Abstract
Death receptor (DR) ligation can lead to divergent signaling pathways causing either caspase-mediated cell death or cell proliferation and inflammation. These variations in cellular fate are determined by adaptor proteins that are recruited to the DR signaling complex. FLICE inhibitory protein (FLIP) is an established inhibitor of caspase-8-mediated apoptosis, and it is also involved in NF-κB activation. However, the molecular mechanism that regulates FLIP within this complex is unknown. In this study, we provide new evidence for the regulation of NF-κB by FLIP through S-nitrosylation, which involves covalent modification of the protein’s cysteine thiol by nitric oxide to form S-nitrosothiol. Point mutations of FLIP at cysteine residues 254 and 259 prevent FLIP S-nitrosylation and its ability to activate NF-κB. The mechanism by which FLIP nitrosylation regulates NF-κB activity involves RIP1 binding and redistribution, whereas TRAF2 binding and distribution are unaffected. We further show that FLIP processing and cleavage is dependent on its nitrosylation status. Collectively, our study reveals a novel pathway for FLIP regulation of NF-κB through protein S-nitrosylation, which is a key posttranslational mechanism controlling DR-mediated cell death and survival. Since increased expression of FLIP and nitric oxide are frequently observed in chemotherapy-resistant tumors, S-nitrosylation of FLIP could be a key mechanism of chemoresistance and tumor growth.
Keywords: FLIP, NF-κB, RIP1, S-nitrosylation, cancer
Introduction
Cellular proliferation, inflammation, and apoptosis are highly regulated cellular processes important in homeostasis. Dysregulation of these processes can cause a multitude of human diseases including cardiovascular disease, autoimmune disorders, and cancer. This occurs frequently in response to aberrant signaling initiated by death receptors (DRs). DRs belong to the tumor necrosis family receptor (TNFR) superfamily, which includes TNF receptors (TNFRs), Fas, and TNF-related apoptosis inducing ligand receptors (TRAIL-Rs).1 DRs have been widely noted to play an important role in cancer development and metastasis.2 DR ligands are pleiotropic cytokines that initiate the formation of signaling complexes on the cytoplasmic tail of the receptors. Contrary to their name, DRs can signal both for cell survival and cell death.3 Receptor ligation results in receptor oligomerization and recruitment of adaptor proteins to the receptor complex, which determines the fate of the cell. Regulation of proteins that are recruited to the signaling complex is equally important in determining the outcome of DR stimulation. An example of a signaling complex that forms at the membrane is the death-inducing signaling complex (DISC), where the adaptor protein FADD and procaspase-8 are recruited, resulting in caspase activation and subsequent cell death. DR-mediated cell survival, however, is mediated by an alternate complex that involves proteins upstream of nuclear factor kappa B (NF-κB), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K) pathways.
Many cancer cells constitutively express DRs, resulting in an increased sensitivity to ligands such as Fas and TRAIL.4 This increased sensitivity was thought to provide a selective opportunity to target cancer cells for apoptosis, sparing normal cells with basal DR expression levels. This has led to the current exploration of adjuvant TRAIL therapies alongside chemotherapy in a wide variety of cancer treatments.5 However, many advanced cancers have gained resistance to apoptotic signaling via the upregulation of apoptotic inhibitor proteins such as cellular inhibitors of apoptosis (cIAPs) and FLICE inhibitor protein (FLIP).6 FLIP, originally noted for its importance in lymphocyte proliferation and development,7 is a known inhibitor of DR-mediated apoptosis.8 Multiple isoforms of FLIP mRNA have been identified, but 2 protein forms, FLIP long (FLIPL) and FLIP short (FLIPS), are most readily observed, each having distinct signaling effects at the DISC.9 FLIPL is a 55-kDa protein that can be a substrate of caspase-8 resulting in truncation from the C-terminal end into a 43-kDa protein, which has been noted to be more active.10,11 FLIPS is a 22-kDa isoform consisting only of the 2 N-terminal death effector domains (DEDs). Both isoforms of FLIP inhibit apoptosis via their ability to form heterodimers with procaspase-8, preventing activation of the caspase cascade.9,12 Elevated FLIP expression levels are directly associated with chemotherapy-resistant cancers as an acquired mechanism to evade DR-mediated apoptosis.9 Additionally, FLIP has been reported in other DR pathways, including the activation of the potent pro-inflammatory transcription factor NF-κB.13,14 FLIP can interact with multiple proteins important in TNF-mediated NF-κB activation, including receptor interacting protein kinase 1 (RIP1),15 TNFR-associated factor 2 (TRAF2),10 and inhibitor kappa B kinase gamma (IKKγ),16 indicating the multiple roles for FLIP in NF-κB signaling. Furthermore, NF-κB promotes FLIP expression in a positive feedback loop.17 These high levels of FLIP can potentially enhance a chronic inflammatory response and promote tumor growth.
Abundant concentrations of DR-ligands are produced in response to infection, inflammation and in response to chemotherapeutic drugs. NF-κB is sequestered by NF-κB inhibitors (IκBs) in the cytosol under normal conditions. Proinflammatory cytokines and DR-ligands activate NF-κB by targeting IKK for phosphorylation and subsequent degradation, allowing NF-κB to translocate to the nucleus to function as a transcription factor. TNFR-mediated activation of NF-κB is initiated by the formation of a cytoplasmic complex containing TNFR superfamily 1A-associated via death domain (TRADD), RIP1, TRAF2, and cIAPs. During TNFR signaling, RIP1 is polyubiquitinated by cIAPs promoting the formation of a pro-survival complex upstream of NF-κB activation.18 RIP is essential for activation of IKKs by this complex.19 Inhibition or loss of cIAPs, as well as the deubiquitination of RIP, causes apoptosis via RIP recruitment to the DISC.20 Therefore, RIP is a pivotal signaling protein in DR-stimulated complex formation. By regulating both cell death and proliferation, it is not surprising that RIP overexpression has also been associated with poor prognosis in cancer.21
Reactive nitrogen and oxygen species are commonly found in pathologies such as chronic inflammation, cardiovascular disease, and advanced tumorigenesis. Activation of inducible nitric oxide synthase (iNOS) and elevated levels of nitric oxide (NO), lead to rapid increases in the S-nitrosylation of proteins. S-nitrosylation is the covalent addition of NO to the thiol side chain of a cysteine residue. This post-translational modification (PTM) can regulate protein interactions and modify downstream signaling. Other PTMs have been reported to significantly alter FLIP stability and function.22,23 We have previously shown that S-nitrosylation of FLIP can prevent its ubiquitination and subsequent degradation, making it a more potent inhibitor of Fas-mediated apoptosis.24
Here we demonstrate the effects of FLIP S-nitrosylation of residues 254 and 259 on NF-κB activation. The underlying mechanism of changes in NF-κB activation is due to the ability of S-nitrosylated FLIP to bind RIP1 following TNF-α stimulation. Additionally, we demonstrate that S-nitrosylation of FLIP regulates its cleavage into shorter forms. Determining the regulation of FLIP signaling is fundamental to understanding its role in disease and is imperative for the future development of effective therapeutic strategies in chemotherapy-resistant cancers.
Results
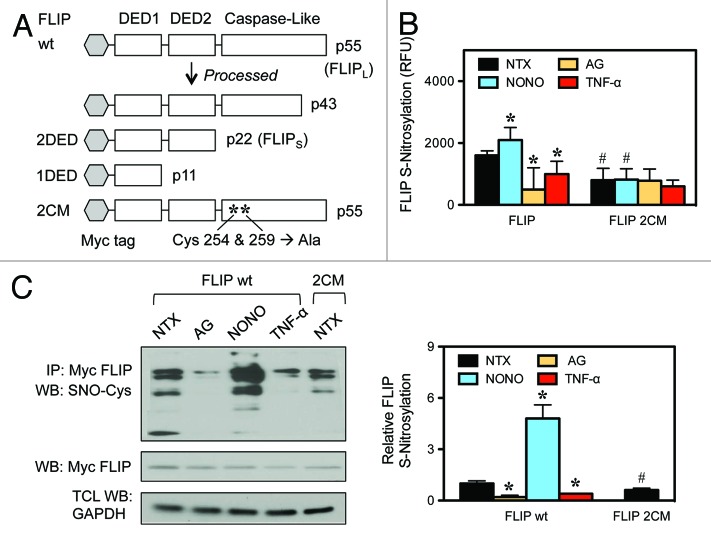
FLIP is S-nitrosylated at residues 254 and 259
The antiapoptotic role of FLIP in DR-mediated apoptotic signaling is firmly established. We have previously reported that FLIP S-nitrosylation prevents its ubiquitination and subsequent degradation, thereby making it a more potent inhibitor of Fas-mediated apoptosis.24 Since DR signaling can also lead to cell survival we tested the hypothesis that S-nitrosylation of FLIP modulates its role as an NF-κB activator. We used our previously described double cysteine mutant of FLIP (2CM), where single point mutations of cysteine residues 254 and 259 to alanines in the caspase-like domain of FLIP prevent S-nitrosylation of FLIP (Fig. 1A).24 Transiently transfected FLIP 2CM showed impaired responsiveness to pharmacological NO supplementation by DPTA NONOate (NONO), as evidenced by the lower overall S-nitrosylation of FLIP 2CM compared with wild-type FLIP (wt), with S-nitrosylation levels comparable to those detectable following the treatment with the iNOS inhibitor aminoguanidine (AG) (Fig. 1B and C). We do not see a complete reduction in S-nitrosylation levels, since there are additional cysteine residues within the FLIP protein sequence that could potentially be S-nitrosylated.
Figure 1. S-nitrosylation of FLIP. (A) A schematic of the domain structure of FLIP isoforms and expression vectors is shown. FLIP wild-type (wt), FLIP death effector domains (2DED), FLIP double cysteine mutant (2CM). (B) HEK-293 cells were transiently transfected with myc-FLIP wt or myc-FLIP 2CM and treated with media alone (NTX), 300 μM AG, 400 μM NONOate for 12 h or 50 ng/mL TNF-α for 30 min. Analysis for S-nitrosylation was determined by fluorescence described in “Materials and Methods”. (C) HEK-293 cells were transiently transfected with myc-FLIP wt or myc-FLIP 2CM. myc-FLIP-expressing cells were treated as in (B). Lysates were coimmunoprecipitated using anti-myc antibodies and assayed for S-nitrocysteine (SNO-Cys) by streptavidin using an S-nitrosylation kit. Membranes were stripped and reprobed with anti-myc antibodies to detect FLIP. Total cell lysates (TCL) were also probed with anti-GAPDH antibodies to control for loading. Data are mean ± SD (n = 3). *P < 0.05 vs. non-treated (NTX) FLIP wt transfected cells. #P < 0.05 vs. FLIP wt transfected cells.
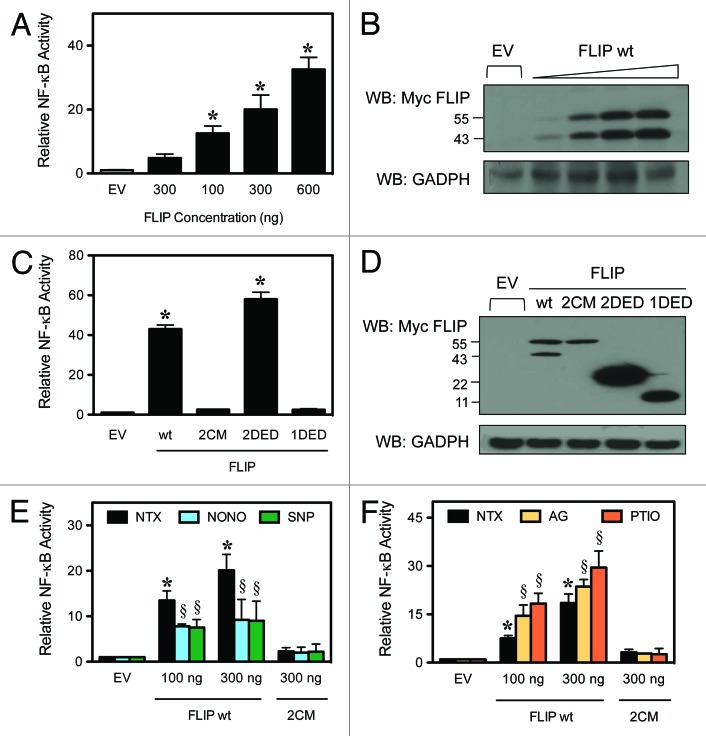
S-nitrosylation of FLIP modulates NF-κB activation
It is well established that FLIP can promote NF-κB activation.10,15,16 While transient transfection of wild-type FLIP activates NF-κB in a dose-dependent manner (Fig. 2A), FLIP 2CM completely lacks any detectable activation of NF-κB (Fig. 2C). This result suggests that the S-nitrosylation of FLIP at residues 254 and 259 is necessary for FLIP-mediated activation of NF-κB. The expression of all FLIP constructs has been verified by immunoblot in comparison to empty vector (EV) (Fig. 2B and D). In agreement with previous results, transient expression of the 2 N-terminal DEDs (2DED), representative of FLIPS, but not one N-terminal DED alone (1DED), independently activates NF-κB (Fig. 2C).10,16 These results indicate that S-nitrosylation of FLIP at cysteine residues 254 and 259 represents a distinct mechanism to activate NF-κB, compared with the DED-mediated response previously reported. To further examine the role of S-nitrosylation of FLIP on NF-κB activation, we performed experiments using NO-modifying reagents. Cells were transiently transfected with FLIP wt (FLIPL) expression constructs, followed by treatment with the NO donor DPTA NONOate (NONO) and sodium nitroprusside (SNP). NO supplementation using either NONO or SNP treatment caused a decrease in FLIP-mediated NF-κB activation, which was independent on the expression level of FLIP, while FLIP 2CM was not affected by NO supplementation and failed to activate NF-κB (Fig. 2E). Although we expected to observe an increase in NF-κB activation, the subunits of NF-κB are also targets for S-nitrosylation, which results in an overall inhibition of its activation by preventing translocation to the nucleus and interfering with DNA binding.25 However, we show the FLIP-mediated induction of NF-κB remains statistically significant compared with control cells (Fig. 2E). Additionally, NF-κB activation promoted by transient transfection of FLIP, but not FLIP 2CM was significantly increased upon global NO inhibition using the NO scavenger 2-(4-carboxy-phenyl)-4,4,5,5 tetramethylimidazoline-1-oxy-3-oxide (PTIO) and the selective iNOS inhibitor AG (Fig. 2F). Thus, our results show that the S-nitrosylation of FLIP is upstream of NF-κB activation and is another S-nitrosylation-mediated mechanism to modulate NF-κB activation.
Figure 2. FLIP-mediated NF-κB activation. HEK-293 cells were transiently transfected with an NF-κB-luciferase and Renilla-luciferase reporter and expression constructs for the FLIP domains, as indicated. Luciferase activity was determined by the dual-luciferase reporter assay. (A) Cells were transiently transfected with increasing amounts of FLIP wt and an NF-κB luciferase reporter plasmid. NF-κB activity was determined by luminescence according to “Materials and Methods”. (B) Total cell lysates were also analyzed by western blot using an anti-myc antibody to verify the increasing expression levels of FLIP. (C) Cells were transiently transfected with empty vector (EV), FLIP wt, FLIP 2CM, FLIP 2DED, or FLIP 1DED and analyzed for NF-κB activity as noted in (A). (D) Total cell lysates were also analyzed by western blotting as above to verify expression of all constructs. (E) Cells were transiently transfected with EV, FLIP wt or FLIP 2CM. Cells treated with media alone, 400 μM NONOate, or 500 μM SNP for 12 h to supplement nitric oxide and analyzed for NF-κB activity as noted in (A). (F) Cells were transiently transfected with EV, FLIP wt, or FLIP 2CM, and cells were treated with media alone, 300 μM AG or 300 μM PTIO for 12 h to inhibit NO production. Lysates were analyzed for NF-κB activity as noted in (A). Data are mean ± SD (n = 3). *P < 0.05 vs. non-treated (NTX) EV-transfected cells. §P < 0.05 vs. non-treated (NTX) control.
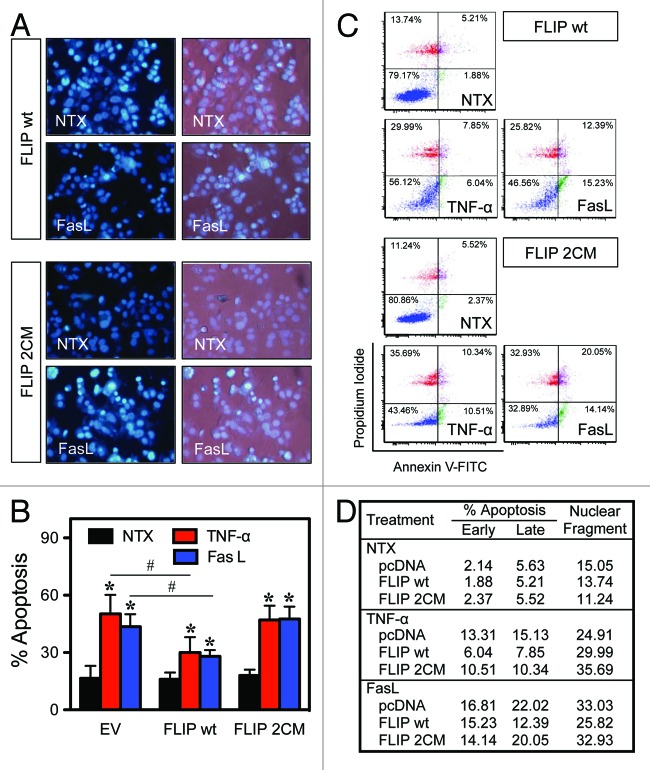
Since DRs signal for NF-κB activation and apoptosis, we also determined the effect of S-nitrosylation of FLIP on apoptosis. Cells were transfected with either an EV, FLIP wt, or FLIP 2CM, followed by treatment with the DR agonist TNF-α or FasL and quantification of apoptosis. Treatment of the cells with TNF-α and FasL causes increased cell death shown by an increase in fragmented nuclei, which is inhibited by FLIP wt, but not FLIP 2CM (Fig. 3A and B). Flow cytometric analysis of apoptosis using annexin V and propidium iodide confirmed the Hoechst DNA fragmentation results (Fig. 3C and D) and indicated that S-nitrosylation of FLIP is important in both proliferative and apoptotic signaling originating from DRs.
Figure 3. S-nitrosylation of FLIP modulates its anti-apoptotic activity. MCF-7 cells were transiently transfected with FLIP wt or FLIP 2CM expression constructs. Twenty-four hours post-transfection, cells were treated with media alone, 50 ng/mL TNF-α, or 200 ng/mL FasL for 16 h, and apoptosis was determined by Hoechst assay. (A) Representative fluorescence micrographs of treated cells stained with the Hoechst dye. (B) Quantitative analysis of 15 fields of view. (C) Cells were similarly treated with TNF-α and FasL for 16 h, and apoptosis was determined by flow cytometry using annexin V and PI as probes. (D) Quantitative analysis of early and late apoptosis (combined percentage of lower and upper quadrants). *P < 0.05 vs. non-treated (NTX) control. #P < 0.05 vs. treated EV-transfected cells.
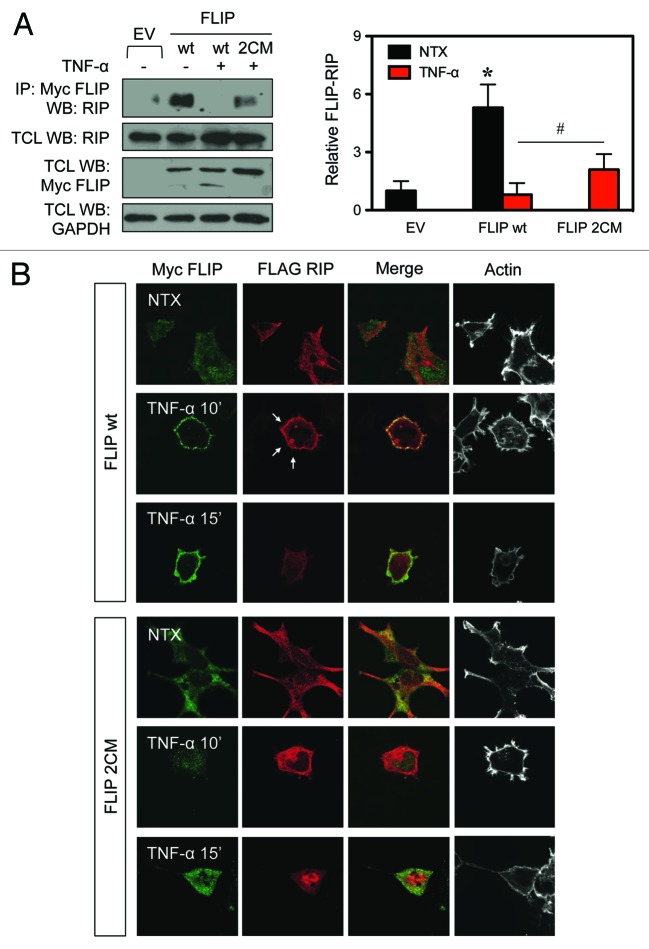
S-nitrosylation of FLIP modulates RIP1 binding and localization
To further delineate the molecular mechanism by which S-nitrosylation of FLIP mediates NF-κB activation, we investigated its interaction with upstream adaptor RIP1, which is a crucial signaling protein upstream of NF-κB activation and known binding partner of FLIP.15 We transiently transfected cells with FLIP wt or FLIP 2CM and tested the interaction with endogenous RIP1. FLIP wt or FLIP 2CM was immunoprecipitated, and the immune complexes were probed for RIP1. FLIP wt binds to RIP1 in the absence of any DR stimulation, and the interaction is disrupted following TNF-α treatment (Fig. 4A). In contrast, FLIP 2CM retained its ability to interact with RIP1 even in cells that were treated with TNF-α (Fig. 4A) at the same level as observed in cells not stimulated by TNF-α (data not shown). This differential binding indicates that the S-nitrosylation of FLIP at residues 254 and 259 inhibits binding to RIP1. Furthermore, FLIP wt, but not FLIP 2CM, is cleaved upon TNF-α treatment, also indicating that S-nitrosylation is necessary for FLIP processing. We next examined the cellular localization of FLIP and RIP1 by confocal microscopy in HEK 293 cells, which express very low levels of endogenous FLIP (data not shown). We observed that in resting cells, RIP1 is distributed in the cytoplasm; however, it is recruited to the membrane in response to TNF-α stimulation. This translocation to the membrane is impaired when the FLIP 2CM mutant is co-expressed (Fig. 4B). These results suggest that the S-nitrosylation of FLIP is essential in regulating its interaction with RIP1, thus affecting localization of RIP1 to the DR complex following receptor ligation, thereby modulating DR signaling.
Figure 4. S-nitrosylation of FLIP modulates its interaction with RIP1 and localization of RIP. (A) HEK-293 cells were transiently transfected with myc-FLIP wt or myc-FLIP 2CM expression constructs. Thirty-six hours post-transfection, cells were either left untreated or treated with 50 ng/mL TNF-α for 30 min. Lysates were co-immunoprecipitated with anti-myc antibodies to purify FLIP and probed for endogenous RIP. Membranes were stripped and reprobed with anti-myc antibodies. Lysates were also probed with anti-myc antibodies to detect FLIP, anti-RIP1, and anti-GAPDH antibodies. (B) HEK-293 cells were transiently transfected with myc-FLIP wt, myc-FLIP 2CM, and FLAG-RIP1 expression constructs as indicated, left untreated, or stimulated with 50 ng/mL TNF-α for 10 or 15 min. Cells were fixed, immunostained with anti-myc and anti-FLAG antibodies and fluorescently labeled secondary antibodies and phalloidin to visualize actin and analyzed by confocal microscopy. White arrows indicate RIP localization to the membrane. *P < 0.05 vs. non-treated (NTX) EV-transfected cells. #P < 0.05 vs. FLIP wt transfected cells.
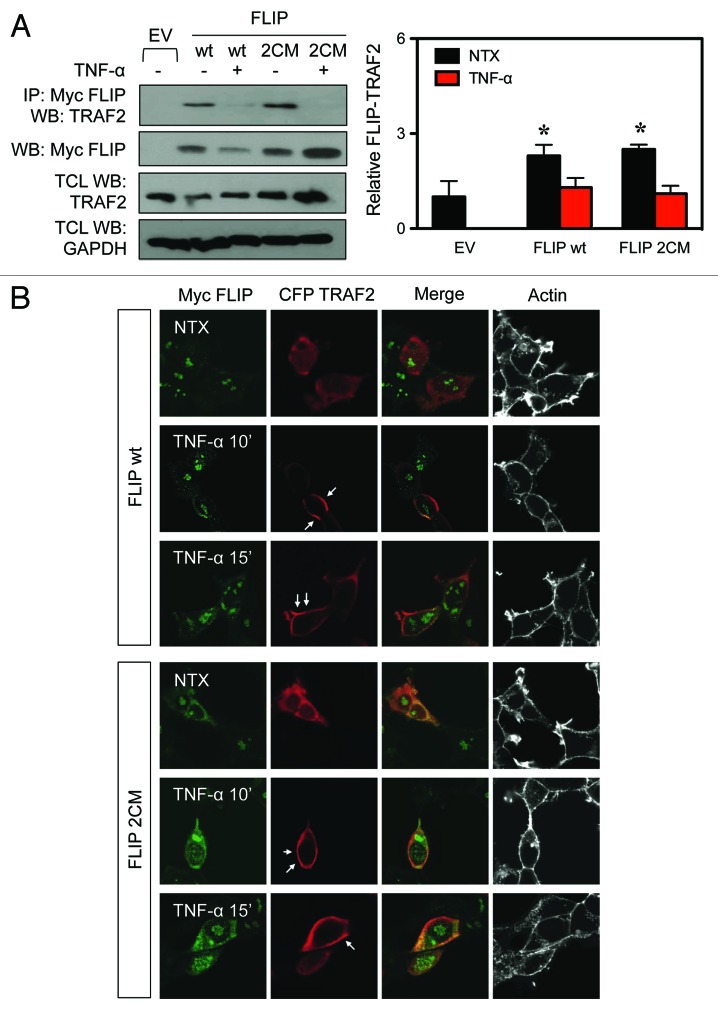
TRAF2 localization and FLIP binding is not dependent on FLIP S-nitrosylation
TRAF2 is another DR signaling complex adaptor protein, which is essential for NF-κB activation.19 Since the S-nitrosylation of FLIP affects its interaction with RIP1, we also examined the interaction of FLIP with TRAF2. We transiently transfected cells with either FLIP wt or FLIP 2CM, immunoprecipitated FLIP, and analyzed the immune complexes for the presence of endogenous TRAF2. However, in contrast to the FLIP interaction with RIP1, its interaction with TRAF2 was not affected when comparing FLIP wt and FLIP 2CM, either in the presence or absence of TNF-α (Fig. 5A). Additionally, we examined the localization of TRAF2 in HEK 293 cells following TNF-α stimulation. We found that TRAF2 is distributed in the cytoplasm in resting cells and translocates to the membrane following receptor stimulation with TNF-α regardless of the presence of FLIP wt or FLIP 2CM (Fig. 5B). Thus, our results indicate that S-nitrosylation of FLIP at residues 254 and 259 does not affect its interaction with TRAF2 and therefore is expected to not cause the effects on NF-κB activation.
Figure 5. TRAF2 binding to FLIP is not dependent on FLIP S-nitrosylation. (A) HEK-293 cells were transiently transfected with myc-FLIP wt or myc-FLIP 2CM expression constructs. Thirty-six hours post-transfection, cells were left untreated or treated with 50 ng/mL TNF-α for 30 min. Lysates were co-immunoprecipitated with anti-myc antibodies to purify FLIP and probed for endogenous TRAF2. Membranes were stripped and reprobed with anti-myc antibodies to detect FLIP. Total cell lysates (TCL) were also probed for myc-FLIP, TRAF2, and GAPDH to demonstrate equal loading. (B) HEK-293 cells were transiently transfected with myc-FLIP wt, myc-FLIP 2CM, and CFP-TRAF2 expression constructs as indicated, left untreated, or stimulated with 50 ng/mL TNF-α for 10 or 15 min. Cells were fixed, immunostained with anti-myc and anti-CFP antibodies and fluorescently labeled secondary antibodies and phalloidin to visualize actin and analyzed by confocal microscopy. White arrows indicate TRAF2 localization to the membrane. *P < 0.05 vs. non-treated (NTX) EV-transfected cells.
S-nitrosylation of FLIP causes processing into shorter forms
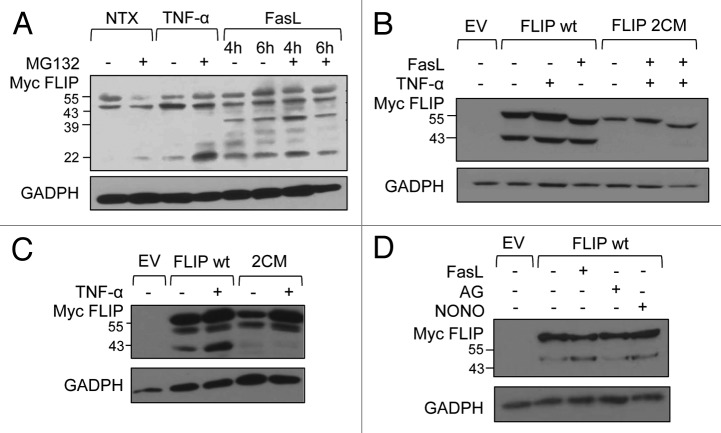
FLIP protein levels and the ratio of FLIPL and FLIPS vary in different cell types. It is also known that FLIPL is rapidly converted into a p43 fragment through proteolytic cleavage.10,11,26 Since different forms of FLIP show distinct effects on NF-κB activation (Fig. 2C), we next tested whether DR ligation causes proteolytic processing of FLIP, which could also impact activation of NF-κB. We used a human lung adenocarcinoma A549 cell line, which expresses a high level of FLIP, and either mock treated the cells, or treated the cells with the DR-ligands TNF-α and FasL for different times in the absence or presence of the proteasome inhibitor MG132. We then probed the lysates for expression of endogenous FLIP. We found that the 55-kDa FLIPL is partially converted in resting cells into the 43-kDa, C-terminally truncated FLIPL (Fig. 6A). Treatment with MG132 also revealed presence of p22 FLIP, which is proteasomally degraded. However, treatment with either TNF-α or FasL enhanced expression or stability of FLIPS, with FasL also promoting further processing of p43 into the p39 version, which is yet another C-terminal processing event. We next tested whether S-nitrosylation could impact proteolytic processing of FLIP. We transiently transfected HEK 293 cells with either FLIP wt or FLIP 2CM, treated cells with FasL or TNF-α, and probed cell lysates for FLIP. While FLIP is partially converted into p43, as observed for endogenous FLIP, FLIP 2CM is protected from processing and remains as p55 FLIPL (Fig. 6B and C). Additionally FLIP processing is enhanced by NO supplementation by treatment with NONO, comparable to FasL treatment (Fig. 6D). This increased processing is also consistent with S-nitrosylation fragments seen in Figure 1C. Thus, our data indicate that S-nitrosylation at residues 254 and 259 is required for the ability of FLIPL to act as a substrate and its proteolytic conversion into p43.
Figure 6. S-nitrosylation of FLIP promotes protein processing into shorter forms. (A) A549 cells were analyzed for endogenous FLIP expression. Cells were treated with 50 ng/mL TNF-α for 30 min or 100 ng/mL FasL for 4 and 6 h in the presence or absence of 1 μM MG132 for 24 h. (B) HEK-293 cells were transiently transfected with EV, myc-FLIP wt, or myc-FLIP 2CM expression constructs. Thirty-six hours post-transfection, cells were left untreated or treated with 50 ng/mL TNF-α for 30 min or 200 ng/mL FasL for 4 h and analyzed by western blot using anti-myc antibodies to detect FLIP. Membranes were stripped and reprobed with anti-GAPDH antibodies to control for equal loading. (C) MCF-7 cells were transiently transfected as in (B) and left untreated or treated with 50 ng/mL TNF-α for 30 min. Total lysates were analyzed by western blot with anti-myc and anti-GAPDH antibodies. (D) MCF-7 cells were transiently transfected with EV or myc-FLIP. Thirty-six hours post-transfection, cells were left untreated or treated with 200 ng/mL FasL, 600 μM AG, or 400 μM NONOate for 4 h. Total lysates were analyzed by western blot with anti-myc and anti-GAPDH antibodies.
Discussion
In this study we demonstrate that S-nitrosylation of FLIP at cysteines 254 and 259 mediate RIP1 binding to induce NF-κB activation. This mechanism is distinct from previously reported FLIP-mediated NF-κB activity, thus FLIP S-nitrosylation provides a novel molecular mechanism of FLIP-mediated NF-κB regulation.10,16 FLIP can interact with multiple proteins upstream of NF-κB, and therefore it is not surprising to discover that differences in FLIP signaling and nitrosylation state can lead to the activation of NF-κB via multiple avenues. An increasing number of studies have been reported examining the inhibition of FLIP using small-molecule inhibitors, chemicals, or siRNA techniques;27-30 however, additional regulation may be found within the cell by PTMs.
FLIP PTMs have been shown to be involved in changes in its stability, binding partner interactions, and localization. FLIP phosphorylation prevents localization to the DISC in TRAIL-mediated DR5 signaling and leads to an increase in TRAIL-mediated apoptosis.22 We have previously reported that FLIP S-nitrosylation prevents ubiquitination and subsequent degradation.24 S-nitrosylation has gained increasing importance as a PTM that affects protein interactions and signaling pathways. The tumor environment frequently has elevated NO levels due to constitutive iNOS activity and redox imbalances, leading to an overall increase in protein nitrosylation. In this study, we demonstrate NF-κB induction by wild-type and nitrosylable FLIP in a dose-dependent manner. However, NF-κB activation is completely abrogated, when testing a mutant of FLIP that cannot be nitrosylated at residues 254 and 259. Rescue of NF-κB activity by FLIPS (FLIP 2DED) indicates there are at least 2 separate mechanisms for FLIP-mediated NF-κB activation, in agreement with previous reports that FLIP isoforms have distinct signaling, because the nitrosylation sites we investigated map within the caspase-like domain and not the DED.9,31 NO supplementation results in an increase in overall protein nitrosylation; however, we observed a decrease in NF-κB activity, contrary to the expected increase from additionally S-nitrosylated FLIP. Others have shown NF-κB activity is inhibited when subunit p50 is S-nitrosylated due to its inability to bind DNA.32 Interestingly, our results show that FLIP significantly induces NF-κB activation even with NO inhibition. FLIP has most notably been reported as an inhibitor of apoptosis by binding procaspase-8, thereby preventing its activation.8,12 We further demonstrate that S-nitrosylation at residues 254 and 259 are necessary to confer this resistance to apoptosis.
It has been previously reported that FLIP interacts specifically with TRAF2 to activate NF-κB.10 We were unable to demonstrate that the S-nitrosylation of FLIP at residues 254 and 259 alters the interaction with TRAF2, indicating that this interaction is not responsible for the change in S-nitrosylated FLIP-mediated NF-κB activation. RIP1 is another essential signaling protein for TNFR-mediated NF-κB activation.33 The ubiquitination state of RIP is mediated by cIAPs, which determine the formation of the DISC or NF-κB signaling complex.20 Our results demonstrate that the S-nitrosylation of FLIP at residues 254 and 259 mediate RIP binding and localization. Our results indicate that the altered interaction between FLIP and RIP1, as a consequence from S-nitrosylation, contributes to the mechanism for FLIP-mediated NF-κB activation. With non-nitrosylable FLIP retaining RIP binding, thus preventing RIP1 recruitment to DRs, NF-κB is unable to be canonically activated. Protein sequestration is a common means for controlling signal transduction. For example, compartmentalization by lipid rafts changes the makeup of TNFR1 and Fas signaling complexes.34
Varying FLIP mRNA transcripts correspond to FLIPL and FLIPS and have been reported to have varying affinity in binding partners and distinctive downstream signaling, in addition to the classical anti-apoptotic function through caspase-8 inhibition.35,36 FLIPL has been shown to be a caspase-8 substrate, resulting in a 43-kDa protein with more robust NF-κB activation.10,11 We identified additional cleavage fragments in FasL- and TNF-α-treated A549 cells, which express high levels of FLIP. Additionally, we demonstrate that FLIPL is being cleaved into smaller fragments by using a myc-tagged expression construct that includes only the exon regions of the coding sequence, thereby preventing the smaller fragments from being mRNA splice variants. Given that different forms of FLIP have different signaling capabilities, this could have dramatic implications in chemotherapy-resistant cancer cells that have elevated levels of FLIP in conjunction with high levels of caspase-8. Ligand-induced cleavage and multiple cleavage truncations of FLIP have not been previously reported, and it is currently unknown if caspase-8 is responsible for the additional cleavage fragments. Additionally, the increases in NO associated with disease could further impact this signaling pathway, since FLIP is subject to nitrosylation. The S-nitrosylation status of FLIP likely affect proteolytical processing of FLIP, since we were unable to detect cleavage in our FLIP 2CM-transfected cells.
Our data could suggest that S-nitrosylation of FLIP leads to an increase in NF-κB activity in 2 distinct ways. First, S-nitrosylation of FLIP results in an increase in FLIP processing, thereby generating a p22 fragment, which is a known inducer of NF-κB.16 The FLIP 2CM was unable to be processed into the p22 fragment and therefore would be unable to induce NF-κB through this mechanism. Second, FLIP S-nitrosylation could lead to the disassociation of binding with RIP1, potentially promoting RIP1’s activation of NF-κB. The FLIP 2CM retained RIP1 binding potentially interfering with RIP1-mediated NF-κB activation. However, there is still a signaling paradox in this system of NO signaling, as TNF-α-stimulation results in an overall decrease in S-nitrosylation of FLIP as well as an overall decrease in NO production (data not shown). The specific details of S-nitrosylation are reported to be exceedingly complex, and the mechanisms of targeted specificity still remains unclear.37 We clearly demonstrate S-nitrosylation of FLIP affects FLIP function; however, it probably does not affect all FLIP interactions.
In summary, many chemotherapy-resistant tumors are insensitive to cell death through elevated levels of FLIP and, consequently, elevated NF-κB activity. Targeted FLIP silencing has been shown to restore sensitivity to DR-mediated apoptosis.38 Studies on FLIP regulation by PTMs and resulting interactions are imperative to better understand its role in normal physiology and pathogenesis of cancer and inflammation. Specifically our results suggest that developing strategies to interfering with FLIP S-nitrosylation could have potential for future targeted therapies for chemotherapy-resistant tumors.
Materials and Methods
Cell lines and reagents
HEK 293, MCF-7, and A549 cells were cultured in Dulbecco modified Eagle medium (Invitrogen) containing 10% fetal bovine serum, 2 mM L-glutamine, 20 mM HEPES, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 5% CO2 environment at 37 °C. Recombinant FasL (SuperFasL), monoclonal antibody against FLIP (Dave-2), and the NO donor dipropylenetriamine (DPTA) NONOate were purchased from Alexis Biochem. The NO donor sodium nitroprusside (SNP), NO inhibitors amino-guanidine (AG), 2-(4-carboxy-phenyl)-4,4,5,5 tetramethylimidazoline-1-oxy-3-oxide (PTIO) were from Sigma. The transfecting agent Lipofectamine Plus was purchased from Invitrogen. Monoclonal anti-CFP (3E6) antibody was purchased from Invitrogen. Anti-myc antibodies were purchased from Millipore and SCBT. Polyclonal antibody against GAPDH was purchased from BD Transduction. Polyclonal antibody against TRAF2 and monoclonal caspase-8 antibody were purchased from Cell Signaling Technology. Polyclonal anti-HA antibody was purchased from Abcam. Polyclonal antibody against FLAG (OctA) was purchased from SCBT. Commercial protease inhibitor mixture was purchased from Roche Applied Science. pRL and pGL vectors were purchased from Promega. pcDNA3-FLIP plasmids were made as described previously.24 pECFP-TRAF2 was made as described previously and provided as a kind gift from J Schmid (Medical University Vienna).39
S-nitrosylation assays
HEK 293 cells were transfected with pcDNA3-FLIP. The following day cells were treated as indicated in figure legend. S-nitrosylated protein detection assay kit was commercially obtained from Cayman Chemical Company and was used according to the manufacturer’s instruction. Briefly, cell pellets were lysed under the protein free thiols blocking condition. Protein S-nitrosothiols were then reduced to yield free thiols, which were covalently labeled with maleimide-biotin. Lysate protein (500 μg) was subjected to the immunoprecipitation as described earlier. Subsequent analysis for S-nitrosylated protein was determined by western blotting using S-nitrosylation detection reagent I (HRP) and by fluorescence reading using S-nitrosylation detection reagent II (fluorescein). For western blot analysis, protein samples were prepared in Laemmli buffer and were resolved under denaturing conditions. For fluorescence reading, immune complexes were further incubated with fluorescein detection agent overnight following the standard immunoprecipitation step. Complexes were rinsed with wash buffer 3 times, resuspended in colorless Laemmli sample buffer, and boiled for 5 minutes. Clear lysates were loaded onto the fluorescence plate and analyzed for fluorescence intensity using a fluorescence plate reader at a 485-nm excitation and a 520-nm emission (FLUOstar OPTIMA, BMG Labtech).
NF-κB gene reporter assay
1.5 × 105 HEK 293 cells/well was seeded on 0.02% gelatin coated 24-well plates. Cells were transfected the following day with 600 ng of empty pcDNA3 vector, the indicated amount of pcDNA3-FLIP plasmids, 100 ng of a multimeric NF-κB pGL2 luciferase vector, and 5 ng of the Renilla pRL-TK vector. Transfections were performed using Lipofectamine 2000 (Invitrogen). Expression of proteins was verified by immunoblotting. Media was changed 6 h after transfection, and cells were grown overnight. Luciferase activity was determined using a dual-luciferase reporter assay system kit (Promega). The activity of the NF-κB reporter luciferase was standardized to that of Renilla luciferase. Within the same experiment, each transfection was performed in quadruplicate, and, where necessary, empty control plasmid was added to keep each transfection receiving the same amount of total DNA. Data shown are representative averages and standard deviations from one of 3 separate experiments performed.
Apoptosis assay
MCF-7 cells were transfected with FLIP wt or FLIP 2CM using Amaxa Nucleofector (Lonza) using recommended manufacturer protocol. Transfected cells were seeded at 15 000 cells per well in a 96-well plate and allowed to recover for 24 h. Cells were treated with 200 ng/mL FasL or 100 ng/mL TNF-α for 16 h. Apoptosis was determined by Hoechst 33342 DNA fragmentation assay and by flow cytometric analysis of annexin V and propidium iodide (PI). In the Hoechst assay, cells were incubated with 10 μg/mL of Hoechst 33342 for 30 min and were visualized under fluorescence microscope (Leica Microsystems). Cells having intensely condensed and/or fragmented nuclei were considered as apoptotic. For flow cytometric analysis, cells were harvested, washed, and stained with annexin V-fluorescein isothiocyanate (FITC) and PI in binding buffer (BioVision) for 5 min at room temperature. Samples were immediately analyzed by FACSAria flow cytometer (Becton Dickinson) using a 488-nm excitation beam and a 530- and 630-nm band-pass filter with BD FACSDiva software (BD Bioscience).
Immunoblotting
Cells were lysed in RIPA lysis buffer containing 1 mM sodium orthovanadate, 100 mM phenylmethylsulfonyl fluoride, and a commercial protease inhibitor mixture (Roche Applied Science) for 20 min on ice. After insoluble debris was pelletted by centrifugation at 14 000 × g for 10 min at 4 °C, the supernatants were collected and determined for protein content using the Bradford method (Bio-Rad). Proteins (60 μg) were resolved on a reducing 10% SDS-polyacrylamide gel and transferred onto PVDF membranes. The transferred membranes were blocked for 1 h in 5% nonfat dry milk in Tris-buffered Tween (25 mM TRIS-HCl, pH 7.4, 125 mM NaCl, 0.05% Tween 20) and incubated with the appropriate primary antibodies. Membranes were washed 3 times with Tris-buffered Tween for 10 min and incubated with horseradish peroxidase-coupled isotype-specific secondary antibodies for 1 h at room temperature. The immune complexes were detected by enhanced chemiluminescence (ECL) detection.
Immunoprecipitation
Cells were lysed in lysis buffer containing 20 mM TRIS-HCl, pH 7.5, 1% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM sodium orthovanadate, 50 mM sodium fluoride, 100 mM phenylmethylsulfonyl fluoride, and a protease inhibitor mixture for 20 min on ice. Immunoprecipitation experiments were performed using Dynabeads (Invitrogen) and the recommended protocol. Immunoblotting was performed as described above.
Immunofluoresence
Cells were seeded on rat type I collagen coated coverslips (5 μg/cm2), fixed with 3.7% paraformaldehyde for 15 min, incubated in 50 mM glycine for 5 min, and permeabilized and blocked with 0.5% saponin, 1.5% BSA, and 1.5% normal goat serum for 30 min. FLIP was immunostained with anti-myc. RIP1 was immunostained with an antibody against FLAG (OctA). TRAF2 was immunostained with antibody against CFP. Secondary Alexa Fluor 405-, 488-, 546-, and 647-conjugated antibodies and phalloidin were used from Invitrogen. Cells were washed with PBS containing 0.5% saponin, and coverslips were mounted using Fluoromount-G (Southern Biotechnology Associates). Cells were viewed with a Zeiss LSM 510 confocal on an AxioImager Z1 microscope using a 63× objective lens.
Statistical analysis
The differences between treatment groups were analyzed using an unpaired Student t test and were considered significant when P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Imaging experiments were performed in the West Virginia University Microscope Imaging Facility, which is supported in part by the Mary Babb Randolph Cancer Center and National Institutes of Health (NIH) grant P20 RR016440. We thank Johannes A Schmid for generously sharing plasmids. This work was financially supported by the American Heart Association (predoctoral fellowship 0715376B to ST) and the NIH Grants R01-HL076340 and R01-HL095579 (to Y.R.) and National Science Foundation Grant EPS-1003907 (to Y.R.).
Glossary
Abbreviations:
- AG
aminoguanidine
- CIAP
cellular inhibitors of apoptosis
- DED
death effector domains
- DISC
death inducing signaling complex
- DR
death receptor
- EV
empty vector
- FLIP
FLICE inhibitory protein
- FLIPL
FLIP long
- FLIPS
FLIP short
- FLIP wt
FLIP wildtype
- FLIP 2CM
FLIP double cysteine mutant
- IKKγ
inhibitor of kappa B kinase gamma
- IκB
nuclear factor kappa B inhibitor
- iNOS
inducible nitric oxide synthase
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor kappa B
- NO
nitric oxide
- NONO
DPTA NONOate
- NTX
non-treatment
- PI3K
phosphatidylinositol 3-kinase
- PTIO
2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide
- PTM
posttranslational modification
- RIP1
receptor interacting protein 1
- SNP
sodium nitroprusside
- TCL
total cell lysates
- TNFR
tumor necrosis family receptor
- TRADD
TNFR superfamily 1A-associated via death domain
- TRAF2
TNFR-associated factor 2
- TRAIL-R
TNF related apoptosis inducing ligand receptor
References
- 1.Baker SJ, Reddy EP. Modulation of life and death by the TNF receptor superfamily. Oncogene. 1998;17:3261–70. doi: 10.1038/sj.onc.1202568. [DOI] [PubMed] [Google Scholar]
- 2.Debatin KM, Krammer PH. Death receptors in chemotherapy and cancer. Oncogene. 2004;23:2950–66. doi: 10.1038/sj.onc.1207558. [DOI] [PubMed] [Google Scholar]
- 3.Budd RC. Death receptors couple to both cell proliferation and apoptosis. J Clin Invest. 2002;109:437–41. doi: 10.1172/JCI0215077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson SB, Oyer R, Spalding AC, Anderson SM, Johnson GL. Increased expression of death receptors 4 and 5 synergizes the apoptosis response to combined treatment with etoposide and TRAIL. Mol Cell Biol. 2000;20:205–12. doi: 10.1128/MCB.20.1.205-212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falschlehner C, Ganten TM, Koschny R, Schaefer U, Walczak H. TRAIL and other TRAIL receptor agonists as novel cancer therapeutics. Adv Exp Med Biol. 2009;647:195–206. doi: 10.1007/978-0-387-89520-8_14. [DOI] [PubMed] [Google Scholar]
- 6.Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–8. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 7.Thome M, Tschopp J. Regulation of lymphocyte proliferation and death by FLIP. Nat Rev Immunol. 2001;1:50–8. doi: 10.1038/35095508. [DOI] [PubMed] [Google Scholar]
- 8.Krueger A, Baumann S, Krammer PH, Kirchhoff S. FLICE-inhibitory proteins: regulators of death receptor-mediated apoptosis. Mol Cell Biol. 2001;21:8247–54. doi: 10.1128/MCB.21.24.8247-8254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wajant H. Targeting the FLICE Inhibitory Protein (FLIP) in cancer therapy. Mol Interv. 2003;3:124–7. doi: 10.1124/mi.3.3.124. [DOI] [PubMed] [Google Scholar]
- 10.Kataoka T, Tschopp J. N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol. 2004;24:2627–36. doi: 10.1128/MCB.24.7.2627-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohrman A, Kataoka T, Cuenin S, Russell JQ, Tschopp J, Budd RC. Cellular FLIP (long form) regulates CD8+ T cell activation through caspase-8-dependent NF-kappa B activation. J Immunol. 2005;174:5270–8. doi: 10.4049/jimmunol.174.9.5270. [DOI] [PubMed] [Google Scholar]
- 12.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schröter M, Burns K, Mattmann C, et al. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–5. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 13.Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, et al. The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol. 2000;10:640–8. doi: 10.1016/S0960-9822(00)00512-1. [DOI] [PubMed] [Google Scholar]
- 14.Sun Q, Zachariah S, Chaudhary PM. The human herpes virus 8-encoded viral FLICE-inhibitory protein induces cellular transformation via NF-kappaB activation. J Biol Chem. 2003;278:52437–45. doi: 10.1074/jbc.M304199200. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhary PM, Eby MT, Jasmin A, Kumar A, Liu L, Hood L. Activation of the NF-kappaB pathway by caspase 8 and its homologs. Oncogene. 2000;19:4451–60. doi: 10.1038/sj.onc.1203812. [DOI] [PubMed] [Google Scholar]
- 16.Golks A, Brenner D, Krammer PH, Lavrik IN. The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappaB activation. J Exp Med. 2006;203:1295–305. doi: 10.1084/jem.20051556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreuz S, Siegmund D, Scheurich P, Wajant H. NF-kappaB inducers upregulate cFLIP, a cycloheximide-sensitive inhibitor of death receptor signaling. Mol Cell Biol. 2001;21:3964–73. doi: 10.1128/MCB.21.12.3964-3973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem. 2008;283:24295–9. doi: 10.1074/jbc.C800128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devin A, Cook A, Lin Y, Rodriguez Y, Kelliher M, Liu Z. The distinct roles of TRAF2 and RIP in IKK activation by TNF-R1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity. 2000;12:419–29. doi: 10.1016/S1074-7613(00)80194-6. [DOI] [PubMed] [Google Scholar]
- 20.Geserick P, Hupe M, Moulin M, Wong WW, Feoktistova M, Kellert B, Gollnick H, Silke J, Leverkus M. Cellular IAPs inhibit a cryptic CD95-induced cell death by limiting RIP1 kinase recruitment. J Cell Biol. 2009;187:1037–54. doi: 10.1083/jcb.200904158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, Hatanpaa KJ, Xie Y, Mickey BE, Madden CJ, Raisanen JM, Ramnarain DB, Xiao G, Saha D, Boothman DA, et al. The receptor interacting protein 1 inhibits p53 induction through NF-kappaB activation and confers a worse prognosis in glioblastoma. Cancer Res. 2009;69:2809–16. doi: 10.1158/0008-5472.CAN-08-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higuchi H, Yoon JH, Grambihler A, Werneburg N, Bronk SF, Gores GJ. Bile acids stimulate cFLIP phosphorylation enhancing TRAIL-mediated apoptosis. J Biol Chem. 2003;278:454–61. doi: 10.1074/jbc.M209387200. [DOI] [PubMed] [Google Scholar]
- 23.Kaunisto A, Kochin V, Asaoka T, Mikhailov A, Poukkula M, Meinander A, Eriksson JE. PKC-mediated phosphorylation regulates c-FLIP ubiquitylation and stability. Cell Death Differ. 2009;16:1215–26. doi: 10.1038/cdd.2009.35. [DOI] [PubMed] [Google Scholar]
- 24.Chanvorachote P, Nimmannit U, Wang L, Stehlik C, Lu B, Azad N, Rojanasakul Y. Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein. J Biol Chem. 2005;280:42044–50. doi: 10.1074/jbc.M510080200. [DOI] [PubMed] [Google Scholar]
- 25.Marshall HE, Hess DT, Stamler JS. S-nitrosylation: physiological regulation of NF-kappaB. Proc Natl Acad Sci U S A. 2004;101:8841–2. doi: 10.1073/pnas.0403034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, Lavrik IN, Eils R. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bijangi-Vishehsaraei K, Saadatzadeh MR, Huang S, Murphy MP, Safa AR. 4-(4-Chloro-2-methylphenoxy)-N-hydroxybutanamide (CMH) targets mRNA of the c-FLIP variants and induces apoptosis in MCF-7 human breast cancer cells. Mol Cell Biochem. 2010;342:133–42. doi: 10.1007/s11010-010-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sánchez-Pérez T, Ortiz-Ferrón G, López-Rivas A. Mitotic arrest and JNK-induced proteasomal degradation of FLIP and Mcl-1 are key events in the sensitization of breast tumor cells to TRAIL by antimicrotubule agents. Cell Death Differ. 2010;17:883–94. doi: 10.1038/cdd.2009.176. [DOI] [PubMed] [Google Scholar]
- 29.Luo A, Wang W, Sima N, Lu Y, Zhou J, Xu G, Yu H, Wang S, Ma D. Short hairpin RNA targeting c-FLIP sensitizes human cervical adenocarcinoma Hela cells to chemotherapy and radiotherapy. Cancer Lett. 2008;271:323–32. doi: 10.1016/j.canlet.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 30.Wilson TR, McEwan M, McLaughlin K, Le Clorennec C, Allen WL, Fennell DA, Johnston PG, Longley DB. Combined inhibition of FLIP and XIAP induces Bax-independent apoptosis in type II colorectal cancer cells. Oncogene. 2009;28:63–72. doi: 10.1038/onc.2008.366. [DOI] [PubMed] [Google Scholar]
- 31.Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem. 2001;276:20633–40. doi: 10.1074/jbc.M101780200. [DOI] [PubMed] [Google Scholar]
- 32.Marshall HE, Stamler JS. Inhibition of NF-kappa B by S-nitrosylation. Biochemistry. 2001;40:1688–93. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- 33.Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/S1074-7613(00)80535-X. [DOI] [PubMed] [Google Scholar]
- 34.Schütze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol. 2008;9:655–62. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 35.Park SJ, Kim YY, Ju JW, Han BG, Park SI, Park BJ. Alternative splicing variants of c-FLIP transduce the differential signal through the Raf or TRAF2 in TNF-induced cell proliferation. Biochem Biophys Res Commun. 2001;289:1205–10. doi: 10.1006/bbrc.2001.6086. [DOI] [PubMed] [Google Scholar]
- 36.Kim DJ, Park C, Oh B, Kim YY. Association of TRAF2 with the short form of cellular FLICE-like inhibitory protein prevents TNFR1-mediated apoptosis. J Mol Signal. 2008;3:2. doi: 10.1186/1750-2187-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tannenbaum SR, White FM. Regulation and specificity of S-nitrosylation and denitrosylation. ACS Chem Biol. 2006;1:615–8. doi: 10.1021/cb600439h. [DOI] [PubMed] [Google Scholar]
- 38.Sharp DA, Lawrence DA, Ashkenazi A. Selective knockdown of the long variant of cellular FLICE inhibitory protein augments death receptor-mediated caspase-8 activation and apoptosis. J Biol Chem. 2005;280:19401–9. doi: 10.1074/jbc.M413962200. [DOI] [PubMed] [Google Scholar]
- 39.Birbach A, Gold P, Binder BR, Hofer E, de Martin R, Schmid JA. Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. J Biol Chem. 2002;277:10842–51. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]