Abstract
Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b), a RING finger E3 ubiquitin-protein ligase, has been demonstrated to play a crucial role in establishing the threshold for T-cell activation and controlling peripheral T-cell tolerance via multiple mechanisms. Accumulating evidence suggests that Cbl-b also regulates innate immune responses and plays an important role in host defense to pathogens. Understanding the signaling pathways regulated by Cbl-b in innate and adaptive immune cells is therefore essential for efficient manipulation of Cbl-b in emerging immunotherapies for human disorders such as autoimmune diseases, allergic inflammation, infections, and cancer. In this article, we review the latest developments in the molecular structural basis of Cbl-b function, the regulation of Cbl-b expression, the signaling mechanisms of Cbl-b in immune cells, as well as the biological function of Cbl-b in physiological and pathological immune responses in animal models and human diseases.
Keywords: Cbl-b, molecular struction, biological function, signaling mechanism, immune disease and therapy
Introduction
Over the last decade, accumulating evidence suggests that ubiquitination of proteins by E3 ligases is a novel and crucial regulation mechanism in innate and adaptive immunity.1,2 The gene of Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b), an E3 ubiquitin-protein ligase and an adaptor protein, was initially cloned and characterized by Keane et al. in 1995.3 Cbl-b belongs to the Cbl family, which consists of c-Cbl and Cbl-3 in addition to Cbl-b and has a broad spectrum of biological functions.
Recent studies using gene-targeting approaches have yielded convincing evidence that Cbl-b negatively regulates the signaling pathways derived from the T-cell receptor (TCR),4,5 B-cell receptor (BCR), CD40,6,7 and FcεR1 (high affinity immunoglobulin epsilon receptor).8 Because of the diversities of substrates of Cbl-b in different cell types, it appears that Cbl-b regulates various signaling pathways in a cell type-dependent manner. In this review, we will summarize the most recent progress on Cbl-b-related studies in immune systems, which encompass Cbl-b structure, regulation of Cbl-b expression, and its role in innate and adaptive immune responses. We will also discuss the potential roles of Cbl-b in various diseases including autoimmune and inflammatory diseases, infection, and cancer.
Genetics, Tissue Distribution, and Subcellular Location of Cbl-b
The mammalian Cbl family of proteins is highly conserved throughout evolution from nematodes to humans and consists of c-Cbl, Cbl-b, and Cbl-3 (Fig. 1). The Cblb gene is located on chromosome 3q13.11 in humans and chromosome 16B5 in the mouse. Cbl-b is abundantly expressed in a variety of immune cells.9,10 In T cells, Cbl-b is predominantly expressed in peripheral T cells, whereas c-Cbl is mainly expressed in thymus, suggesting a distinct role of c-Cbl and Cbl-b in T-cell development and tolerance induction.11 In T cells, Cbl-b is located in the cytoplasm but can be translocated to the plasma membrane upon TCR stimulation.12
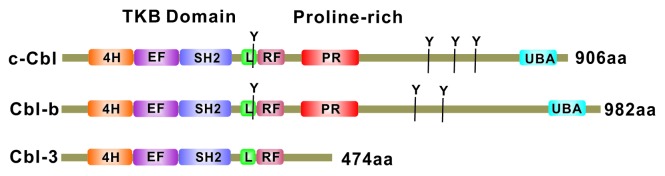
Figure 1. Functional domains of the Cbl family (c-Cbl, Cbl-b, and Cbl-3) in mammals. All three (3) members of the Cbl family of proteins share a highly homologous N-terminal region that serves as the structural platform for direct binding to specific pY-containing peptide motifs in activated PTKs and is accordingly referred to as the tyrosine kinase-binding (TKB) domain; this domain is assembly of a 4-helical (4H) bundle, an EF hand domain, and a variant SH2 domain. The TKB domain is followed by a highly conserved helical linker (L) domain and a RING (really interesting new gene) finger (RF) domain, which bind to ubiquitin-conjugating enzymes (E2). The proline-rich motifs (PR) bind to SH3 domain containing signaling and endocytic proteins. Induced tyrosine phosphorylation sites (major sites at Y700, Y731, and Y774 in c-Cbl) recruit SH2 domain-containing signaling proteins. The leucine zipper (LZ)/ubiquitin-associated (UBA) domain near the C-terminus is involved in ubiquitin binding and dimerization. Cbl-c lacks most of the C-terminal regions except for a short PR region for potential interactions with SH3 domain-containing proteins.
Structural Components of Cbl-b and Their Functions
The Cbl family of ubiquitin ligases in mammals share highly conserved regions in their N-terminal halves, which encompass their TKB (protein tyrosine-kinase-binding), linker (L), and RING (really interesting new gene) finger (RF) domains (Fig. 1). The unique feature of the TKB domain is that it recognizes specific substrates of Cbl-b, which is achieved by binding to proteins containing specific phosphorylated tyrosine-containing motifs, such as Syk and Zap-70, and a range of receptor tyrosine kinases.6,13 Interaction of proteins with the TKB domain of Cbl is mediated by 3 distinct subdomains consisting of a 4-helix bundle (4H), a calcium-binding EF hand, and a variant SH2 domain, all 3 of which are functionally required to form a unique PTB (phosphotyrosine-binding) module.14 SH2 domain within the TKB recognizes tyrosine-phosphorylated proteins for ubiquitin conjugation.15 A highly conserved α-helix of the L domain plays an important role in maintaining E3 activity.16,17 The crystal structure shows that the L region contacts the TKB, RF, and E2 ubiquitin-conjugating enzymes.16 The RF domain has intrinsic E3 ubiquitin ligase activity and binds to ubiquitin-E2 for the transfer of ubiquitin to specific substrates.18-20 Recent studies also indicate that the phosphorylation of Y363, located in the L region between TKB and RF domains, regulates the E3 activity of Cbl-b by 2 mechanisms: one is to remove the masking of the RF domain from the TKB domain, and the other is to form a surface to enhance binding affinity to E2s.21,22 Consistent with this finding, the equivalent tyrosine in c-Cbl, i.e., Y371, has been shown to regulate its E3 ubiquitin ligase activity in a similar fashion.23
In contrast, the C-terminal regions of this family of proteins are less conserved. The proline-rich (PR) domain in the C-terminus of c-Cbl and Cbl-b refers to a PX(P/A)XXR motif that binds to SH3 domains of the CIN85/RUK (regulator of ubiquitous kinase)/CD2AP (C2-associated protein) family of proteins.24,25 The tyrosine residues at the C termini of c-Cbl and Cbl-b are phosphorylated by protein tyrosine kinases (PTKs) following stimulation of a diverse array of cell surface receptors.26,27 c-Cbl can bind Vav-family guanine nucleotide exchange factors (GEFs), the p85 regulatory subunit of phosphoinositide 3-kinase (PI 3K), and the Crk-family of adaptor proteins that link Cbl proteins to C3G through interactions with phospho-Y700, Y731, and Y774, respectively.28-31 Likewise, Cbl-b is phosphorylated at Y655 and Y709 upon TCR stimulation. However, the phosphorylation of Cbl-b is weaker compared with that of c-Cbl in response to this stimulation.10,32 It is noted that the Y731EAM motif provides c-Cbl a docking site for the SH2 domains of p85, thus enabling c-Cbl to function as a positive regulator of PI3K activity.33-36 Indeed, Akt and c-Cbl Y737 (mouse) are highly phosphorylated in lineage-negative bone-marrow (BM) cells upon stimulation with stem cell factor (SCF) or FLT3 ligand, whereas c-Cbl−/− BM cells display defective phosphorylation of Akt, possibly due to loss of c-Cbl, which results in the uncoupling of PI3K p85 from the membrane.37 The absence of this p85-binding motif in Cbl-b highlights a potentially important divergence in the role and mode of action of these 2 highly similar regulatory proteins.33 In support of this notion, Cbl-b has been documented to inhibit Pten inactivation via Nedd4 (neuronal precursor of cell expressed developmentally downregulated gene 4) independently of its E3 ubiquitin ligase activity in T cells.12
Both c-Cbl and Cbl-b have an ubiquitin-associated (UBA) domain at their C terminal end, which interacts with ubiquitin and ubiquitin-like domains of proteins such as Nedd8,38,39 and is present in a variety of proteins involved in ubiquitin-mediated processes. Moreover, c-Cbl and Cbl-b also can form homo- and hetero-dimers through interaction between their UBA domains. Cbl-b dimerization is regulated by ubiquitin binding and requires tyrosine phosphorylation of Cbl-b and ubiquitination of Cbl-b substrates.40 However, Cbl-b, rather than c-Cbl, constitutively coimmunoprecipitates with high molecular weight ubiquitinated proteins.41 Furthermore, the UBA domain of Cbl-b has a much greater affinity for polyubiquitin chains than for monoubiquitin, and inhibits a variety of ubiquitin-mediated processes, such as degradation of ubiquitinated proteins.41,42 This difference in ubiquitin-binding reflects distinct regulatory functions of Cbl-b from c-Cbl.
Biochemical Function and the Expression Modification of Cbl-b
The ubiquitin–protein ligase system consists of 3 classes of enzymes known as ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin–protein ligases (E3). The ubiquitination reaction is initiated when 76-amino acid ubiquitin is activated by E1. A thioester bond forms between the active cysteine residue of E1 and the C terminus of ubiquitin in an ATP-dependent reaction. Following ubiquitin activation, activated ubiquitin is transferred to E2 in another ATP-dependent reaction. As an E3 ubiquitin-protein ligase, Cbl-b transfers ubiquitin from specific E2 to the ε-amino group of a lysine (K) residue on the protein substrate. (Fig. 2). The fate of the tagged substrate depends on the number of ubiquitin molecules added (mono-ubiquitin vs. poly-ubiquitin) as well as the K residue involved in the formation of the polyubiquitination chains. Generally, proteins polyubiquitinated through K48 are degraded in the 26S proteasome,43,44 whereas mono-ubiquitination (or multi-ubiquitination) usually marks membrane proteins for endocytosis and subsequent degradation in lysosomes.45 Polyubiquitination through K11, 29, 63 may endow substrate proteins new functions,46 which serves as a signal for functional modification of the substrate, including transcriptional regulation,47-49 ubiquitination-dependent processing of precursor proteins,50 and kinase activation.51 Much less is known about the precise function and topology of unconventional polyubiquitin chains linked through K6, K11, K27, K29, or K33,52 which may target proteins for degradation.53 Given that Cbl-b interacts with many proteins in various immune cells,10,54-61 Cbl-b is thought to play important roles in maintaining the homeostasis of the immune system through elaborate signal transduction pathways.
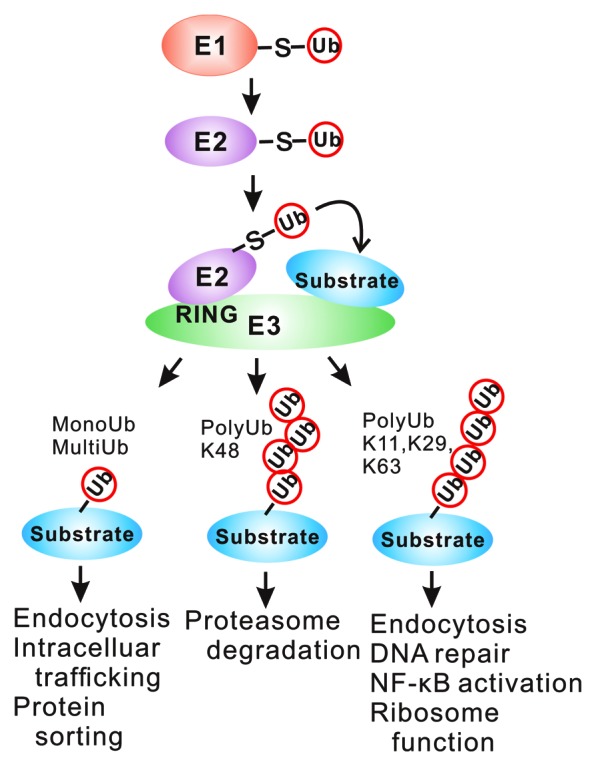
Figure 2. Overview of the ubiquitin pathway utilized by RING type E3 ligases. Three types of enzyme are required for substrate ubiquitination: ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin–protein ligase (E3) enzymes. The E1-E2-E3 cascade mediates ubiquitination of the substrate with the substrate specificity provided by the E3 enzyme. The substrate protein can be tagged with just one ubiquitin, or polyubiquitin chains, which determines the fate of target proteins. –S–, thiolester bond.
The tyrosine and serine residues of Cbl-b are phosphorylated upon stimulation of an vast array of cell-surface receptors, including the TCR,10,40,56,59,62-66 a process that is essential for Cbl-b function. Cbl-b also undergoes ubiquitination upon CD28 costimulation in T cells, resulting in its proteasomal degradation. CD28 costimulation potentiates TCR-induced Cbl-b degradation, whereas CTLA-4-B7 interaction is required for Cbl-b re-expression.67,68 Thus, CD28 and CTLA-4 tightly regulate Cbl-b expression, which is critical for establishing the threshold for T-cell activation and tolerance induction. The proteasomal degradation of Cbl-b may be mediated by Nedd4, which has been shown to target Cbl-b for ubiquitination and degradation,69 and PKC-θ, which phosphorylates Cbl-b at Ser282 in the TKB domain, facilitating Cbl-b ubiquitination.59 In keeping with this, it was reported that Nedd4 promotes adaptive T-cell responses in vitro and in vivo.12,69
Signaling Pathway of Cbl-b in Innate and Adaptive Immune Cells
Genetic and biochemical studies have shown that Cbl family proteins, including those from Drosophila and Caenorhabditis elegans, attenuate intracellular signaling induced by the engagement of cell surface receptors.57 Cbl-b plays a negative regulatory role by targeting proteins for ubiquitination or by interacting with other proteins via its PR region, TKB domain, or UBA domain. For example, Cbl-b interacts with phospho-tyrosine-containing proteins via its TKB domain,10,13 E2-ubiquitin complexes via its RF domain,20,70 binds to SH3 domain-containing proteins via its PR region,57 SH2 domain-containing proteins via its C-terminal tyrosine residues,10 and polyubiquitinated proteins via its UBA domain.40,41 It has been documented that dimerization of Cbl-b is required for the binding of Cbl-b to poly-ubiquitin but not for mono-ubiquitin.9,10,41,56,57,71-73 Thus, Cbl-b plays an important regulatory role in innate and adaptive immune cells through its involvement with many signaling pathways.
Cbl-b in innate immune responses
The responses of innate immune cells to extracellular matrix proteins, cytokines, pathogens, cellular damage, and many other stimuli are regulated by a complex network of intracellular signal transduction pathways, most of which are either initiated or modulated by Src-family or Syk tyrosine kinases present in innate cells.74 Cbl-b has been implicated in the major signaling pathways of macrophages, dendritic cells, natural killer (NK) cells, and NKT cells and mast cells in innate immunity.
Integrins are critical for the migration and function of macrophages during inflammation, and Cbl-b plays an important role in integrin signal transduction. Cbl-b deficiency facilitates activation of β2-integrin leukocyte function-associated antigen-1 (LFA-1) and LFA-1-mediated inflammatory cell recruitment. Cblb−/− mice display increased macrophage recruitment in thioglycollate-induced peritonitis, and Cblb−/− bone marrow-derived mononuclear phagocytes (BMDMPs) show increased adhesion to endothelial cells resulting from activation of LFA-1, which mediates adhesion of BMDMPs to ICAM-1. Cbl-b deficiency also results in increased phosphorylation of T758 in the β2-chain, thereby enhancing the association between 14–3-3β protein and the β2-chain, leading to activation of LFA-1.75 Cbl-b is also implicated in the Toll-like receptor (TLR)-triggered PI3K-RapL-integrin-α(M), CD11b activation pathways.76 The initial study that suggests that Cbl-b is involved in innate immune responses came from evidence that Cbl-b participates in acute lung injury by negatively regulating TLR4 signaling in mouse monocytes. Loss of Cbl-b markedly aggravates acute lung inflammation and leads to 100% lethality upon polymicrobial sepsis induction.76 However, no additional studies by other investigators verified this finding. Subsequently, Cbl-b was shown to target MyD88 and TRIF, which is potentiated by activating the tyrosine kinases Src and Syk in macrophages upon TLR stimulation including TLR4.77 However, Nrdp1 has also been shown to ubiquitinate MyD88 and TBK-1 in macrophages upon TLR4 ligation.78 Since these studies did not examine macrophages from mice lacking Cbl-b, or expressing a Cbl-b RF mutation, it is currently unknown whether Cbl-b is indeed the E3 ubiquitin ligase for MyD88 in a physiological setting. Therefore, the physiological substrates for Cbl-b in innate immune cells are largely unknown.
Cbl-b also plays an important regulatory role in dendritic cells, NK, NKT cells, and mast cells. Cbl-b functions not only as a negative regulator of signaling, but also as a positive modulator of TNF receptor superfamily signaling. TRANCE and CD40L-mediated Akt activation is defective in Cblb−/− dendritic cells,35 suggesting that Cbl-b positively regulates these pathways. It was reported that Cbl-b may target CARMA1, a critical signaling molecule in NF-κB activation, for mono-ubiquitination in NKT cells.79 Ubiquitin conjugation to CARMA1 disrupts its complex formation with Bcl10 without affecting its protein stability, suggesting that this process is mediated by a proteolysis-independent mechanism.79 It has been recently reported that a novel inhibitory role of Cbl-b in the regulation of NK cell functions via TAM receptors. Releasing the inhibition imposed by this TAM/Cbl-b pathway would render NK cells capable of rejecting tumor metastases.80 Furthermore, Cbl-b negatively regulates IgE-mediated activation of mast cells as well as the activation and tissue infiltration of macrophages.81,82 The molecular mechanism by which Cbl-b inhibits IgE-mediated mast cell activation remains to be defined.
Signaling pathways of Cbl-b in B cells
Direct target molecules of Cbl-b such as tyrosine kinase Syk, phospholipase C-gamma2 (PLC-γ2), p85/PI3K, Rho-family GTP-GDP exchange factor Vav, and growth-factor receptor-bound protein-2 (Grb2) are involved in BCR signaling during the normal response course.83 Syk and its substrate BLNK (also called SLP65) and Cbl-interacting protein of 85 kDa (CIN85) are key components of the BCR-associated primary transducer module required for the onset and progression phases of BCR signal transduction. Syk-mediated complex formation consisting of Vav, Btk, BLNK, and PLC-γ2 is required for effective downstream signaling including MAPKs, Akt, Ca2+ influx, and NF-κB activation.83 CIN85 also interacts with SH2-containing inositol phosphatase 1 (SHIP-1), an inositol 5-phosphatase expressed in hemopoietic cells, which acts by hydrolysing the 5-phosphates from PtdIns(3,4,5)P(3) and Ins(1,3,4,5)P(4), thereby negatively regulating the PI3K pathway.84 Thus, multiple signaling pathways of Cbl-b are coordinating in response to BCR stimulation.
Studies using Cblb−/− mice have yielded more definitive results that support the notion that Cbl-b is a negative regulator of BCR signaling.6 Cblb−/− B cells display sustained phosphorylation of Igα, Syk, and PLC-γ2 in response to BCR stimulation, which leads to prolonged Ca2+ mobilization and increases extracellular signal-regulated kinase (ERK) and JNK phosphorylation, and surface expression of the activation marker, CD69.6 This heightened BCR signaling is possibly mediated by ubiquitination and proteasomal degradation of Syk and Igα by Cbl-b. In accordance with these data, B cell-specific ablation of both c-Cbl and Cbl-b (Cbl−/−Cblb−/−) results in enhanced tyrosine phosphorylation of Syk, PLC-γ2, and Vav, and Ca2+ mobilization and substantial attenuation of tyrosine phosphorylation of adaptor protein BLNK.7
Cbl-b is also involved in the germinal center formation. Loss of Cbl-b restores Ig class switching and germinal center formation in Vav1 mutant mice in response to an in vivo viral challenge.85 Genetic inactivation of Cbl-b also rescues impaired antiviral IgG production rather than germinal center formation in Cd28−/− mice.85 It has been shown that Grb2 is degraded in a Cbl-b-dependent fashion and plays an important role in germinal center formation in the spleen.55 Ablation of Grb2 in B cells results in enhanced BCR signaling, and Grb2−/− B cells do not form germinal centers in the spleen after antigen stimulation.86 Therefore, it is assumed that the Cbl-b/Grb2 signaling pathway might play an important role in germinal center formation. In addition, we have previously shown that Cblb−/− mice display enhanced thymus-dependent antibody responses and germinal center formation, whereas introduction of CD40 deficiency abolishes these effects.87 Cbl-b selectively downmodulates CD40-induced activation of NF-κB and JNK. Cbl-b associates with TRAF-2 upon CD40 ligation and inhibits the recruitment of TRAF-2 to CD40. These data suggest that Cbl-b attenuates CD40-mediated NF-κB and JNK activation, thereby suppressing B-cell responses.87
Signaling pathways of Cbl-b in T cells
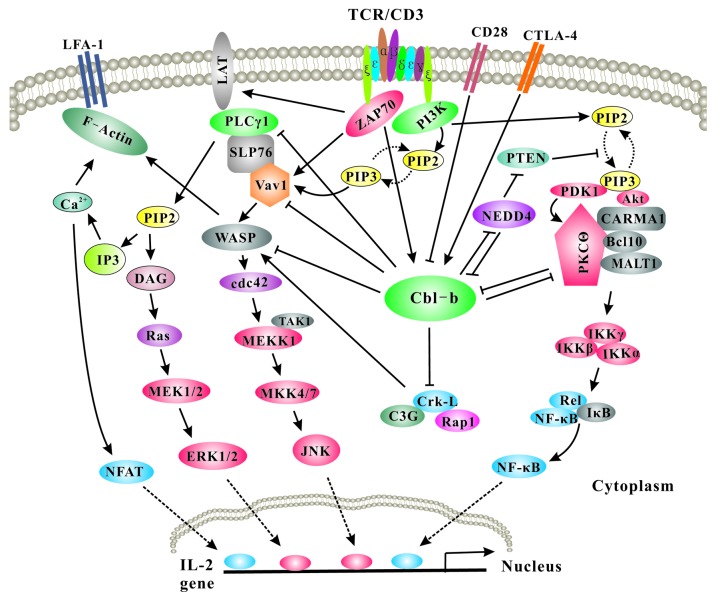
Cbl-b acts as a gatekeeper that prevents excessive T-cell activation initiated by the engagement of TCR, thus setting the threshold for T-cell activation and regulating peripheral T-cell tolerance. The signaling pathways that are regulated by Cbl-b in T cells have been more extensively studied than those of other immune cells (Fig. 3).
Figure 3. Model of Cbl-b action on T-cell activation Upon TCR stimulation, Pten is inactivated via Nedd4, which targets Pten for K63-linked polyubiquitination, and this process is inhibited by Cbl-b. Inactivation of Pten leads to the accumulation of PtdIns(3,4,5)P3, which recruits PDK-1, Vav-1, and Akt to the plasma membrane via its interaction with the PH domains within these molecules. Therefore, Cbl-b inhibits Vav-dependent activation of WASP, which leads to actin reorganization and TCR clustering. In addition, Vav1 links PKC-θ to PDK-1, the former coupling IKKs, to the CBM complex. Activated Akt also facilitates the formation of the CBM complex possibly by phosphorylating CARMA1. Thus, Cbl-b inhibits NF-κB activation via PKC-θ and Akt. One of the important outcomes for Akt is that Akt can phosphorylates Foxo1/3a, which excludes them from the nucleus, thus inhibiting Foxp3 expression. In anergic T cells, Cbl-b targets PLC-γ1 and PKC-θ for ubiquitination, thus promoting T-cell anergy induction. The expression of Cbl-b in T cells is controlled by CD28 and CTLA-4. CD28 costimulation induces Cbl-b ubiquitination and proteasomal degradation, which is possibly mediated by Nedd4 and PKC-θ. In contrast, CTLA-4-B7 interaction induces Cbl-b expression.
In naϊve T cells, Cbl-b regulates CD28-dependent T-cell activation by selectively restraining the TCR-mediated Vav1–Wiscott Aldrich syndrome protein (WASP) signaling pathway.4,88-90 Loss of Cbl-b in T cells frees TCR-triggered receptor clustering, lipid raft aggregation, and sustained tyrosine phosphorylation from the requirement for CD28 costimulation. The Rho family GDP/GTP exchange factor, Vav1, GTPases Rac1, CDC42, and RhoA, and CDC42-associated WASP constitute a signaling pathway that links antigen receptor engagement to cytoskeletal reorganization, receptor clustering and cap formation, and effective T-cell activation.91 Vav1 is optimally tyrosine phosphorylated by co-stimulation of TCR/CD28. Loss of Cbl-b results in hyper-activation of Vav1 upon TCR stimulation and uncouples the requirement for optimal Vav activation from CD28 costimulation. Further study suggests that Cbl-b suppresses the activation of Vav, thus attenuating the extent of actin reorganization and TCR clustering via a CDC42/WASP-dependent mechanism.92 Introduction of the Cbl-b deficiency into a Vav1−/− background relieves the functional defects of Vav1−/− T cells and causes spontaneous autoimmunity.90 In further support of the notion that Cbl-b regulates the CD28 dependence of T-cell activation, T cells deficient for Cbl-b do not require CD28 costimulation for T-cell proliferation and IL-2 production, and Cbl-b deficiency fully restores defective T-cell proliferation, IL-2 production, and T cell-dependent antibody responses in Cd28−/− mice.88 WASP is a key regulator of actin dynamics during cell motility and adhesion. The studies using Cblb−/−Wasp−/− and Cblb−/−Vav1−/−Wasp−/− mice reveal that WASP deficiency abrogates hyper-T-cell responses and TCR clustering.90 WASP phosphorylation at tyrosine 291 results in recruitment of Cbl-b, which, together with c-Cbl, ubiquitinates WASP at lysine residues 76 and 81, located at the WASP WH1 domain. Disruption of WASP ubiquitination causes WASP accumulation and alters actin dynamics and the formation of actin-dependent structures.93 Taken together, these data suggest that Cbl-b negatively regulates the Vav–WASP signaling pathway downstream of CD28 but upstream or at the level of WASP. In addition to deregulated actin reorganization and TCR clustering, the loss of Cbl-b selectively results in aberrant activation of NF-κB upon TCR ligation, which is mediated by Akt and PKC-θ 94.
In an effort to define the molecular mediator(s) that regulates Vav activation in T cells, Cbl-b was suggested to promote the ubiquitination of p85, the regulatory subunit of PI3K, through an interaction with the C-terminal PR domain, resulting in the inhibition of the binding of p85 to TCRζ and CD28, thus attenuating the activation of the downstream targets Vav and Akt.57 However, this finding, although well-cited, has not been independently verified by other investigators. Rather, our recent study revealed that Cbl-b does not regulate PI3K but rather inhibits the ubiquitin ligase activity of Nedd4, which targets Pten for K63-linked polyubiquitination, thus suppressing inactivation of Pten. Cbl-b may exert its effect on Pten by impeding the binding of Pten to Nedd4, which is independent of its E3 ubiquitin ligase activity.12
Cbl-b also plays a negative role in Crk-L-C3G-mediated Rap1 and LFA-1 activation in T cells. Cbl-b affects the association between Crk-L and C3G, rather than the stability of Crk-L by ubiquitinating Crk-L. In Cblb−/− T cells, the interaction between Crk-L and C3G, and the activity of the small GTPase Rap1, are increased. Cblb−/− T cells also display increased adhesion and cell surface binding to ICAM-1 by the enhanced clustering of LFA-1 in response to TCR stimulation.95 By contrast, ICOS upregulation, germinal center (GC) formation, and production of IFN-γ and IL-4 are under the control of signaling pathways independent of Cbl-b-regulated Vav1 activity.85
In addition to the above signaling pathways regulated by Cbl-b in primary naïve T cells, Cbl-b also ubiquitinates PLCγ 1 and PKC-θ in anergic T cells, attenuating the activation of PLCγ 1 and PKC-θ, which suppresses calcium mobilization and the activation of transcription factors that lead to IL-2 production.59,69,70 Therefore, Cbl-b appears to be crucial for the induction of T-cell anergy which we will discuss below.
Roles of Cbl-b in Immune-Related Diseases
Cbl-b in tolerance induction
E3 ubiquitin ligase Cbl-b is involved in maintaining a balance between immunity and tolerance by functioning as a gatekeeper.88,89 It has been demonstrated that CD28 and CTLA-4 may regulate the threshold for T-cell activation by controlling Cbl-b expression.67,68 In support of this notion, Cbl-b has been shown to be a key mediator involved in T-cell anergy induction in vitro and in vivo.70,96 In addition, CD4+CD25− effector T cells from Cblb−/− mice are resistant to TGF-β, Cblb−/− and wild-type CD4+CD25+ regulatory T cells.97 Furthermore, Cbl-b has been shown to facilitate the conversion of naïve CD4+CD25− T cells into inducible CD4+CD25+Foxp3+ T cells (iTregs) via a Foxo1/3a-dependent mechanism.98 Using both in vitro and in vivo approaches, we demonstrated that the T-cell activation threshold regulated by Cbl-b determines the fate of iTregs, and that this process is mediated by an Akt-2-dependent mechanism.99 These results suggest that Cbl-b regulates peripheral T-cell tolerance by multiple mechanisms.
Cbl-b in autoimmunity and allergic airway inflammation
Cbl-b has been implicated in various diseases in a range of animal models. Cblb−/− mice,89 Cbl-b RF mutant mice,20 C-terminal-truncated Cbl-b in rats,100 and c-Cbl/Cbl-b double mutant mice (Cbl−/−Cblb−/−) mice4 all develop spontaneous autoimmunity or are highly susceptible to experimental autoimmune encephalomyelitis (EAE) (a model of a human demyelinating disease, multiple sclerosis [MS])101 and murine collagen-induced arthritis (CIA) (a mouse model of rheumatoid arthritis).102,103 Mice with B cell-specific Cbl−/−Cblb−/− mutations also develop a systemic lupus erythematosus (SLE)-like autoimmune disease,7 further indicating that Cbl-b is essential for promoting immune tolerance. The importance of Cbl-b in peripheral T-cell tolerance is further supported by the fact that Cbl-b deficiency exacerbates disease development (exocrine pancreatitis) in mice deficient for AIRE (autoimmune regulator), which is essential for clonal deletion in the thymus.104 In further support of this notion, Cbl-b deficiency subsequently precipitates type 1 diabetes in most 3A9 TCR:insHEL double transgenic mice.105 In a mouse model of allergic asthma, we recently found that Cblb−/− mice display increased airway inflammation upon OVA/alum immunization, which is due to aberrant Th2 and Th9 responses. At the molecular level, Cbl-b was found to target Stat6, a transcription factor involved in both Th2 and Th9 cell differentiation, for ubiquitination and proteasomal degradation.106
Prominent autoimmune phenotypes in mice with Cbl-b (or Cbl plus Cbl-b) deletion have prompted analyses of polymorphisms/mutations of Cbl-b in animal models and human patients with autoimmune diseases. Polymorphisms of Cbl-b have been found in some autoimmune diseases, such as rat type 1 diabetes (T1D),100,107,108 human MS,109 SLE,110 and asthma.111 A nonsense mutation in Cbl-b has been identified from the Komeda diabetes-prone (KDP) rat, and wild-type Cbl-b significantly suppresses development of the KDP phenotype.100,107 Furthermore, one SNP in exon 12 of the Cbl-b gene was significantly demonstrated to be associated with T1D in a large Danish T1D study of 480 families,108 although further verification should be performed using large, well-characterized populations. A recent genome-wide associated study (GWAS) indicated an association of CBLB gene variants with MS, which was confirmed in 1775 cases and 2005 controls.109 These data together with data that mice lacking the ortholog are prone to EAE88 strongly support the involvement of Cbl-b in MS development. Consistent with this finding, a significant association between the 2126(A/G) SNP of Cbl-b gene and SLE was detected,110 suggesting that Cbl-b may contribute to the deregulated activation of T lymphocytes observed in SLE. A Cbl-b D454A variant associated asthma was found the in asthmatic children by whole-exome sequencing.111
Cbl-b, a target for tumor immunotherapy
Although genetic inactivation of Cbl-b clearly has detrimental consequences, e.g., sensitizing the mice to develop autoimmunity, these mice do have the enviable ability to spontaneously reject various types of solid and hematopoietic tumors and viruses. Cbl-b deficiency in mice elicits an efficient and spontaneous rejection of xenografted TC1, EL4, and E.G7 tumorigenic cell lines,92,112 and shows a markedly lower incidence of skin cancer than the wild-type control cohort upon chronic exposure to UV-B light.112 In support of these observations, Cblb−/− mice crossed to an ataxia telangiectasia mutated-deficient background (Atm−/−) exhibits a significantly reduced incidence and delayed onset of spontaneous T-cell lymphomas compared with Cblb+/+Atm−/− controls.92 The enhanced anti-tumor immunity in Cblb−/− mice has been ascribed to increased activity of CD8+ T cells.113 Indeed, transfer of siRNA Cbl-b-silenced CD8+ T lymphocytes augments tumor vaccine efficacy in a B16 melanoma model.114,115 Thus, abrogating Cbl-b expression in effector T cells may improve the efficacy of adoptive therapy of some human malignancies. The recent report that an inhibitory role of Cbl-b on rejecting tumor metastases of NK cell functions would give rise to potential therapeutic effect specific for Cbl-b to tumor metastases.80
Perspective
Although the roles of Cbl-b in adaptive immunity have been extensively studied, the involvement of Cbl-b in innate immunity and infection has only recently been appreciated. Studies using various animal models of immune diseases will unveil the potential cellular and molecular mechanisms for Cbl-b in these disease processes, and determine whether Cbl-b is a drug target for the treatment of immune-related diseases, such as autoimmune/inflammatory diseases, infectious diseases and tumors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We apologize to those colleagues whose work was not cited due to space constraints. This work is supported by the Special Funds for the Major State Basic Research Program of China (973 Program) (2009CB521702) and a grant from the National Institutes of Health (R01AI090901 to J.Z.).
Glossary
Abbreviations:
- Cbl-b
Casitas B-lineage lymphoma proto-oncogene-b
- TCR
T-cell receptor
- BCR
B-cell receptor
- FcεR
high affinity immunoglobulin epsilon receptor
- TKB
protein tyrosine-kinase-binding
- RF
really interesting new gene finger
- PTB
phosphotyrosine-binding
- PTKs
protein tyrosine kinases
- GEFs
guanine nucleotide exchange factors
- PI3K
phosphoinositide 3-kinase
- BM
bone-marrow
- Nedd4
neuronal precursor of cell expressed developmentally down-regulated gene 4
- UBA
ubiquitin associated region
- NK
natural killer
- TLR
Toll-like receptor
- LFA-1
β2-integrin leukocyte function-associated antigen-1
- BMDMPs
bone marrow-derived mononuclear phagocytes
- PLC-γ2
phospholipase C-gamma2
- Grb2
growth-factor receptor-bound protein-2
- CIN85
Cbl-interacting protein of 85 kDa
- SHIP-1
SH2-containing inositol phosphatase
- 1 WASP
Wiscott Aldrich Syndrome protein
- iTregs
inducible CD4+CD25+Foxp3+ T cells
- EAE
experimental autoimmune encephalomyelitis
- MS
multiple sclerosis
- CIA
murine collagen-induced arthritis
- SLE
systemic lupus erythematosus
- AIRE
autoimmune regulator
- T1D
type 1 diabetes
- GWAS
genome-wide associated study
Reference
- 1.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–7. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 2.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 3.Keane MM, Rivero-Lezcano OM, Mitchell JA, Robbins KC, Lipkowitz S. Cloning and characterization of cbl-b: a SH3 binding protein with homology to the c-cbl proto-oncogene. Oncogene. 1995;10:2367–77. [PubMed] [Google Scholar]
- 4.Naramura M, Jang IK, Kole H, Huang F, Haines D, Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat Immunol. 2002;3:1192–9. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 5.Shamim M, Nanjappa SG, Singh A, Plisch EH, LeBlanc SE, Walent J, Svaren J, Seroogy C, Suresh M. Cbl-b regulates antigen-induced TCR down-regulation and IFN-gamma production by effector CD8 T cells without affecting functional avidity. J Immunol. 2007;179:7233–43. doi: 10.4049/jimmunol.179.11.7233. [DOI] [PubMed] [Google Scholar]
- 6.Sohn HW, Gu H, Pierce SK. Cbl-b negatively regulates B cell antigen receptor signaling in mature B cells through ubiquitination of the tyrosine kinase Syk. J Exp Med. 2003;197:1511–24. doi: 10.1084/jem.20021686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitaura Y, Jang IK, Wang Y, Han YC, Inazu T, Cadera EJ, Schlissel M, Hardy RR, Gu H. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity. 2007;26:567–78. doi: 10.1016/j.immuni.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu X, Miah SM, Hatani T, Okazaki M, Hori-Tamura N, Yamamura H, Hotta H, Sada K. Selective inhibition of Fcepsilon RI-mediated mast cell activation by a truncated variant of Cbl-b related to the rat model of type 1 diabetes mellitus. J Biochem. 2005;137:711–20. doi: 10.1093/jb/mvi088. [DOI] [PubMed] [Google Scholar]
- 9.Bustelo XR, Crespo P, López-Barahona M, Gutkind JS, Barbacid M. Cbl-b, a member of the Sli-1/c-Cbl protein family, inhibits Vav-mediated c-Jun N-terminal kinase activation. Oncogene. 1997;15:2511–20. doi: 10.1038/sj.onc.1201430. [DOI] [PubMed] [Google Scholar]
- 10.Elly C, Witte S, Zhang Z, Rosnet O, Lipkowitz S, Altman A, Liu YC. Tyrosine phosphorylation and complex formation of Cbl-b upon T cell receptor stimulation. Oncogene. 1999;18:1147–56. doi: 10.1038/sj.onc.1202411. [DOI] [PubMed] [Google Scholar]
- 11.Liu YC, Gu H. Cbl and Cbl-b in T-cell regulation. Trends Immunol. 2002;23:140–3. doi: 10.1016/S1471-4906(01)02157-3. [DOI] [PubMed] [Google Scholar]
- 12.Guo H, Qiao G, Ying H, Li Z, Zhao Y, Liang Y, Yang L, Lipkowitz S, Penninger JM, Langdon WY, et al. E3 Ubiquitin Ligase Cbl-b Regulates Pten via Nedd4 in T Cells Independently of Its Ubiquitin Ligase Activity. Cell reports. 2012;1:472–82. doi: 10.1016/j.celrep.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Elly C, Qiu L, Altman A, Liu YC. A direct interaction between the adaptor protein Cbl-b and the kinase zap-70 induces a positive signal in T cells. Current biology: CB. 1999;9:203–6. doi: 10.1016/S0960-9822(99)80090-6. [DOI] [PubMed] [Google Scholar]
- 14.Meng W, Sawasdikosol S, Burakoff SJ, Eck MJ. Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase. Nature. 1999;398:84–90. doi: 10.1038/18050. [DOI] [PubMed] [Google Scholar]
- 15.Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, et al. Protein tyrosine kinase regulation by ubiquitination: Critical roles of Cbl-family ubiquitin ligases. Biochimica et biophysica acta. 2013;1833:122–39. doi: 10.1016/j.bbamcr.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/S0092-8674(00)00057-X. [DOI] [PubMed] [Google Scholar]
- 17.Kassenbrock CK, Anderson SM. Regulation of ubiquitin protein ligase activity in c-Cbl by phosphorylation-induced conformational change and constitutive activation by tyrosine to glutamate point mutations. J Biol Chem. 2004;279:28017–27. doi: 10.1074/jbc.M404114200. [DOI] [PubMed] [Google Scholar]
- 18.Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paolino M, Thien CB, Gruber T, Hinterleitner R, Baier G, Langdon WY, Penninger JM. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. J Immunol. 2011;186:2138–47. doi: 10.4049/jimmunol.1003390. [DOI] [PubMed] [Google Scholar]
- 21.Ryan PE, Sivadasan-Nair N, Nau MM, Nicholas S, Lipkowitz S. The N terminus of Cbl-c regulates ubiquitin ligase activity by modulating affinity for the ubiquitin-conjugating enzyme. J Biol Chem. 2010;285:23687–98. doi: 10.1074/jbc.M109.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobashigawa Y, Tomitaka A, Kumeta H, Noda NN, Yamaguchi M, Inagaki F. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci U S A. 2011;108:20579–84. doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dou H, Buetow L, Hock A, Sibbet GJ, Vousden KH, Huang DT. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–92. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–7. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 25.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–90. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 26.Kowanetz K, Szymkiewicz I, Haglund K, Kowanetz M, Husnjak K, Taylor JD, Soubeyran P, Engstrom U, Ladbury JE, Dikic I. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J Biol Chem. 2003;278:39735–46. doi: 10.1074/jbc.M304541200. [DOI] [PubMed] [Google Scholar]
- 27.Ettenberg SA, Rubinstein YR, Banerjee P, Nau MM, Keane MM, Lipkowitz S. cbl-b inhibits EGF-receptor-induced apoptosis by enhancing ubiquitination and degradation of activated receptors. Mol Cell Biol Res Commun. 1999;2:111–8. doi: 10.1006/mcbr.1999.0157. [DOI] [PubMed] [Google Scholar]
- 28.Lupher ML, Jr., Rao N, Eck MJ, Band H. The Cbl protooncoprotein: a negative regulator of immune receptor signal transduction. Immunol Today. 1999;20:375–82. doi: 10.1016/S0167-5699(99)01484-X. [DOI] [PubMed] [Google Scholar]
- 29.Rao N, Dodge I, Band H. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J Leukoc Biol. 2002;71:753–63. [PubMed] [Google Scholar]
- 30.Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- 31.Tsygankov AY, Teckchandani AM, Feshchenko EA, Swaminathan G. Beyond the RING: CBL proteins as multivalent adapters. Oncogene. 2001;20:6382–402. doi: 10.1038/sj.onc.1204781. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, DeYoung SM, Hwang JB, O’Leary EE, Saltiel AR. The roles of Cbl-b and c-Cbl in insulin-stimulated glucose transport. J Biol Chem. 2003;278:36754–62. doi: 10.1074/jbc.M300664200. [DOI] [PubMed] [Google Scholar]
- 33.Hunter S, Burton EA, Wu SC, Anderson SM. Fyn associates with Cbl and phosphorylates tyrosine 731 in Cbl, a binding site for phosphatidylinositol 3-kinase. J Biol Chem. 1999;274:2097–106. doi: 10.1074/jbc.274.4.2097. [DOI] [PubMed] [Google Scholar]
- 34.Ueno H, Sasaki K, Honda H, Nakamoto T, Yamagata T, Miyagawa K, Mitani K, Yazaki Y, Hirai H. c-Cbl is tyrosine-phosphorylated by interleukin-4 and enhances mitogenic and survival signals of interleukin-4 receptor by linking with the phosphatidylinositol 3′-kinase pathway. Blood. 1998;91:46–53. [PubMed] [Google Scholar]
- 35.Arron JR, Vologodskaia M, Wong BR, Naramura M, Kim N, Gu H, Choi Y. A positive regulatory role for Cbl family proteins in tumor necrosis factor-related activation-induced cytokine (trance) and CD40L-mediated Akt activation. J Biol Chem. 2001;276:30011–7. doi: 10.1074/jbc.M100414200. [DOI] [PubMed] [Google Scholar]
- 36.Thien CB, Langdon WY. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J. 2005;391:153–66. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18:341–52. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Oved S, Mosesson Y, Zwang Y, Santonico E, Shtiegman K, Marmor MD, Kochupurakkal BS, Katz M, Lavi S, Cesareni G, et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J Biol Chem. 2006;281:21640–51. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt MH, Dikic I. Ubiquitin and NEDD8: brothers in arms. Science's STKE: signal transduction knowledge environment. 2006;2006:pe50. doi: 10.1126/stke.3622006pe50. [DOI] [PubMed] [Google Scholar]
- 40.Peschard P, Kozlov G, Lin T, Mirza IA, Berghuis AM, Lipkowitz S, Park M, Gehring K. Structural basis for ubiquitin-mediated dimerization and activation of the ubiquitin protein ligase Cbl-b. Mol Cell. 2007;27:474–85. doi: 10.1016/j.molcel.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 41.Davies GC, Ettenberg SA, Coats AO, Mussante M, Ravichandran S, Collins J, Nau MM, Lipkowitz S. Cbl-b interacts with ubiquitinated proteins; differential functions of the UBA domains of c-Cbl and Cbl-b. Oncogene. 2004;23:7104–15. doi: 10.1038/sj.onc.1207952. [DOI] [PubMed] [Google Scholar]
- 42.Zhou ZR, Gao HC, Zhou CJ, Chang YG, Hong J, Song AX, Lin DH, Hu HY. Differential ubiquitin binding of the UBA domains from human c-Cbl and Cbl-b: NMR structural and biochemical insights. Protein science: a publication of the Protein Society. 2008;17:1805–14. doi: 10.1110/ps.036384.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–8. doi: 10.1016/S0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 44.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, Kolaczkowska A, Devaux F, Panwar SL, Hallstrom TC, Jacq C, Moye-Rowley WS. Transcriptional regulation by Lge1p requires a function independent of its role in histone H2B ubiquitination. J Biol Chem. 2005;280:2759–70. doi: 10.1074/jbc.M408333200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–40. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 49.Fan Y, Xie P, Zhang T, Zhang H, Gu D, She M, Li H. Regulation of the stability and transcriptional activity of NFATc4 by ubiquitination. FEBS Lett. 2008;582:4008–14. doi: 10.1016/j.febslet.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 50.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 51.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–62. doi: 10.1016/S0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 52.Woelk T, Sigismund S, Penengo L, Polo S. The ubiquitination code: a signalling problem. Cell Div. 2007;2:11. doi: 10.1186/1747-1028-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng S, Xu Z, Lipkowitz S, Longley JB. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005;105:226–32. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- 55.Ettenberg SA, Magnifico A, Cuello M, Nau MM, Rubinstein YR, Yarden Y, Weissman AM, Lipkowitz S. Cbl-b-dependent coordinated degradation of the epidermal growth factor receptor signaling complex. J Biol Chem. 2001;276:27677–84. doi: 10.1074/jbc.M102641200. [DOI] [PubMed] [Google Scholar]
- 56.Ettenberg SA, Keane MM, Nau MM, Frankel M, Wang LM, Pierce JH, Lipkowitz S. cbl-b inhibits epidermal growth factor receptor signaling. Oncogene. 1999;18:1855–66. doi: 10.1038/sj.onc.1202499. [DOI] [PubMed] [Google Scholar]
- 57.Fang D, Wang HY, Fang N, Altman Y, Elly C, Liu YC. Cbl-b, a RING-type E3 ubiquitin ligase, targets phosphatidylinositol 3-kinase for ubiquitination in T cells. J Biol Chem. 2001;276:4872–8. doi: 10.1074/jbc.M008901200. [DOI] [PubMed] [Google Scholar]
- 58.Yasuda T, Tezuka T, Maeda A, Inazu T, Yamanashi Y, Gu H, Kurosaki T, Yamamoto T. Cbl-b positively regulates Btk-mediated activation of phospholipase C-gamma2 in B cells. J Exp Med. 2002;196:51–63. doi: 10.1084/jem.20020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gruber T, Hermann-Kleiter N, Hinterleitner R, Fresser F, Schneider R, Gastl G, Penninger JM, Baier G. PKC-theta modulates the strength of T cell responses by targeting Cbl-b for ubiquitination and degradation. Sci Signal. 2009;2:ra30. doi: 10.1126/scisignal.2000046. [DOI] [PubMed] [Google Scholar]
- 60.Sattler M, Pride YB, Quinnan LR, Verma S, Malouf NA, Husson H, Salgia R, Lipkowitz S, Griffin JD. Differential expression and signaling of CBL and CBL-B in BCR/ABL transformed cells. Oncogene. 2002;21:1423–33. doi: 10.1038/sj.onc.1205202. [DOI] [PubMed] [Google Scholar]
- 61.Oshikawa G, Nagao T, Wu N, Kurosu T, Miura O. c-Cbl and Cbl-b ligases mediate 17-allylaminodemethoxygeldanamycin-induced degradation of autophosphorylated Flt3 kinase with internal tandem duplication through the ubiquitin proteasome pathway. J Biol Chem. 2011;286:30263–73. doi: 10.1074/jbc.M111.232348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–48. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 63.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 64.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Körner R, Greff Z, Kéri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–48. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 65.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heibeck TH, Ding SJ, Opresko LK, Zhao R, Schepmoes AA, Yang F, Tolmachev AV, Monroe ME, Camp DG, 2nd, Smith RD, et al. An extensive survey of tyrosine phosphorylation revealing new sites in human mammary epithelial cells. J Proteome Res. 2009;8:3852–61. doi: 10.1021/pr900044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Gál I, Vermes C, Alegre ML, Chong AS, Chen L, Shao Q, Adarichev V, Xu X, Koreny T, et al. Cutting edge: Cbl-b: one of the key molecules tuning CD28- and CTLA-4-mediated T cell costimulation. J Immunol. 2004;173:7135–9. doi: 10.4049/jimmunol.173.12.7135. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Bárdos T, Li D, Gál I, Vermes C, Xu J, Mikecz K, Finnegan A, Lipkowitz S, Glant TT. Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J Immunol. 2002;169:2236–40. doi: 10.4049/jimmunol.169.5.2236. [DOI] [PubMed] [Google Scholar]
- 69.Yang B, Gay DL, MacLeod MK, Cao X, Hala T, Sweezer EM, Kappler J, Marrack P, Oliver PM. Nedd4 augments the adaptive immune response by promoting ubiquitin-mediated degradation of Cbl-b in activated T cells. Nat Immunol. 2008;9:1356–63. doi: 10.1038/ni.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, et al. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167–77. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 71.Szymkiewicz I, Kowanetz K, Soubeyran P, Dinarina A, Lipkowitz S, Dikic I. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J Biol Chem. 2002;277:39666–72. doi: 10.1074/jbc.M205535200. [DOI] [PubMed] [Google Scholar]
- 72.Jozic D, Cárdenes N, Deribe YL, Moncalián G, Hoeller D, Groemping Y, Dikic I, Rittinger K, Bravo J. Cbl promotes clustering of endocytic adaptor proteins. Nat Struct Mol Biol. 2005;12:972–9. doi: 10.1038/nsmb1000. [DOI] [PubMed] [Google Scholar]
- 73.Moncalián G, Cárdenes N, Deribe YL, Spínola-Amilibia M, Dikic I, Bravo J. Atypical polyproline recognition by the CMS N-terminal Src homology 3 domain. J Biol Chem. 2006;281:38845–53. doi: 10.1074/jbc.M606411200. [DOI] [PubMed] [Google Scholar]
- 74.Lowell CA. Src-family and Syk kinases in activating and inhibitory pathways in innate immune cells: signaling cross talk. Cold Spring Harb Perspect Biol. 2011;3:a002352. doi: 10.1101/cshperspect.a002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi EY, Orlova VV, Fagerholm SC, Nurmi SM, Zhang L, Ballantyne CM, Gahmberg CG, Chavakis T. Regulation of LFA-1-dependent inflammatory cell recruitment by Cbl-b and 14-3-3 proteins. Blood. 2008;111:3607–14. doi: 10.1182/blood-2007-07-103077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bachmaier K, Toya S, Gao X, Triantafillou T, Garrean S, Park GY, Frey RS, Vogel S, Minshall R, Christman JW, et al. E3 ubiquitin ligase Cblb regulates the acute inflammatory response underlying lung injury. Nat Med. 2007;13:920–6. doi: 10.1038/nm1607. [DOI] [PubMed] [Google Scholar]
- 77.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734–42. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 78.Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nat Immunol. 2009;10:744–52. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- 79.Kojo S, Elly C, Harada Y, Langdon WY, Kronenberg M, Liu YC. Mechanisms of NKT cell anergy induction involve Cbl-b-promoted monoubiquitination of CARMA1. Proc Natl Acad Sci U S A. 2009;106:17847–51. doi: 10.1073/pnas.0904078106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, Jamieson AM, Langdon WY, Ikeda F, Fededa JP, et al. The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature. 2014;507:508–12. doi: 10.1038/nature12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qu X, Sada K, Kyo S, Maeno K, Miah SM, Yamamura H. Negative regulation of FcepsilonRI-mediated mast cell activation by a ubiquitin-protein ligase Cbl-b. Blood. 2004;103:1779–86. doi: 10.1182/blood-2003-07-2260. [DOI] [PubMed] [Google Scholar]
- 82.Hirasaka K, Kohno S, Goto J, Furochi H, Mawatari K, Harada N, Hosaka T, Nakaya Y, Ishidoh K, Obata T, et al. Deficiency of Cbl-b gene enhances infiltration and activation of macrophages in adipose tissue and causes peripheral insulin resistance in mice. Diabetes. 2007;56:2511–22. doi: 10.2337/db06-1768. [DOI] [PubMed] [Google Scholar]
- 83.Ying H, Li Z, Yang L, Zhang J. Syk mediates BCR- and CD40-signaling integration during B cell activation. Immunobiology. 2011;216:566–70. doi: 10.1016/j.imbio.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buchse T, Horras N, Lenfert E, Krystal G, Korbel S, Schumann M, Krause E, Mikkat S, Tiedge M. CIN85 interacting proteins in B cells-specific role for SHIP-1. Molecular & cellular proteomics: MCP. 2011;10:M110 006239. doi: 10.1074/mcp.M110.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krawczyk CM, Jones RG, Atfield A, Bachmaier K, Arya S, Odermatt B, Ohashi PS, Penninger JM. Differential control of CD28-regulated in vivo immunity by the E3 ligase Cbl-b. J Immunol. 2005;174:1472–8. doi: 10.4049/jimmunol.174.3.1472. [DOI] [PubMed] [Google Scholar]
- 86.Jang IK, Cronshaw DG, Xie LK, Fang G, Zhang J, Oh H, Fu YX, Gu H, Zou Y. Growth-factor receptor-bound protein-2 (Grb2) signaling in B cells controls lymphoid follicle organization and germinal center reaction. Proc Natl Acad Sci U S A. 2011;108:7926–31. doi: 10.1073/pnas.1016451108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiao G, Lei M, Li Z, Sun Y, Minto A, Fu YX, Ying H, Quigg RJ, Zhang J. Negative regulation of CD40-mediated B cell responses by E3 ubiquitin ligase Casitas-B-lineage lymphoma protein-B. J Immunol. 2007;179:4473–9. doi: 10.4049/jimmunol.179.7.4473. [DOI] [PubMed] [Google Scholar]
- 88.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ, Jang IK, Gutkind JS, Shevach E, Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–20. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 89.Bachmaier K, Krawczyk C, Kozieradzki I, Kong YY, Sasaki T, Oliveira-dos-Santos A, Mariathasan S, Bouchard D, Wakeham A, Itie A, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–6. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 90.Krawczyk C, Bachmaier K, Sasaki T, Jones RG, Snapper SB, Bouchard D, Kozieradzki I, Ohashi PS, Alt FW, Penninger JM. Cbl-b is a negative regulator of receptor clustering and raft aggregation in T cells. Immunity. 2000;13:463–73. doi: 10.1016/S1074-7613(00)00046-7. [DOI] [PubMed] [Google Scholar]
- 91.Snapper SB, Rosen FS, Mizoguchi E, Cohen P, Khan W, Liu CH, Hagemann TL, Kwan SP, Ferrini R, Davidson L, et al. Wiskott-Aldrich syndrome protein-deficient mice reveal a role for WASP in T but not B cell activation. Immunity. 1998;9:81–91. doi: 10.1016/S1074-7613(00)80590-7. [DOI] [PubMed] [Google Scholar]
- 92.Chiang JY, Jang IK, Hodes R, Gu H. Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J Clin Invest. 2007;117:1029–36. doi: 10.1172/JCI29472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reicher B, Joseph N, David A, Pauker MH, Perl O, Barda-Saad M. Ubiquitylation-dependent negative regulation of WASp is essential for actin cytoskeleton dynamics. Mol Cell Biol. 2012;32:3153–63. doi: 10.1128/MCB.00161-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qiao G, Li Z, Molinero L, Alegre ML, Ying H, Sun Z, Penninger JM, Zhang J. T-cell receptor-induced NF-kappaB activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol Cell Biol. 2008;28:2470–80. doi: 10.1128/MCB.01505-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang W, Shao Y, Fang D, Huang J, Jeon MS, Liu YC. Negative regulation of T cell antigen receptor-mediated Crk-L-C3G signaling and cell adhesion by Cbl-b. J Biol Chem. 2003;278:23978–83. doi: 10.1074/jbc.M212671200. [DOI] [PubMed] [Google Scholar]
- 96.Heissmeyer V, Macián F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255–65. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 97.Wohlfert EA, Callahan MK, Clark RB. Resistance to CD4+CD25+ regulatory T cells and TGF-beta in Cbl-b-/- mice. J Immunol. 2004;173:1059–65. doi: 10.4049/jimmunol.173.2.1059. [DOI] [PubMed] [Google Scholar]
- 98.Harada Y, Harada Y, Elly C, Ying G, Paik JH, DePinho RA, Liu YC. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207:1381–91. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qiao G, Zhao Y, Li Z, Tang PQ, Langdon WY, Yang T, Zhang J. T cell activation threshold regulated by E3 ubiquitin ligase Cbl-b determines fate of inducible regulatory T cells. J Immunol. 2013;191:632–9. doi: 10.4049/jimmunol.1202068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yokoi N, Komeda K, Wang HY, Yano H, Kitada K, Saitoh Y, Seino Y, Yasuda K, Serikawa T, Seino S. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet. 2002;31:391–4. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
- 101.Zamvil SS, Steinman L. The T lymphocyte in experimental allergic encephalomyelitis. Annu Rev Immunol. 1990;8:579–621. doi: 10.1146/annurev.iy.08.040190.003051. [DOI] [PubMed] [Google Scholar]
- 102.Kim WU, Lee WK, Ryoo JW, Kim SH, Kim J, Youn J, Min SY, Bae EY, Hwang SY, Park SH, et al. Suppression of collagen-induced arthritis by single administration of poly(lactic-co-glycolic acid) nanoparticles entrapping type II collagen: a novel treatment strategy for induction of oral tolerance. Arthritis Rheum. 2002;46:1109–20. doi: 10.1002/art.10198. [DOI] [PubMed] [Google Scholar]
- 103.Park MJ, Park HS, Oh HJ, Lim JY, Yoon BY, Kim HY, Cho ML, Cho SG. IL-17-deficient allogeneic bone marrow transplantation prevents the induction of collagen-induced arthritis in DBA/1J mice. Exp Mol Med. 2012;44:694–705. doi: 10.3858/emm.2012.44.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Teh CE, Daley SR, Enders A, Goodnow CC. T-cell regulation by casitas B-lineage lymphoma (Cblb) is a critical failsafe against autoimmune disease due to autoimmune regulator (Aire) deficiency. Proc Natl Acad Sci U S A. 2010;107:14709–14. doi: 10.1073/pnas.1009209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hoyne GF, Flening E, Yabas M, Teh C, Altin JA, Randall K, Thien CB, Langdon WY, Goodnow CC. Visualizing the role of Cbl-b in control of islet-reactive CD4 T cells and susceptibility to type 1 diabetes. J Immunol. 2011;186:2024–32. doi: 10.4049/jimmunol.1002296. [DOI] [PubMed] [Google Scholar]
- 106.Qiao G, Ying H, Zhao Y, Liang Y, Guo H, Shen H, Li Z, Solway J, Tao E, Chiang YJ, et al. E3 Ubiquitin Ligase Cbl-b Suppresses Proallergic T Cell Development and Allergic Airway Inflammation. Cell reports. 2014;6:709–23. doi: 10.1016/j.celrep.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yokoi N, Hayashi C, Fujiwara Y, Wang HY, Seino S. Genetic reconstitution of autoimmune type 1 diabetes with two major susceptibility genes in the rat. Diabetes. 2007;56:506–12. doi: 10.2337/db06-1027. [DOI] [PubMed] [Google Scholar]
- 108.Bergholdt R, Taxvig C, Eising S, Nerup J, Pociot F. CBLB variants in type 1 diabetes and their genetic interaction with CTLA4. J Leukoc Biol. 2005;77:579–85. doi: 10.1189/jlb.0904524. [DOI] [PubMed] [Google Scholar]
- 109.Sanna S, Pitzalis M, Zoledziewska M, Zara I, Sidore C, Murru R, Whalen MB, Busonero F, Maschio A, Costa G, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42:495–7. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Doníz-Padilla L, Martínez-Jiménez V, Niño-Moreno P, Abud-Mendoza C, Hernández-Castro B, González-Amaro R, Layseca-Espinosa E, Baranda-Cándido L. Expression and function of Cbl-b in T cells from patients with systemic lupus erythematosus, and detection of the 2126 A/G Cblb gene polymorphism in the Mexican mestizo population. Lupus. 2011;20:628–35. doi: 10.1177/0961203310394896. [DOI] [PubMed] [Google Scholar]
- 111.DeWan AT, Egan KB, Hellenbrand K, Sorrentino K, Pizzoferrato N, Walsh KM, Bracken MB. Whole-exome sequencing of a pedigree segregating asthma. BMC Med Genet. 2012;13:95. doi: 10.1186/1471-2350-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Loeser S, Loser K, Bijker MS, Rangachari M, van der Burg SH, Wada T, Beissert S, Melief CJ, Penninger JM. Spontaneous tumor rejection by cbl-b-deficient CD8+ T cells. J Exp Med. 2007;204:879–91. doi: 10.1084/jem.20061699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stromnes IM, Blattman JN, Tan X, Jeevanjee S, Gu H, Greenberg PD. Abrogating Cbl-b in effector CD8(+) T cells improves the efficacy of adoptive therapy of leukemia in mice. J Clin Invest. 2010;120:3722–34. doi: 10.1172/JCI41991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hinterleitner R, Gruber T, Pfeifhofer-Obermair C, Lutz-Nicoladoni C, Tzankov A, Schuster M, Penninger JM, Loibner H, Lametschwandtner G, Wolf D, et al. Adoptive transfer of siRNA Cblb-silenced CD8+ T lymphocytes augments tumor vaccine efficacy in a B16 melanoma model. PLoS One. 2012;7:e44295. doi: 10.1371/journal.pone.0044295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lutz-Nicoladoni C, Wallner S, Stoitzner P, Pircher M, Gruber T, Wolf AM, Gastl G, Penninger JM, Baier G, Wolf D. Reinforcement of cancer immunotherapy by adoptive transfer of cblb-deficient CD8+ T cells combined with a DC vaccine. Immunol Cell Biol. 2012;90:130–4. doi: 10.1038/icb.2011.11. [DOI] [PubMed] [Google Scholar]