Abstract
Nasopharyngeal carcinoma (NPC) is one of the most common malignancies in southern China and Southeast Asia, with the highest metastasis rate among head and neck cancers. The mechanisms underlying NPC progression remain poorly understood. Genome-wide expression profiling on 18 NPC vs. 18 noncancerous nasopharyngeal tissues together with GeneGo pathway analysis and expression verification in NPC cells and tissues revealed a potential role of urokinase-type plasminogen activator receptor (uPAR) in NPC progression, which has not been investigated in NPC. We then observed that uPAR expression is increased in poorly differentiated, highly metastatic NPC cells compared with lowly metastatic cells or differentiated NPC cells. In vitro studies demonstrated that uPAR regulates NPC cell growth, colony formation, migration, and invasion and promotes the epithelial–mesenchymal transition (EMT). Additional tumor xenograft and spontaneous metastasis experiments revealed that uPAR promotes NPC cell growth and metastasis in vivo. The JAK–STAT pathway is involved in uPAR-regulated signaling in NPC cells as determined by immunoblotting. Moreover, uPAR-mediated growth and motility is partially abolished upon treatment with the Jak1/Jak2 inhibitor INCB018424. We suppressed uPA expression in uPAR-overexpressing NPC cells and found that uPAR-mediated cellular growth and motility is not exclusively dependent on uPA. In summary, uPAR is a significant regulator of NPC progression and could serve as a promising therapeutic target.
Keywords: uPAR, nasopharyngeal carcinoma, JAK, STAT, genome-wide expression profiling, tumor growth, metastasis
Introduction
Nasopharyngeal carcinoma (NPC) is one of the most common malignancies in South China1 and Southeast Asia.2-4 NPC occurs in the epithelial lining of the nasopharynx and has the highest metastasis rate among head and neck cancers.5,6 Thus, the majority of patients present with regional lymph node or even distant metastasis at the time of diagnosis.7-9 Distant metastasis is the primary cause of treatment failure.10 However, the molecular mechanisms underlying NPC progression and metastasis are not fully understood.
Urokinase-type plasminogen activator receptor (uPAR) is a glycosyl phosphatidylinositol (GPI)-anchored membrane protein with multiple functions.11 It consists of 283 amino acids divided into 3 similarly sized homologous domains (each with approximately 90 amino acids).12,13 Upon the binding of the extracellular protease urokinase-type plasminogen activator (uPA) and its zymogen form pro-uPA to cell surface, uPAR activates uPA to generate serine protease plasmin, which degrades a range of extracellular matrix (ECM) components and activates matrix metalloproteases (MMPs).14,15 In addition to regulating ECM proteolysis, uPAR also serves as a signaling receptor that interacts with vitronectin, integrins, and numerous other proteins. This interaction activates many intracellular signaling pathways, further promoting cellular proliferation, migration, and invasion.16,17 Although increased uPAR expression correlates with a poor prognosis in other malignancies,18,19 the role of uPAR in NPC progression and metastasis remains unknown.
The evolutionarily conserved Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathway mediates the cellular response to cytokines and growth factors.20 Earlier studies have shown that the JAK–STAT pathway is crucial for developmental processes, growth control, and the maintenance of homeostasis in various cells and tissues.21 Researchers have discovered that JAK–STAT signaling dysregulation is significantly associated with tumorigenesis.22 It has been reported that the JAK–STAT pathway is activated by uPAR in human aortic vascular smooth muscle cells23 and human kidney epithelial tumor cells.24 Despite these findings, evidence for the association between uPAR and the JAK–STAT pathway in human cancers remains inconclusive.
In the present study, we hypothesize that uPAR significantly influences NPC progression. We assess this hypothesis by comparing differentially expressed genes in NPC vs. non-cancerous nasopharyngeal tissues and subsequent validation. Moreover, our GeneGo pathway analysis suggests that the JAK–STAT pathway was likely implicated in uPAR signaling in NPC. Next, various in vitro and in vivo studies were conducted to test our hypothesis. We determined that uPAR is critically involved in NPC cell growth and motility. Moreover, uPAR may activate the JAK–STAT signaling pathway and the epithelial–mesenchymal transition (EMT) in NPC cells.
Results
uPAR is identified as a promising candidate gene for NPC progression
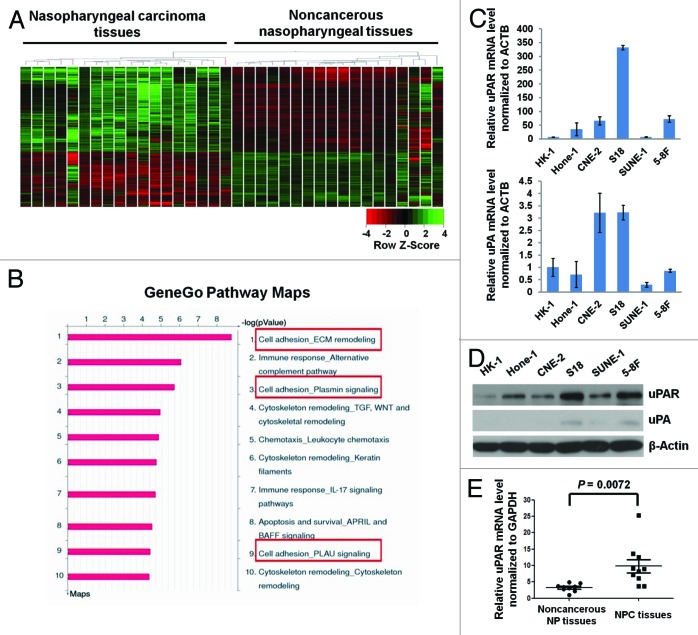
The unsupervised clustering of the genome-wide expression profiling data clearly distinguishes the NPC tissues from the non-cancerous tissues (Fig. 1A), thereby supporting the use of these data for further pathway analyses. Subsequently, 2992 genes were identified as differentially expressed in NPC tissues compared with the non-cancerous tissues (fold change, FC > 2, P < 0.05). Among these 2992 genes, uPA ranked highest of all the upregulated genes sorted in ascending order according to P values (Table S1). Moreover, uPAR expression was also upregulated in NPC tissues (FC = 3.34 and P = 7.52 × 10−5; data not shown).
Figure 1. uPAR expression is elevated in NPC tissues, and the highest expression is observed in highly metastatic cells. (A) A heat map showing the expression pattern of 41 091 genes in NPC vs. non-cancerous nasopharyngeal tissues derived from unsupervised clustering analysis. Red or green reflects low or high expression, respectively, as indicated in the scale bar (Raw Z score). (B) The pathway maps derived from the GeneGo Metacore analysis of microarray data sets of 2992 differentially expressed genes. The top 10 scored (log transformed P value) pathways affected by these genes are displayed. All the framed 3 pathways cover uPA–uPAR signaling. (C) The relative uPAR and uPA mRNA levels (normalized to ACTB) in NPC cells as assessed by quantitative real-time PCR. All data were compared with the control (uPA expression in HK-1 cells). Column, mean; error bar, ± SD (from triplicate replications). (D) uPAR and uPA protein levels in NPC cells determined by immunoblotting. Note, the highest expression of uPAR protein was observed in the highly metastatic S18 and 5-8F cell lines. (E) The relative uPAR mRNA levels (normalized to GAPDH) in 10 NPC vs. 9 non-cancerous nasopharyngeal tissue samples determined by real-time PCR. P values were calculated using the Student t test.
To further investigate the signaling pathways potentially associated with these differentially expressed genes, we analyzed the correlations among these 2992 genes using the GeneGo Metacore software. The pathway maps derived from the Metacore analysis represent the top 10 scored (log transformed P values) pathways affected by these genes (Fig. 1B). Interestingly, 3 of the pathways involved uPAR signaling, including ECM remodeling, Plasmin signaling, and PLAU (uPA) signaling pathway (Fig. 1B). These findings suggest that uPAR signaling is likely involved in NPC progression.
We also explored uPA and uPAR expression in the well-established NPC cell lines. Interestingly, uPAR mRNA expression levels were much higher when compared with uPA (Fig. 1C). Increased uPAR protein expression was observed in the poorly differentiated cell lines (Hone-1, CNE-2, S18, SUNE-1, 5-8F) compared with the well-differentiated, low-metastasis HK-1 cells (Fig. 1D). In addition, the highly metastatic cell lines (S18 and 5-8F) expressed higher levels of uPAR protein compared with their lowly metastatic parental lines (CNE-2 and SUNE-1, respectively) (Fig. 1D). To further confirm these findings, 10 NPC patient samples and 9 non-cancerous nasopharyngeal tissue samples were used to evaluate uPAR mRNA levels. Consistent with our previous results, uPAR mRNA was significantly elevated in NPC patients (Fig. 1E).
Several studies have reported that elevated uPAR expression regulates tumor cell migration, invasion, proliferation, and survival independent of uPA.25-29 These results are consistent with our findings regarding low uPA expression in NPC cell lines (Fig. 1C and D). We therefore hypothesized that uPAR plays a critical role in NPC progression, potentially independent of uPA.
uPAR suppression inhibits NPC cell growth, colony formation, migration, and invasion
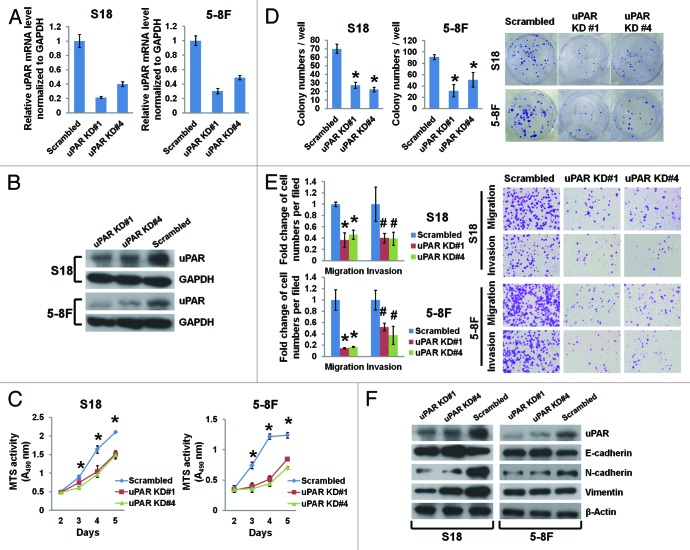
To explore the causal role of uPAR in NPC cell growth and motility, we stably expressed either uPAR-targeted shRNAs (uPAR KD#1 and #4) or a scrambled non-target shRNA in S18 and 5-8F cells. The shRNA suppression efficiency of uPAR mRNA and protein levels was validated by real-time PCR and immunoblotting (Fig. 2A and B). We observed that uPAR knockdown significantly impeded NPC cell growth (Fig. 2C) and colony formation (Fig. 2D). Additionally, uPAR knockdown reduced the number of migratory and invasive cells in vitro (Fig. 2E).
Figure 2. uPAR suppression affects the EMT and inhibits growth, colony formation, migration, and invasion in NPC cells. The highly metastatic S18 and 5-8F cells were transduced with lentiviruses expressing scrambled shRNA or shRNAs targeting uPAR at different loci (KD#1 and KD#4). uPAR mRNA levels (normalized to GAPDH) determined by quantitative real-time PCR (A) and uPAR protein levels determined by immunoblotting (B) in uPAR knockdown cells were both significantly reduced. (C) uPAR suppression impedes the growth of S18 and 5-8F cells as determined by MTS assays. *P < 0.01 for uPAR KD#1 and KD#4 compared with the scrambled controls. (D) uPAR suppression reduces the number of colonies formed with S18 and 5-8F cells. *P < 0.0001 relative to the scrambled controls. (E) uPAR suppression remarkably attenuates S18 and 5-8F cell migration and invasion as evaluated by the Transwell assays. *P < 0.001, #P < 0.05 compared with the scrambled controls. Photomicrographs are 100× (right panel). (F) Immunoblotting of EMT markers reveals elevated E-cadherin expression as well as decreased N-cadherin and vimentin expression upon uPAR suppression in NPC cells (vimentin expression in 5-8F cells was not affected by uPAR suppression). The data are presented as the mean ± SD of triplicate replicates.
uPAR overexpression promotes NPC cell growth, colony formation, migration, and invasion
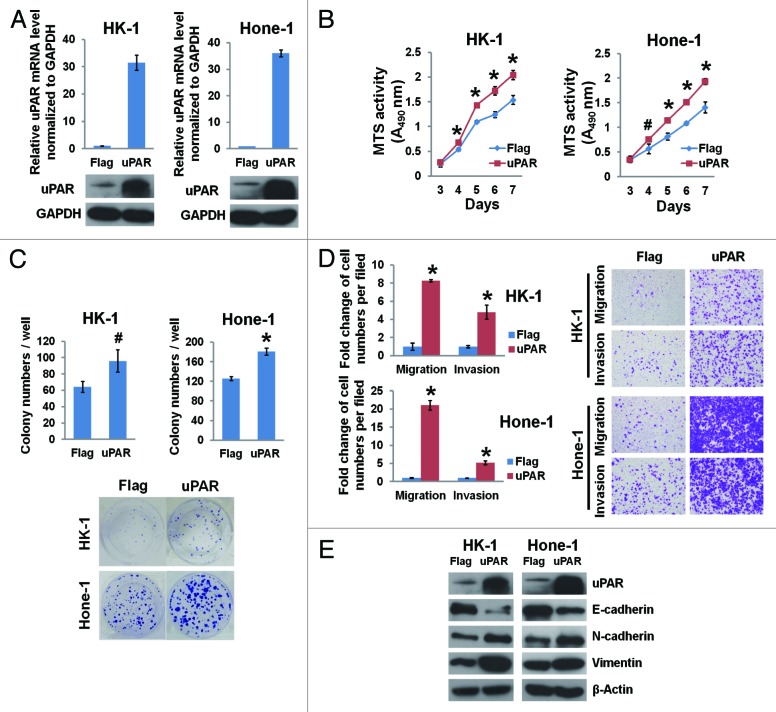
We also generated cell lines (HK-1 and Hone-1) stably overexpressing uPAR; uPAR mRNA and protein expression was determined by real-time PCR and immunoblotting (Fig. 3A). Increased uPAR levels in NPC cells resulted in accelerated growth (Fig. 3B), increased colony formation (Fig. 3C), and enhanced migration and invasion capabilities (Fig. 3D).
Figure 3. uPAR overexpression promotes NPC cellular growth, colony formation, migration, invasion, and the EMT. The lowly metastatic HK-1 and Hone-1 cells were transduced with lentiviruses expressing uPAR cDNA or a Flag sequence as a control. (A) The efficacy of uPAR overexpression in NPC cells was determined by quantitative real-time PCR and immunoblotting. (B) uPAR overexpression accelerates the growth of HK-1 and Hone-1 cells as determined by MTS assays. *P < 0.01, #P < 0.05 compared with the Flag controls. (C) uPAR overexpression increases the number of colonies formed with HK-1 and Hone-1 cells. #P < 0.05, *P = 0.00029 relative to the Flag controls. (D) uPAR overexpression notably enhances NPC cell migration and invasion as determined by the Transwell assays. *P < 0.002 relative to the Flag controls. Photomicrographs are 100× (right panel). (E) Immunoblotting of EMT markers indicates reduced E-cadherin levels as well as increased N-cadherin and vimentin levels in the uPAR-overexpressing NPC cells. The data are presented as the mean ± SD of triplicate replicates.
uPAR promotes the epithelial–mesenchymal transition in NPC cells
The epithelial–mesenchymal transition (EMT) is a crucial process implicated in the development and differentiation of numerous tissues and organs, and it also promotes cancer progression through a variety of mechanisms.30 Given the increased expression of mesenchymal markers and loss of epithelial markers, the EMT endows cancer cells with enhanced migratory and invasive capabilities.31,32 In our study, we observed that uPAR suppression in highly metastatic NPC cells (S18 and 5-8F) results in upregulation of the epithelial marker E-cadherin as well as downregulation of the mesenchymal markers N-cadherin and vimentin (Fig. 2F). On the other hand, in lowly metastatic NPC cells (HK-1 and Hone-1) overexpressing uPAR, E-cadherin protein levels were downregulated, whereas N-cadherin and vimentin protein levels were upregulated (Fig. 3E).
uPAR promotes NPC cell growth and metastasis in vivo
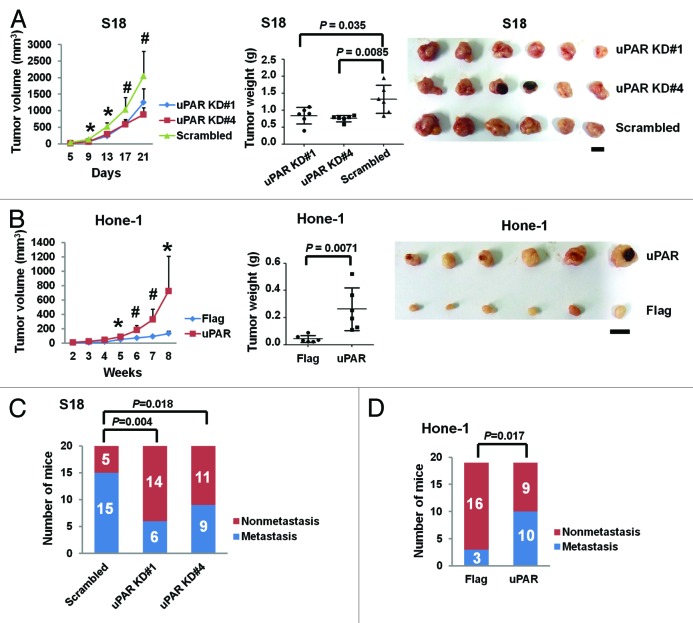
To further confirm that uPAR is crucial for in vivo regulation of NPC growth and metastasis, we utilized an animal xenograft model and lymph node (LN) metastasis model. The tumor growth curves derived from the xenograft experiments indicate that uPAR knockdown impedes NPC cell growth in nude mice (Fig. 4A, left). In contrast, uPAR overexpression promotes tumor growth (Fig. 4B, left). The final tumor weights and the photographs of isolated tumors are shown in Figure 4A and B (middle and right), respectively.
Figure 4. uPAR expression promotes the growth and metastasis of NPC tumors in vivo. (A) The uPAR knockdown S18 (#1 and #4) cells and the scrambled control cells were subcutaneously injected into nude mice. The growth curve (left) indicates S18 growth suppression upon uPAR knockdown in vivo. The terminal tumor weights (middle) are also decreased compared with the scrambled controls. The photographs of isolated tumors are displayed on the right. *P < 0.001, #P < 0.05 for uPAR KD cells relative to the scrambled control (Student t test). The results are presented as the mean ± SD, n = 6 per group. Scale bar, 1 cm. (B) The Hone-1 cells expressing uPAR cDNA display increased growth rates (left) and terminal tumor weights (middle) compared with the Flag control in vivo. Photograph of isolated tumors are displayed on the right. *P < 0.05, #P < 0.01 relative to the Flag control (Student t test). The results are presented as the mean ± SD, n = 6 per group. Scale bar, 1 cm. (C) The in vivo metastasis rates of uPAR knockdown S18 cells. The proportion of popliteal LN metastases was significantly reduced upon uPAR knockdown in S18 cells (n = 20 per group). (D) The in vivo metastasis rates of uPAR-overexpressing Hone-1. The metastasis rate was significantly increased upon uPAR overexpression in Hone-1 cells (n = 19 per group). P values were calculated using a chi-square test.
The spontaneous metastasis experiments indicate that the popliteal LN metastasis rate was significantly reduced from 75% (15/20) to 30% (6/20) or 45% (9/20) via suppression of uPAR expression in NPC cells (Fig. 4C). However, uPAR overexpression enhances metastasis as the metastasis rate increased from 16% (3/19) to 53% (10/19) (Fig. 4D).
The JAK–STAT pathway is implicated in uPAR signaling in NPC cells
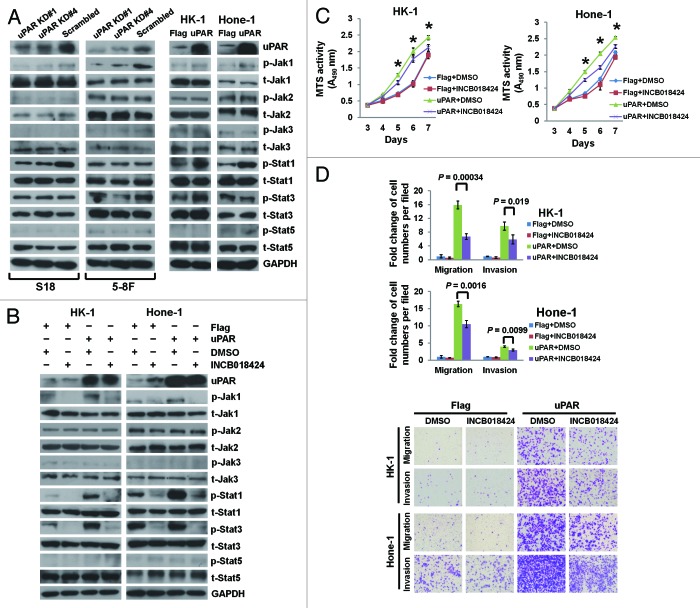
The PLAU signaling pathway network derived from the Metacore connectivity analysis suggests that the Jak1–Stat1 pathway is likely implicated in uPAR downstream signaling (Fig. S1). Therefore, we first examined the phosphorylated and total protein levels of Jak1 and Stat1 in different NPC cell lines. The highest expression of p-Jak1 and p-Stat1 proteins was observed in S18 cells with the highest expression level of uPAR (Fig. S2). Subsequently, we determined the expression of various proteins in the JAK–STAT pathway after uPAR downregulation and overexpression by immunoblotting. uPAR knockdown in S18 and 5-8F cells reduced p-Jak1 levels. In addition, we observed decreased p-Stat1 and p-Stat3 in S18 and 5-8F cells, respectively (Fig. 5A, left). On the contrary, uPAR overexpression in HK-1 and Hone-1 cells upregulated p-Jak1 expression and subsequently enhanced the expression of both p-Stat1 and p-Stat3 in HK-1 cells and p-Stat1 in Hone-1 cells (Fig. 5A, right). Additional JAK–STAT pathway components, including Jak2, Jak3, and Stat5, were also detected; however, the phosphorylated and total protein levels were unaltered (Fig. 5A). The GeneGo pathway network analysis also implicated AKT and ERK in uPA–uPAR signaling (Fig. S1). AKT and ERK are also reported to be involved in uPAR signaling.33-36 However, immunoblotting analyses showed that uPAR expression exerted no demonstrable effect on AKT or ERK in NPC cells (Fig. S3).
Figure 5. uPAR promotes NPC cellular growth, migration, and invasion in part through activation of the JAK–STAT pathway. (A) Immunoblotting was performed with antibodies related to the JAK–STAT pathway in whole-cell lysates from S18 and 5-8F cells expressing shRNAs targeting uPAR or scrambled control (left panel) or HK-1 and Hone-1 cells expressing uPAR or Flag sequences (right panel). (B) Immunoblots of whole-cell lysates from HK-1 and Hone-1-expressing uPAR or Flag sequences after pretreatment with 3 μM INCB018424 (Jak1/Jak2 inhibitor) for 48 h. (C) The growth curves of NPC cells expressing uPAR or Flag sequences as assessed by the MTS assay after pretreatment with INCB018424. *P < 0.05 for uPAR-overexpressing cells treated with INCB018424 compared with DMSO treatment. (D) Migration and invasion in NPC cells expressing uPAR or Flag sequences as evaluated by the Transwell assays after pretreatment with INCB018424. Photomicrographs are 100× (below). The data are presented as the mean ± SD of triplicate replicates. P values were calculated using the Student t test.
uPAR promotes NPC cell growth and motility in part through JAK–STAT pathway activation
INCB018424, a potent Jak1/Jak2 inhibitor, effectively suppresses p-Jak1 expression in NPC cells without affecting p-Jak2 and t-Jak2 (Fig. 5B). Moreover, INCB018424-mediated p-Jak1 suppression results in reduced Stat1 and Stat3 phosphorylation in HK-1 cells as well as reduced Stat1 phosphorylation in Hone-1 cells (Fig. 5B). In vitro functional assays indicate that enhanced NPC cell growth, migration, and invasion caused by uPAR overexpression are partially suppressed by INCB018424 treatment (Fig. 5C and D). Nevertheless, the EMT induced by uPAR overexpression is not reversed by inhibition of the JAK–STAT pathway in NPC cells (Fig. S4). In addition, the highly metastatic S18 cells with the highest level of uPAR displayed impaired migration and invasion upon suppression of p-Jak1 and p-Stat1 after treatment with INCB018424, whereas the motility of its parental cell line CNE-2, which expressed lower level of uPAR protein, was not affected by the treatment (Fig. S5). Although E-cadherin protein levels were upregulated upon inhibition of JAK–STAT pathway, N-cadherin and vimentin protein levels were not altered both in S18 and CNE-2 cells (Fig. S5A, middle).
uPAR-promoted cellular growth and motility occurs partially independently of uPA
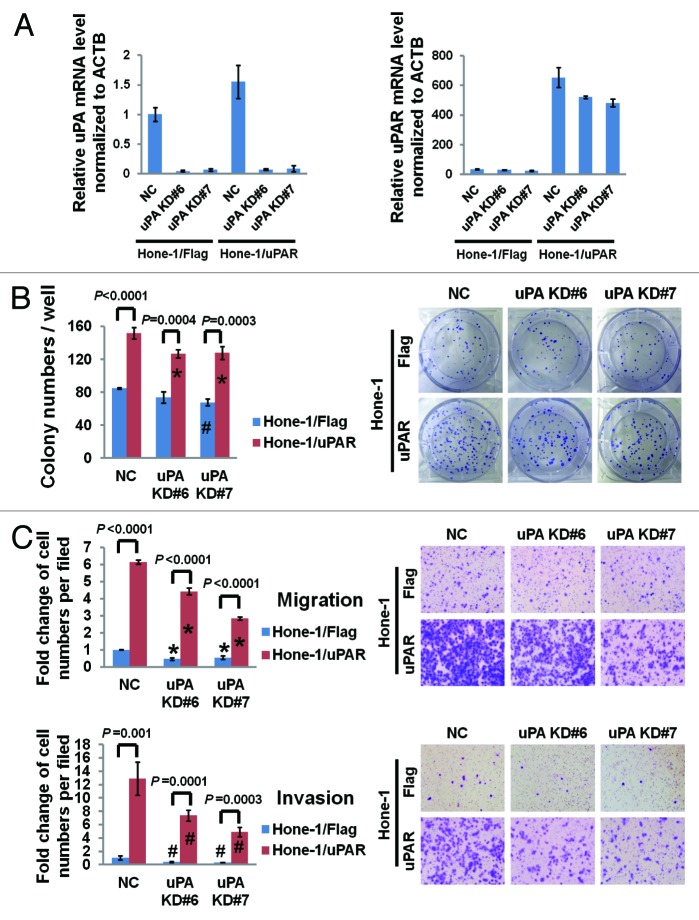
To further clarify the extent that uPAR depends on uPA in NPC cell regulation, we used siRNAs to suppress uPA expression (uPA KD#6 and #7) in uPAR-overexpressing Hone-1 cells. The uPA suppression efficacy was determined by real-time PCR detection of uPA mRNA levels (Fig. 6A). Interestingly, we found that uPA expression increased by approximately 50% after uPAR overexpression in Hone-1, whereas uPAR expression also partially decreased (in Hone-1/uPAR cells) after uPA downregulation (Fig. 6A). Colony formation was slightly reduced upon uPA suppression in both the uPAR-overexpressing Hone-1 cells and the Flag control cells (Fig. 6B), indicating that uPA may play a limited role in NPC cell growth. With regard to cellular motility, the reduction of migratory and invasive cells was more pronounced in uPAR-overexpressing cells after uPA knockdown compared with the control (Fig. 6C). However, despite the noticeably impaired motility upon uPA suppression, the uPAR-overexpressing cells still displayed significantly enhanced migration and invasion compared with the Flag control cells (Fig. 6C). Similarly, we suppressed uPA expression in CNE-2 and S18 and observed more obvious impairment of migration and invasion in S18 compared with CNE-2 (Fig. S6). Moreover, S18 still kept stronger motility than CNE-2 even lacking of uPA (Fig. S6). Taken together, we may deduce that uPAR-mediated regulation of NPC cell migration and invasion is not exclusively dependent on uPA. Additionally, immunoblotting revealed that uPA downregulation had a mild effect on uPAR-induced EMT in Hone-1 (Fig. S7A); suppression of uPA in S18 cells resulted in a mild alteration of E-cadherin and vimentin levels, which was not observed in CNE-2 cells (Fig. S7B). All of these data suggested that uPA could only induce a mild effect of uPAR-induced EMT in NPC cells.
Figure 6. uPAR promotes NPC cell colony formation and motility partially independent of uPA. (A) The relative uPA and uPAR mRNA levels (normalized to ACTB) in uPAR-overexpressing Hone-1 cells after uPA knockdown. The data are compared with Hone-1/Flag uPA KD NC. (B) The colony formation assay was performed to determine the effect of uPA knockdown on the growth of uPAR overexpressing Hone-1 and control cells. *P < 0.05 relative to Hone-1/uPAR uPA KD NC cells, #P = 0.002 compared with Hone-1/Flag uPA KD NC cells. (C) The impact of uPA knockdown on the migration and invasion of uPAR overexpressing Hone-1 cells as determined by the Transwell assays. *P < 0.005, #P < 0.05 relative to parental uPA knockdown NC controls. Photomicrographs are 100× (right panel). The data are presented as the mean ± SD of triplicate replicates.
Discussion
uPAR is overexpressed in numerous human cancers, including solid tumors, leukemias, and lymphomas.17 Increased uPAR expression frequently indicates poor prognosis. Moreover, numerous in vitro and in vivo models have shown that uPAR regulates cancer cell proliferation, migration, and invasion through the induction of extracellular matrix proteolysis and cell signaling.16,17 However, minimal research has been conducted to elucidate the role of uPAR in human NPC. Coincidently, our data from the genome-wide expressing profiling of 18 NPC tissues vs. 18 non-cancerous nasopharyngeal tissues and subsequent GeneGo Metacore pathway analysis indicates that uPAR signaling is potentially implicated in NPC progression. Additional in vitro and in vivo functional studies confirmed that uPAR promotes NPC cancer cell growth, migration, invasion, and metastasis, and activation of JAK/STAT signaling is probably one of the most significant pathways involved in this process.
In addition to JAK-STAT, several other pathways, including Src, ERK/MAPK, PI3K-AKT and the Rho family small GTPase Rac, are also implicated in uPAR-induced downstream signaling.17 Different pathways are likely to be simultaneously activated by uPAR, and may have crosstalk with each other. For instance, Src activity was found to be essential for activation of both ERK and Rac by uPAR in human breast (MCF-7) and colon (BE) cancer cells.37,38 Although our study demonstrated that AKT and ERK are not affected upon uPAR expression, other untested pathways are probably involved in uPAR-induced signalings besides JAK–STAT pathway in NPC cells. This speculation is based on our results that suppression of JAK–STAT signaling failed to completely reverse the enhanced motility induced by uPAR overexpression in Hone-1.
Many of studies have demonstrated that uPAR could activate cellular signaling and induce cell motility in the absence of uPA through the binding of vitronectin,27,28,38 suggesting that uPA may not be indispensable to activate signaling. Moreover, uPAR functionally interacts with a group of proteins to activate various intracellular signalings encompassing receptor tyrosine kinases, integrins, G-protein-coupled receptors, caveolin, and so on.16 Results of our study revealed that uPA do have an effect on motility of NPC cells, especially in uPAR-overexpressing cells. However, the enhanced motility promoted by uPAR expression cannot be completely abolished upon knockdown of uPA, suggesting that uPAR may interact with other ligands or transmembrane proteins to activate intracellular signaling, further to regulate motility of NPC cells.
uPAR induces the EMT in breast cancer36,39 and medulloblastoma cells,40 thereby promoting cancer cell migration, invasion, and metastasis. In our study, we also observed that uPAR promotes the EMT in NPC cell lines, thus we hypothesize that the EMT may be a possible mechanism utilized by uPAR to regulate NPC cell migration and invasion. However, inhibition of the JAK–STAT pathway by INCB018424 does not alter the expression of EMT marker proteins, indicating that JAK–STAT signaling is not involved in uPAR-induced EMT in NPC cells. A mild effect of uPA on the uPAR-induced EMT was also observed in our study, further confirming that uPAR was able to promote NPC cellular motility in a way partially dependent on uPA.
Given the influential role of the uPA/uPAR system in the progression of numerous malignancies, uPA/uPAR targeting is a promising therapeutic approach. Several strategies have been suggested for targeting the uPA/uPAR system, including the reduction of uPA or uPAR expression, inhibition of interactions between uPAR and uPA/vitronectin/integrins, suppression of uPA activity, and utilization of recombinant toxins to target uPAR-upregulated cells.41 Moreover, these approaches have demonstrated preclinical efficacy by impeding growth, invasion, or metastasis in prostate, breast, thyroid, and other cancers in vitro and in vivo.42-45 Another benefit of uPA/uPAR targeting is that the therapy is predicted to have tolerable side effects, because uPAR signaling is not essential for fertility or survival under physiological conditions.42 Our study is the first to provide solid evidence for uPAR as a potential therapeutic target in NPC.
In conclusion, we demonstrate that uPAR plays a crucial role in regulating NPC cell growth, motility, and metastasis via the JAK–STAT pathway and EMT induction. However, many questions remain: Does uPAR expression correlate with the clinical stages of NPC in human tissues? Does uPAR serve as a prognostic marker in NPC patients? Which ligand and co-receptor are involved in uPAR-mediated activation of the JAK–STAT pathway? What role does uPAR-induced proteolysis play in NPC progression? We hope future research efforts will explore these important questions and build a solid foundation for clinically targeting uPAR signaling in the prevention and treatment of NPC.
Materials and methods
Cell culture
The human nasopharyngeal carcinoma cell lines HK-1, Hone-1, CNE-2, and its highly metastatic clone S18,46,47 and SUNE-1 and its highly metastatic clone 5-8F,48 were maintained in Dulbecco modified Eagle medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS; Gibco) in a humidified atmosphere containing 5% CO2 at 37 °C. HK-1 is a well-differentiated squamous carcinoma cell line, whereas the other cell lines are poorly differentiated and represent the majority of patients in endemic areas.
Antibodies and reagents
The following antibodies were used: rabbit anti-uPAR from Abgent (AP8156c); rabbit anti-ERK1/2 from Proteintech (51068-1-AP); rabbit anti-uPA and rabbit anti-p-ERK1 (Thr202)/ERK2 (Thr185) from Epitomics (P27361); rabbit anti-phospho-Jak1 (pY1022/1023) from Abcam (ab138005); rabbit anti-p-Jak2 (pYpY1007/1008; 44–426G), rabbit anti-p-Stat3 (pY705; 44–380G), and rabbit anti-p-Stat5 (pY694; 44–390G) from Invitrogen; rabbit anti-E-cadherin (#3195), N-cadherin (#4061), vimentin (#5741), total-Jak1 (#3344), t-Jak2 (#3230), t-Jak3 (#8827), t-Stat1 (#9175), t-Stat5 (#9358), t-AKT (#4691), p-Jak3(Tyr980/981; #5031), p-Stat1(Tyr701; #9167), GAPDH (glyceraldehyde-3-phosphate dehydrogenase; #2118), and β-Actin (#4967) as well as mouse anti-t-Stat3 (#9139) and p-AKT(Ser473; #4051) from Cell Signaling Technology. The anti-mouse (W4021) and anti-rabbit (W4011) peroxidase-conjugated secondary antibodies were obtained from Promega. The Jak1/Jak2 inhibitor INCB018424 was purchased from Selleck (S1378).
Cell proliferation assay
The colorimetric MTS assays (CellTiter 96 Aqueous One Solution Cell Proliferation Assay; Promega; G3582) were performed to determine cancer cell growth and viability as previously reported.47,49 Briefly, 1 × 103 cells/well were seeded in triplicate on 96-well culture plates. Parallel culture plates were harvested at various times post-seeding, and 20 μL MTS solution was added to each well. The solution was incubated with the cells for an additional hour, and the optical density (OD) value was measured at 490 nm. All experiments were independently repeated at least thrice.
Colony-formation assay
Cells were seeded in triplicate in 6-well culture plates at a density of 200 cells/well (for S18 and 5-8F) or 500 cells/well (for HK-1 and Hone-1). The culture medium was subsequently changed every 3 d. After 2 wk, the resulting colonies were fixed with methanol and stained with 0.1% crystal violet. Colonies that contained greater than 50 cells were counted.50 All experiments were independently repeated at least thrice.
Migration and invasion assays (Transwell assays)
The migration and invasion assays were performed using cell culture inserts with 8-μm pore transparent polyethylene terephthalate filters (Becton Dickinson) coated with (for invasion assays) or without (for migration assays) Matrigel. Cells suspended in 200 μL serum-free DMEM were added to the insert; 800 μL DMEM containing 10% FBS was added to the bottom chamber. After incubation for 24 h at 37 °C, the cells on the upper filter were removed, and the cells that migrated or invaded the lower surface of the membrane were fixed in methanol and stained with crystal violet. Five optical fields (100× magnification) from each filter with triplicate inserts were randomly selected to calculate the number of migrated or invaded cells. For CNE-2, S18, and 5-8F, 3 × 104 cells were plated into each insert; for HK-1 and Hone-1, 8 × 104 cells were plated. The experiments were performed independently thrice.
Immunoblotting
The cells were lysed in RIPA buffer (100 mM TRIS-HCl, 300 mM NaCl, 2% NP40, 0.5% sodium deoxycholate) supplemented with the cOmplete Mini Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail (Roche). Protein concentrations were assayed using the BCA™ Protein Assay Kit (Pierce Biotechnology). Equal amounts of protein mixed with sample loading buffer (5×; Beyotime) were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). After blocking with 5% skim milk or BSA in Tris-buffered saline tween-20 (TBST), the membranes were incubated with the primary antibodies overnight at 4 °C and then the horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. The protein bands were visualized using a chemiluminescence kit (Pierce). GAPDH and β-Actin were used as controls for equal protein loading.
Quantitative real-time PCR
Total RNA was extracted from cells using the TRIzol reagent (Invitrogen) and subjected to reverse transcription using the iScript™ cDNA Synthesis Kit (Bio-Rad). The threshold cycle (Ct) value of each sample was determined using the SsoFast EvaGreen supermix (Bio-Rad) on a CFX96 real-time PCR detection system (Bio-Rad). GAPDH or ACTB served as the normalization genes for these studies. The relative expression levels of the target genes were calculated as two power values of ΔCt (the Ct of GAPDH or ACTB minus the Ct of the target gene). The sequence of the PCR primers used for uPA, uPAR, HPRT, GAPDH, and ACTB amplification are as follows:
uPA forward: 5′-GCCACACACT GCTTCATTGA-3′;
uPA reverse: 5′-TATACATCGA GGGCAGGCAG-3′;
uPAR forward: 5′-GCCTTACCGA GGTTGTGTGT-3′;
uPAR reverse: 5′-CATCCAGGCA CTGTTCTTCA-3′;
GAPDH forward: 5′-CTCATGACCA CAGTCCATGC-3′;
GAPDH reverse: 5′-CAGTGAGCTT CCCGTTCAG-3′;
ACTB (human) forward: 5′-CATCCGCAAA GACCTGTACG-3′;
ACTB (human) reverse: 5′-CCTGCTTGCT GATCCACATC-3′.
All PCR amplification reactions were performed in triplicate using optimized conditions recommended by the manufacturer.
siRNA transfection
The targets of the uPA siRNAs (Qiagen) are 5′-CCGCATGACT TTGACTGGAA T-3′ (KD#6) and 5′-GAGCTGGTGT CTGATTGTTA A-3′ (KD#7). The negative control siRNA (NC) was purchased from GenePharma. Transfections were performed using the Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer’s instruction. Briefly, the siRNAs and transfection reagent diluted in Opti-MEM Medium (Invitrogen) were mixed and incubated at room temperature for 5 min. Next, the siRNA–reagent complex was added to the cells, which had already grown to 60% confluency on a 6-well plate. The gene-silencing efficacy was evaluated 48 h after transfection using real-time PCR and immunoblotting. For the colony formation and migration/invasion assays, the cells were seeded in 6-well plates or transwell inserts at 72 h post-transfection.
Lentiviral transduction studies
Lentivirus-expressing uPAR shRNAs were produced by cotransfection of 293T cells with pLKO.1 vectors carrying the shRNAs (MISSON shRNA plasmid DNAs were purchased from Sigma-Aldrich; the TRC numbers for the uPAR KD#1 and KD#4 plasmids are TRCN0000306814 and TRCN0000296118, respectively.) and the MISSION lentiviral packaging mix (Sigma-Aldrich) using the X-tremeGENE HP DNA transfection reagent (Roche). A MISSION non-target shRNA control vector served as the scrambled control (Sigma-Aldrich, SH002). Infectious lentiviruses were harvested, concentrated using the Lentivirus concentration PL liquid (Biomiga), and filtered through 0.45-μm filters (Millipore). Next, S18 and 5-8F cells were transduced with the lentiviral particles and then cultured in medium containing 5 μg/ml puromycin (Sigma-Aldrich) for 5 d to select for successfully transduced cells. Real-time PCR and immunoblotting were performed to evaluate uPAR-knockdown efficiency.
To generate the cell lines (HK-1 and Hone-1) stably overexpressing uPAR, the uPAR coding sequence was cloned into a lentiviral vector using the Gateway recombination cloning system (Invitrogen) according to the manufacturer’s instructions. A homologous vector carrying a Flag sequence served as the control. The lentiviruses were packaged and transduced into cells as described above. The pMDLg-PRRE, pMD2.G, and pRSV-REV packaging plasmids were used for the overexpression studies, and 2 μg/mL puromycin was used for selection. Real-time PCR and immunoblotting were also performed to evaluate the efficiency of uPAR overexpression.
Animal experiments
Female athymic mice (BALB/c nu/nu; Guangdong Medical Laboratory Animal Center) were purchased at 4–5-wk-of-age and maintained under a specific pathogen-free environment. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Sun Yat-Sen University Cancer Center.
For the tumor xenograft experiments, the tumor cells (1 × 106 cells/tumor in 100 μL DMEM) were subcutaneously injected into the right flanks of the nude mice. The tumor width (W) and length (L) were measured every 4 d (S18) or weekly (Hone-1). The mice were euthanized 3 (S18) or 8 wk (Hone-1) after inoculation, and the tumors were isolated and weighed. The tumor volume was calculated as volume = π/6 × L × W2.51
The spontaneous lymph node (LN) metastasis experiments were conducted as previously reported.46,47 Briefly, 1 × 105 cells in 20 μL DMEM were subcutaneously injected into the footpad of the left hind limb of each mouse to generate a primary tumor. After 4 wk (S18) or 16 wk (Hone-1), the experiments were terminated, and the popliteal LNs of the left hind feet were isolated and preserved in RNAlater solution (Invitrogen). The primary tumor weight was measured and calculated by subtracting the weight of the contralateral foot without the tumor from the weight of the foot carrying the tumor. The LNs were homogenized in TRIzol for total RNA extraction using the Bullet Blender (Next Advance). Reverse transcription and real-time PCR were performed to assess metastasis using specific primers for human HPRT that do not cross-react with the corresponding mouse gene.52 The following human and mouse primers were used:
HPRT forward: 5′-TTCCTTGGTC AGGCAGTATA ATCC-3′;
HPRT reverse: 5′-AGTCTGGCTT ATATCCAACA CTTCG-3′;
ACTB (universal for human and mouse) forward: 5′- CAATGAGCTG CGTGTGGC-3′;
ACTB (universal for human and mouse) reverse: 5′-CGTACATGGC TGGGGTGTT-3′.
Patients and tissue samples
The human tissue samples used in the genome-wide expression profiling and quantitative real-time PCR were obtained from the Department of Nasopharyngeal Carcinoma, Sun Yat-sen University Cancer Center (SYSUCC); written informed consent from the patients and approval from the Institutional Clinical Ethics Review Board at SYSUCC were obtained. Twenty-eight qualified primary NPC samples from patients with undifferentiated nasopharyngeal and 27 non-cancerous nasopharyngeal tissues from patients with non-cancerous nasopharyngeal chronic inflammation were collected from December 2007 to April 2010. The patient median age was 46 y (range, 19–77) for the NPC patients and 45 y (range, 18–78) for the non-cancerous cohort. Almost one-third of the patients were female. The samples were collected prior to the administration of anti-cancer treatment. Eighteen NPC tissues and 18 non-cancerous tissues were used for genome-wide expression profiling, and the remaining tissues were used to validate uPAR expression levels.
Genome-wide expression profiling, pathway analysis and statistical analysis
Genome-wide expression profiling of 18 primary NPC tumors vs. 18 non-cancerous nasopharyngeal tissues was performed using the Agilent 4 × 44 K human whole-genome microarray that contains 41 091 human genes or transcripts, and this study was entrusted to Shanghai Biochip Co, Ltd. In brief, total RNA was extracted from the samples using the TRIzol reagent (Invitrogen) and purified using an RNeasy kit (Qiagen). Next, the RNA amount and quality was evaluated. Qualified RNA was reverse transcribed into cDNA, which was subsequently transcribed into cRNA and labeled with fluorescent dyes. The labeled cRNA was purified, mixed with hybridization buffer and hybridized to the microarrays according to the manufacturer’s recommendations. Finally, the arrays were washed and scanned using the Agilent microarray scanner. One-way ANOVA was used to identify genes differentially expressed between the primary NPC and non-cancerous nasopharyngeal tissues. The gene expression profiling statistical analyses were performed in the R statistical environment using the Bioconductor project version 1, and a P value < 0.05 was considered statistically significant. All microarray data have been submitted to the Gene Expression Omnibus with an accession number GSE53819.
In total, 2992 genes achieving the criteria of FC > 2 and P value < 0.05 were identified as differentially expressed between primary NPC tumors and non-cancerous nasopharyngeal tissues. Unsupervised clustering analysis of the expression of 41091 genes in all 36 cases demonstrated that samples used in this study were suitable for subsequent analyses, given that NPC and control cases clustered as 2 distinct groups in accordance with their original histological types (Fig. 1A).
Furthermore, potential signaling pathways involved with these 2992 differentially expressed genes were explored using the system biology tool MetaCore from GeneGo (GeneGo Inc). The GeneGo pathway maps (Fig. 1B) and networks (Fig. S1) for these genes were obtained based on the microarray data sets.
For additional statistical analyses, the Student t test and chi-square test were used for comparisons with the 2 independent groups of data. A P value < 0.05 was recognized as statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by grants from The National Natural Science Foundation of China (No. 81030043 and No. 81272340), the National High Technology Research and Development Program of China (863 Program) (No. 2012AA02A501), and the Sci-Tech Project Foundation of Guangdong Province (No. 2012B031800364).
References
- 1.Cao SM, Simons MJ, Qian CN. The prevalence and prevention of nasopharyngeal carcinoma in China. Chin J Cancer. 2011;30:114–9. doi: 10.5732/cjc.010.10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kataki AC, Simons MJ, Das AK, Sharma K, Mehra NK. Nasopharyngeal carcinoma in the Northeastern states of India. Chin J Cancer. 2011;30:106–13. doi: 10.5732/cjc.010.10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adham M, Kurniawan AN, Muhtadi AI, Roezin A, Hermani B, Gondhowiardjo S, Tan IB, Middeldorp JM. Nasopharyngeal carcinoma in Indonesia: epidemiology, incidence, signs, and symptoms at presentation. Chin J Cancer. 2012;31:185–96. doi: 10.5732/cjc.011.10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarmiento MP, Mejia MB. Preliminary assessment of nasopharyngeal carcinoma incidence in the Philippines: a second look at published data from four centers. Chin J Cancer. 2014;33:159–64. doi: 10.5732/cjc.013.10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad A, Stefani S. Distant metastases of nasopharyngeal carcinoma: a study of 256 male patients. J Surg Oncol. 1986;33:194–7. doi: 10.1002/jso.2930330310. [DOI] [PubMed] [Google Scholar]
- 6.Lee AW, Poon YF, Foo W, Law SC, Cheung FK, Chan DK, Tung SY, Thaw M, Ho JH. Retrospective analysis of 5037 patients with nasopharyngeal carcinoma treated during 1976-1985: overall survival and patterns of failure. Int J Radiat Oncol Biol Phys. 1992;23:261–70. doi: 10.1016/0360-3016(92)90740-9. [DOI] [PubMed] [Google Scholar]
- 7.Hsu MM, Tu SM. Nasopharyngeal carcinoma in Taiwan. Clinical manifestations and results of therapy. Cancer. 1983;52:362–8. doi: 10.1002/1097-0142(19830715)52:2<362::AID-CNCR2820520230>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 8.Huang CJ, Leung SW, Lian SL, Wang CJ, Fang FM, Ho YH. Patterns of distant metastases in nasopharyngeal carcinoma. Kaohsiung J Med Sci. 1996;12:229–34. [PubMed] [Google Scholar]
- 9.Wei WI, Mok VW. The management of neck metastases in nasopharyngeal cancer. Curr Opin Otolaryngol Head Neck Surg. 2007;15:99–102. doi: 10.1097/MOO.0b013e3280148a06. [DOI] [PubMed] [Google Scholar]
- 10.Lee AW, Sze WM, Au JS, Leung SF, Leung TW, Chua DT, Zee BC, Law SC, Teo PM, Tung SY, et al. Treatment results for nasopharyngeal carcinoma in the modern era: the Hong Kong experience. Int J Radiat Oncol Biol Phys. 2005;61:1107–16. doi: 10.1016/j.ijrobp.2004.07.702. [DOI] [PubMed] [Google Scholar]
- 11.Ploug M, Rønne E, Behrendt N, Jensen AL, Blasi F, Danø K. Cellular receptor for urokinase plasminogen activator. Carboxyl-terminal processing and membrane anchoring by glycosyl-phosphatidylinositol. J Biol Chem. 1991;266:1926–33. [PubMed] [Google Scholar]
- 12.Ploug M, Ellis V. Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins. FEBS Lett. 1994;349:163–8. doi: 10.1016/0014-5793(94)00674-1. [DOI] [PubMed] [Google Scholar]
- 13.Kjaergaard M, Hansen LV, Jacobsen B, Gardsvoll H, Ploug M. Structure and ligand interactions of the urokinase receptor (uPAR) Front Biosci. 2008;13:5441–61. doi: 10.2741/3092. [DOI] [PubMed] [Google Scholar]
- 14.Ellis V, Pyke C, Eriksen J, Solberg H, Danø K. The urokinase receptor: involvement in cell surface proteolysis and cancer invasion. Ann N Y Acad Sci. 1992;667:13–31. doi: 10.1111/j.1749-6632.1992.tb51591.x. [DOI] [PubMed] [Google Scholar]
- 15.Carmeliet P, Moons L, Lijnen R, Baes M, Lemaître V, Tipping P, Drew A, Eeckhout Y, Shapiro S, Lupu F, et al. Urokinase-generated plasmin activates matrix metalloproteinases during aneurysm formation. Nat Genet. 1997;17:439–44. doi: 10.1038/ng1297-439. [DOI] [PubMed] [Google Scholar]
- 16.Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–43. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 17.Smith HW, Marshall CJ. Regulation of cell signalling by uPAR. Nat Rev Mol Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 18.Jacobsen B, Ploug M. The urokinase receptor and its structural homologue C4.4A in human cancer: expression, prognosis and pharmacological inhibition. Curr Med Chem. 2008;15:2559–73. doi: 10.2174/092986708785909012. [DOI] [PubMed] [Google Scholar]
- 19.Rasch MG, Lund IK, Almasi CE, Hoyer-Hansen G. Intact and cleaved uPAR forms: diagnostic and prognostic value in cancer. Front Biosci. 2008;13:6752–62. doi: 10.2741/3186. [DOI] [PubMed] [Google Scholar]
- 20.Stark GR, Darnell JE., Jr. The JAK–STAT pathway at twenty. Immunity. 2012;36:503–14. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison DA. The Jak/STAT pathway. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vera J, Rateitschak K, Lange F, Kossow C, Wolkenhauer O, Jaster R. Systems biology of JAK–STAT signalling in human malignancies. Prog Biophys Mol Biol. 2011;106:426–34. doi: 10.1016/j.pbiomolbio.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Dumler I, Weis A, Mayboroda OA, Maasch C, Jerke U, Haller H, Gulba DC. The Jak/Stat pathway and urokinase receptor signaling in human aortic vascular smooth muscle cells. J Biol Chem. 1998;273:315–21. doi: 10.1074/jbc.273.1.315. [DOI] [PubMed] [Google Scholar]
- 24.Koshelnick Y, Ehart M, Hufnagl P, Heinrich PC, Binder BR. Urokinase receptor is associated with the components of the JAK1/STAT1 signaling pathway and leads to activation of this pathway upon receptor clustering in the human kidney epithelial tumor cell line TCL-598. J Biol Chem. 1997;272:28563–7. doi: 10.1074/jbc.272.45.28563. [DOI] [PubMed] [Google Scholar]
- 25.Nusrat AR, Chapman HA., Jr. An autocrine role for urokinase in phorbol ester-mediated differentiation of myeloid cell lines. J Clin Invest. 1991;87:1091–7. doi: 10.1172/JCI115070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei Y, Waltz DA, Rao N, Drummond RJ, Rosenberg S, Chapman HA. Identification of the urokinase receptor as an adhesion receptor for vitronectin. J Biol Chem. 1994;269:32380–8. [PubMed] [Google Scholar]
- 27.Kjøller L, Hall A. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J Cell Biol. 2001;152:1145–57. doi: 10.1083/jcb.152.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen CD, Ferraris GM, Andolfo A, Cunningham O, Sidenius N. uPAR-induced cell adhesion and migration: vitronectin provides the key. J Cell Biol. 2007;177:927–39. doi: 10.1083/jcb.200612058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasi F, Sidenius N. The urokinase receptor: focused cell surface proteolysis, cell adhesion and signaling. FEBS Lett. 2010;584:1923–30. doi: 10.1016/j.febslet.2009.12.039. [DOI] [PubMed] [Google Scholar]
- 30.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 31.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–37. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ossowski L, Aguirre-Ghiso JA. Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr Opin Cell Biol. 2000;12:613–20. doi: 10.1016/S0955-0674(00)00140-X. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Aguirre Ghiso J, Estrada Y, Ossowski L. EGFR is a transducer of the urokinase receptor initiated signal that is required for in vivo growth of a human carcinoma. Cancer Cell. 2002;1:445–57. doi: 10.1016/S1535-6108(02)00072-7. [DOI] [PubMed] [Google Scholar]
- 35.Gondi CS, Kandhukuri N, Dinh DH, Gujrati M, Rao JS. Down-regulation of uPAR and uPA activates caspase-mediated apoptosis and inhibits the PI3K/AKT pathway. Int J Oncol. 2007;31:19–27. [PMC free article] [PubMed] [Google Scholar]
- 36.Lester RD, Jo M, Montel V, Takimoto S, Gonias SL. uPAR induces epithelial-mesenchymal transition in hypoxic breast cancer cells. J Cell Biol. 2007;178:425–36. doi: 10.1083/jcb.200701092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen DH, Webb DJ, Catling AD, Song Q, Dhakephalkar A, Weber MJ, Ravichandran KS, Gonias SL. Urokinase-type plasminogen activator stimulates the Ras/Extracellular signal-regulated kinase (ERK) signaling pathway and MCF-7 cell migration by a mechanism that requires focal adhesion kinase, Src, and Shc. Rapid dissociation of GRB2/Sps-Shc complex is associated with the transient phosphorylation of ERK in urokinase-treated cells. J Biol Chem. 2000;275:19382–8. doi: 10.1074/jbc.M909575199. [DOI] [PubMed] [Google Scholar]
- 38.Smith HW, Marra P, Marshall CJ. uPAR promotes formation of the p130Cas-Crk complex to activate Rac through DOCK180. J Cell Biol. 2008;182:777–90. doi: 10.1083/jcb.200712050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo M, Lester RD, Montel V, Eastman B, Takimoto S, Gonias SL. Reversibility of epithelial-mesenchymal transition (EMT) induced in breast cancer cells by activation of urokinase receptor-dependent cell signaling. J Biol Chem. 2009;284:22825–33. doi: 10.1074/jbc.M109.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta R, Chetty C, Bhoopathi P, Lakka S, Mohanam S, Rao JS, Dinh DE. Downregulation of uPA/uPAR inhibits intermittent hypoxia-induced epithelial-mesenchymal transition (EMT) in DAOY and D283 medulloblastoma cells. Int J Oncol. 2011;38:733–44. doi: 10.3892/ijo.2010.883. [DOI] [PubMed] [Google Scholar]
- 41.Mazar AP. The urokinase plasminogen activator receptor (uPAR) as a target for the diagnosis and therapy of cancer. Anticancer Drugs. 2001;12:387–400. doi: 10.1097/00001813-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Sidenius N, Blasi F. The urokinase plasminogen activator system in cancer: recent advances and implication for prognosis and therapy. Cancer Metastasis Rev. 2003;22:205–22. doi: 10.1023/A:1023099415940. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Cozzi PJ. Targeting uPA/uPAR in prostate cancer. Cancer Treat Rev. 2007;33:521–7. doi: 10.1016/j.ctrv.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Baldini E, Sorrenti S, D’Armiento E, Di Matteo FM, Catania A, Ulisse S. The urokinase plasminogen activating system in thyroid cancer: clinical implications. G Chir. 2012;33:305–10. [PubMed] [Google Scholar]
- 45.LeBeau AM, Duriseti S, Murphy ST, Pepin F, Hann B, Gray JW, VanBrocklin HF, Craik CS. Targeting uPAR with antagonistic recombinant human antibodies in aggressive breast cancer. Cancer Res. 2013;73:2070–81. doi: 10.1158/0008-5472.CAN-12-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian CN, Berghuis B, Tsarfaty G, Bruch M, Kort EJ, Ditlev J, Tsarfaty I, Hudson E, Jackson DG, Petillo D, et al. Preparing the “soil”: the primary tumor induces vasculature reorganization in the sentinel lymph node before the arrival of metastatic cancer cells. Cancer Res. 2006;66:10365–76. doi: 10.1158/0008-5472.CAN-06-2977. [DOI] [PubMed] [Google Scholar]
- 47.Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY, Ooi A, Peng LX, Lu WH, Zhang Z, Petillo D, et al. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 2011;71:3162–72. doi: 10.1158/0008-5472.CAN-10-3557. [DOI] [PubMed] [Google Scholar]
- 48.Song LB, Yan J, Jian SW, Zhang L, Li MZ, Li D, Wang HM. [Molecular mechanisms of tumorgenesis and metastasis in nasopharyngeal carcinoma cell sublines] Ai Zheng. 2002;21:158–62. [PubMed] [Google Scholar]
- 49.Li XJ, Peng LX, Shao JY, Lu WH, Zhang JX, Chen S, Chen ZY, Xiang YQ, Bao YN, Zheng FJ, et al. As an independent unfavorable prognostic factor, IL-8 promotes metastasis of nasopharyngeal carcinoma through induction of epithelial-mesenchymal transition and activation of AKT signaling. Carcinogenesis. 2012;33:1302–9. doi: 10.1093/carcin/bgs181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang Y, Zhong Z, Huang Y, Deng W, Cao J, Tsao G, Liu Q, Pei D, Kang T, Zeng YX. Stem-like cancer cells are inducible by increasing genomic instability in cancer cells. J Biol Chem. 2010;285:4931–40. doi: 10.1074/jbc.M109.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–54. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 52.Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.