-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hagan JP, O’Neill BL, Stewart CL, Kozlov SV, Croce CM. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]