Abstract
Epidemiological studies and clinical trials revealed that chronic consumption coffee is associated with the inhibition of several metabolic diseases as well as reduction in overall and cause-specific mortality. We show that both natural and decaffeinated brands of coffee similarly rapidly trigger autophagy in mice. One to 4 h after coffee consumption, we observed an increase in autophagic flux in all investigated organs (liver, muscle, heart) in vivo, as indicated by the increased lipidation of LC3B and the reduction of the abundance of the autophagic substrate sequestosome 1 (p62/SQSTM1). These changes were accompanied by the inhibition of the enzymatic activity of mammalian target of rapamycin complex 1 (mTORC1), leading to the reduced phosphorylation of p70S6K, as well as by the global deacetylation of cellular proteins detectable by immunoblot. Immunohistochemical analyses of transgenic mice expressing a GFP–LC3B fusion protein confirmed the coffee-induced relocation of LC3B to autophagosomes, as well as general protein deacetylation. Altogether, these results indicate that coffee triggers 2 phenomena that are also induced by nutrient depletion, namely a reduction of protein acetylation coupled to an increase in autophagy. We speculate that polyphenols contained in coffee promote health by stimulating autophagy.
Keywords: acetyl-coenzyme A, acetylation, mTOR, macroautophagy
Introduction
Large epidemiological studies involving large cohorts of individuals have demonstrated that coffee consumption is inversely associated with total and cause-specific mortality, both in males and in females. The consumption of coffee is associated with a reduction of cancer, heart disease, respiratory disease, stroke, diabetes, and infections (both in males and females).1-3 These effects are dose-dependent (with a plateau of 6 cups per day) and do not depend on caffeine content, because both decaffeinated and caffeinated coffee were similarly associated with improved health.4,5 Independent surveys revealed that coffee consumption might have protective effects, among many, on highly penetrant tumors such as endometrial,6,7 mammary8 hepatocellular,9 colorectal,10 and prostatic cancer.11 Although such studies cannot differentiate between causal and associational findings, they do suggest that chronic coffee consumption might have broad health-improving effects.
One of the general cell biological phenomena that has been attributed a global health-promoting and anti-aging property is autophagy,12-14 a lysosomal degradation pathway responsible for the selective renewal of cytoplasmic organelles.15,16 In macroautophagy (here referred to as “autophagy”), portions of the cytoplasm are sequestered in 2-membraned vesicles, the autophagosomes, which later fuse with lysosomes for the degradation of the luminal content by lysosomal hydrolases. Since autophagy preferentially targets damaged macromolecules (such as unfolded and aggregated proteins) and organelles (such as dysfunctional mitochondria),17 it contributes to ridding the cytoplasm of aged structures and hence potentially “rejuvenates” non-nuclear portions of the cell.18,19 Autophagy has also been suggested to participate in hormesis,20 which consists in the adaptation of cells to low levels of stress, rendering them resistant to otherwise lethal effects of intense stress.21,22 Several studies indicate that autophagy may act as a tumor-suppressive mechanism.23-26 Based on these premises, we investigated the possibility that coffee might induce autophagy in vivo in mice. Here, we report that both natural and decaffeinated coffee similarly induce a broad organism-wide autophagic response.
Results and Discussion
Chronic administration of non-toxic doses of coffee induces autophagy in mice
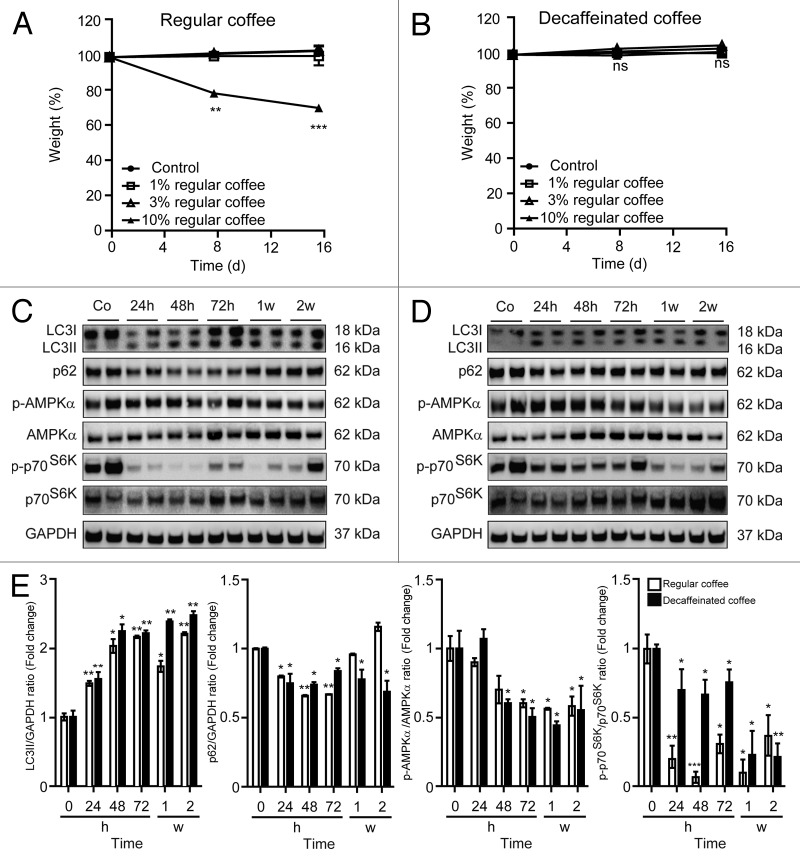
Continuous administration of 1 or 3% coffee in the drinking water for 16 d did not affect the body weight of C57Bl/6 mice. In contrast, 10% of caffeinated (but not of decaffeinated) coffee did cause weight reduction over this period, suggesting some caffeine-associated effects (Fig. 1A and B). As a result, we evaluated the capacity of chronic treatment with 3% coffee to induce autophagy in vivo. This dose is well within the range of that absorbed by coffee-consuming persons. Three percent of either caffeinated or decaffeinated coffee induced similar signs of increased autophagic flux in the liver (Fig. 1C–E), heart (Fig. S1), and muscle (Fig. S2), namely lipidation of microtubule-associated protein 1 light chain 3 β (LC3B), leading to an increase in its electrophoretic mobility in sodium dodecyl sulfate PAGE (SDS-PAGE), generating the isoform II of LC3B, accompanied by a decrease in the overall abundance of the autophagic substrate sequestosome-1 (p62/SQSTM1) (Fig. 1C–E). These results were observable as early as 24 h after beginning of the treatment and lasted for the entire 2-wk experiment, indicating that chronic exposure to coffee can induce autophagy irrespective from its caffeine content.
Figure 1. Long-term administration of regular and decaffeinated coffee at a dose not affecting body weight induces autophagy in the liver. (A and B). Effect of 2 wk administration of regular (A) or decaffeinated coffee (B) on body weight. C57BL/6 mice were administrated with the indicated doses of regular coffee (A) or decaffeinated coffee (B) diluted in drinking water, which was provided to mice ad libitum. The dose not affecting body weight (3% w/v) was designated for investigating pro-autophagic effects. Results from n = 3 independent experiments. (C and D). Immunoblotting analysis of long-term coffee administration on autophagy regulation in liver. Administration of both regular (C) and decaffeinated coffee (D) for up to 2 wk (w) resulted in activation of autophagy, as measured by LC3 lipidation and p62/SQSTM1 degradation (quantified in E). Autophagy activation was associated with a decrease in mTORC1 activity, as measured by the phosphorylation of p70s6k, but not to an activation of AMPK (quantified in E). GAPDH levels were monitored to ensure equal loading. Representative images are reported in (CandD). Results from n = 3 independent experiments are presented as fold change ± SEM *P < 0.05; **P < 0.01; ***P < 0.005 (unpaired, 2-tailed Student t test), compared with untreated mice.
Short-term administration of coffee induces autophagy in vivo
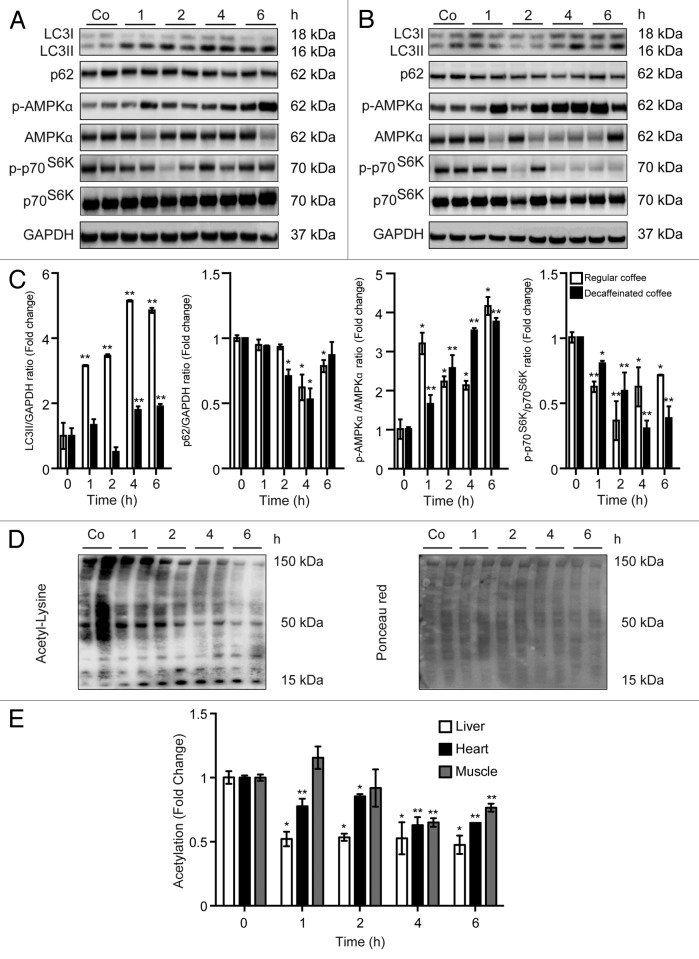
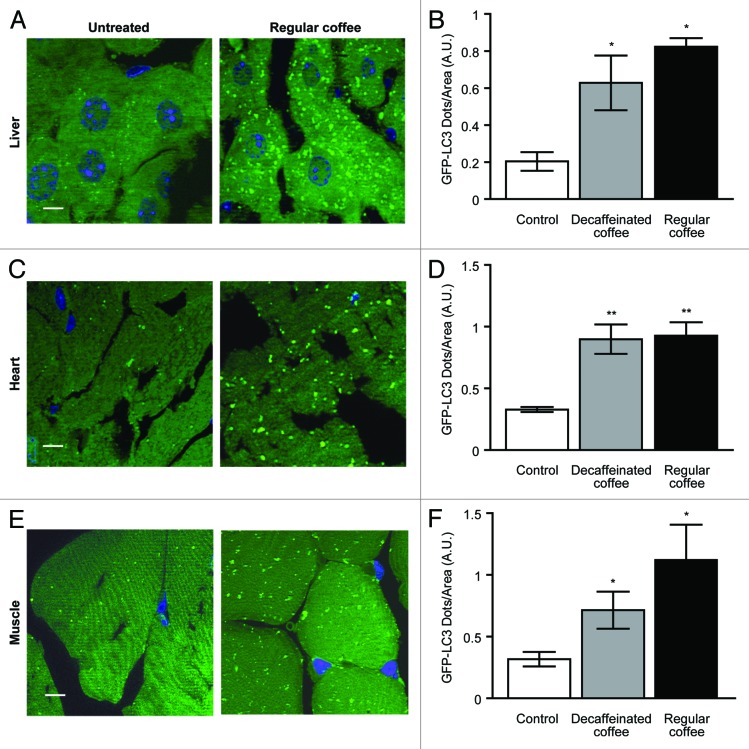
We next determined the kinetics with which coffee can stimulate autophagy in vivo. For this, coffee was administered by gavage to C57BL/6 mice, and autophagy was quantified by immunoblot determination of the conversion of native LC3B-I into lipidated, membrane-associated LC3B-II, as well as by measurements of p62/SQSTM1 degradation. Irrespective of the absence or presence of caffeine, coffee induced a rapid LC3B-I to LC3B-II conversion and a depletion of p62/SQSTM1 in the liver (Fig. 2A and B). Similarly, both regular and decaffeinated coffee induced signs of autophagy in the heart (Fig. S3) and in the muscle (Fig. S4). The kinetics of LC3B-I to LC3B-II conversion were similar in all organs. The depletion of p62/SQSTM1 occurred earlier (1 h) in the heart (Fig. S3) than in the liver (Fig. 2) and in the skeletal muscle (Fig. S4), where it was detectable only at 4 h, thus confirming an organ-specific and transcription-dependent behavior of p62, as recently described.29 To further evaluate the capacity of coffee to induce autophagy in vivo, we next took advantage of transgenic mice expressing a chimeric GFP–LC3 protein under the control of the ubiquitous CAG promoter.30 Microscopic detection of the GFP-dependent fluorescent signal allowed for the detection of coffee-induced autophagy, as GFP-LC3 relocated from a mostly diffuse distribution (in untreated animals) to cytoplasmic puncta (in treated animals). The accumulation of GFP-LC3+ puncta was observed in liver, muscle, and heart 4 h after administration of either caffeinated or decaffeinated coffee (Fig. 3), confirming the notion that coffee does induce a significant induction of autophagy in multiple organs.
Figure 2. Short-term administration of both regular coffee and decaffeinated coffee induces autophagy accompanied by a reduction in global acetylation levels of proteins in the liver. (A and B). Immunoblotting analysis of short-term coffee administration on autophagy regulation in liver. Gavage of both regular coffee (A) and decaffeinated coffee (B) resulted in an activation of autophagy, although at different extent and timing, as measured by LC3 lipidation and p62 degradation (quantified in C). In both cases, autophagy induction was accompanied by an activation of AMPK and by a reduction in the activity of mTORC1, as measured by the phosphorylation of its substrate p70s6k (quantified in C). Representative images are depicted in (AandB). Results from n = 3 independent experiments are presented as fold change ± SEM *P < 0.05; **P < 0.01; (unpaired, 2-tailed Student t test), compared with untreated mice. (D and E) Immunoblot detection of protein acetylation in mice administered with regular coffee. Coffee administration (by gavage) resulted in a significant drop in the overall acetylation levels of proteins in liver, heart, and muscle (quantified in E) in a range of time of 1 to 6 h depending on the tissue. Panels in (D) refer to liver. Ponceau red staining was used to monitor equal loading of the lanes. Results from n = 3 independent experiments are presented as fold change ± SEM *P < 0.05; **P < 0.01 (unpaired, 2-tailed Student t test), compared with untreated mice.
Figure 3. Autophagy induction mediated by short-term coffee administration is confirmed for liver, heart, and muscle from GFP-LC3 transgenic mice. GFP-LC3-expressing mice were administered with regular and decaffeinated coffee by gavage and analyzed for autophagy induction after 4 h. (A–F). Fluorescence microscopic analysis of liver (A), heart (C), and muscle (E) revealed a significant increase in the number of LC3II puncta per area of cells in all tissues (quantified in B, D, andF). Representative images of tissues from untreated vs. regular coffee-treated mice are depicted in (A, C, and E) (bar scale: 10 µm). Results from 3 independent experiments are presented as GFP-LC3 dots/area (means ± SEM). *P < 0.05; **P < 0.01; (unpaired, 2-tailed Student t test), compared with untreated mice.
Coffee ingestion causes global deacetylation of proteins and hence mimics caloric restriction
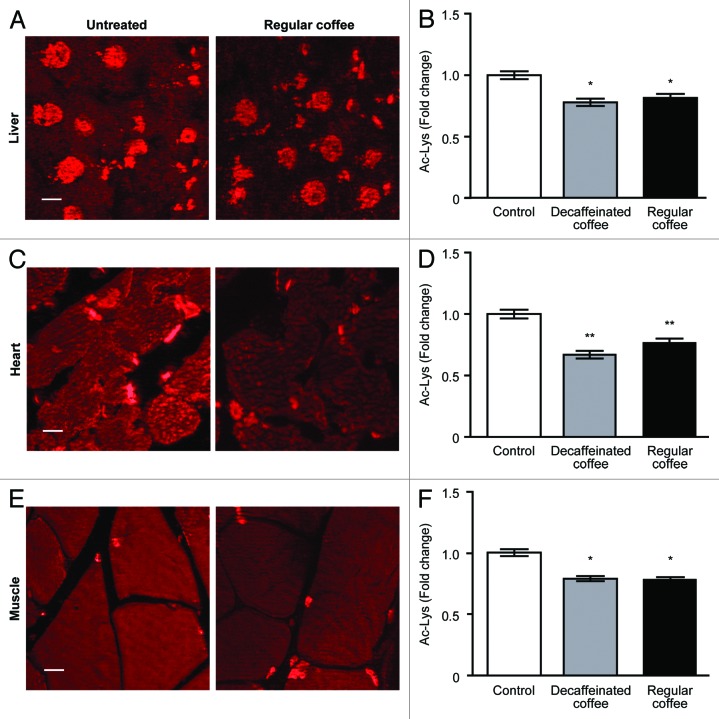
To characterize the mechanisms through which coffee induces autophagy, we determined its capacity to activate prominent energy sensors such as AMP-activated protein kinase (AMPK) and mammalian target of rapamycin complex 1 (mTORC1). While acute (within hours) stimulation with caffeinated or decaffeinated coffee did induce the activating phosphorylation of AMPK (Fig. 2A–C), a more protracted exposure (days to weeks) failed to do so and actually reduced AMPK phosphorylation (Fig. 1C–E). Thus, the present data suggest that AMPK cannot be responsible for autophagy induced by long-term coffee administration. However, both acute and chronic administration of coffee did reduce the phosphorylation of the mTORC1 substrate p70S6K (Figs. 1C–E and 2A–C), suggesting that this nutrient sensor might be involved in pro-autophagic signaling. Coffee contains multiple polyphenols,31,32 and a range of chemically different phenolic compounds have been demonstrated capable to stimulate the deacetylation of cellular proteins, correlating with their pro-autophagic activity.33 We therefore investigated the possibility that in vivo treatment with coffee would reduce the acetylation of cellular proteins using 2 distinct methodologies designed to detect acetyl-lysine-modified proteins. First, extracts from liver (Fig. 2D and E), myocardium (Fig. S3), and skeletal muscle (Fig. S4) revealed a reduction in protein acetylation upon coffee treatment. This effect was detectable as soon as 1 h after coffee gavage in the liver and in the heart and 4 h post-gavage in the skeletal muscle (Fig. 2E). Second, the deacetylation of proteins was detectable by immunofluorescence staining with antibodies specific for acetyl-lysine-modified proteins, revealing a reduction in protein acetylation both in the cytoplasm and in nuclei from hepatocytes, cardiomyocytes, and skeletal myocytes 4 h post-gavage (Fig. 4). Again, these effects were similar for caffeinated and decaffeinated coffee. Altogether, these results support the notion that autophagy induction by coffee is linked to the deacetylation of cellular proteins as well as to the inhibition of mTORC1.
Figure 4. Reduction in global protein acetylation levels mediated by short-term coffee administration in liver, heart, and muscle. Livers, hearts, and muscles from C57Bl/6 mice were analyzed 4 h after treatment with regular and decaffeinated coffee to determine protein acetylatation by immunofluorescence. Fluorescence microscope analysis of liver (A), heart (C), and muscle (E) revealed a significant decrease in the acetylation of proteins in all tissues (quantified in B, D, andF). Representative images of untreated vs. regular coffee-treated mice are depicted in (A, C, and E) (bar scale: 10 µm). Results from n = 3 independent experiments are presented as acetylated lysine intensity fold change ± SEM, considering the values of tissues from untreated mice as 1. *P < 0.05; **P < 0.01; (unpaired, 2-tailed Student t test), compared with untreated mice.
Concluding remarks
The data presented in this paper unequivocally demonstrate that coffee is a potent, rapid inducer of autophagy in multiple tissues in vivo in mice. This effect is independent of caffeine content. Although caffeine has been shown to inhibit mTORC1 and to induce autophagy in hepatocytes in vivo, thereby reducing intrahepatic lipid content and stimulating β-oxidation, as well as counteracting hepatosteatosis,34 caffeine is apparently not required for coffee-induced autophagy. Rather, other coffee components that are not eliminated during the decaffeination process, presumably polyphenols, must be responsible for these effects. Autophagy induced by coffee is accompanied by the inhibition of mTORC1, which represses autophagy in conditions of nutrient availability (particularly amino acids and lipids).35,36 Several polyphenols have been shown to induce autophagy, correlating with their capacity to reduce the acetylation levels of cellular proteins.33,37,38 In theory, the reduction of acetylation levels may be explained by the depletion of the sole donor of acetyl groups, acetyl coenzyme A,39,40 the suppression of the activity of acetyltransferases (for instance by spermidine or C646),21,39 or the activation of deacetylatses (such as sirtuin 1, which can be activated by resveratrol).37 Irrespective of the precise mechanism causing protein deacetylation, this phenomenon is broadly associated with the induction of autophagy by starvation39 or by several pharmacological inducers including spermidine and resveratrol.41 In the present report, we extend the list of agents able to trigger deacetylation reactions to coffee, correlating with its capacity to stimulate autophagy.
Based on the present data, the hypothesis that coffee reduces general mortality by inducing autophagy warrants further experimental and clinical scrutiny.
Material and Methods
Reagents
The results presented refer to a single commercial brand (Belle France) of 100% regular and decaffeinated hydro soluble coffee. Results were confirmed using other 2 commercial brands (Nestle, Lavazza). Concentrations are expressed in % (weight per volume).
Mouse experiments and tissue processing
Six-weeks-aged female C57BL/6 mice (Charles River Laboratory) and transgenic C57BL/6 mice expressing the fusion protein GFP-LC3 under the control of CAG (cytomegalovirus immediate-early [CMVie] enhancer and chicken β-actin promoter) promoter, were bred and maintained according to both the FELASA and the European Community regulations for animal use in research (2010/63UE) as well as the local Ethics Committee for Animal Welfare (project number: 2012–065, 2012–067). Mice were housed in a temperature-controlled environment with 12-h light/dark cycles and received food and water ad libitum. For long-term coffee experiments, mice were first administered with 1%, 3% and 10% w/v of regular and decaffeinated coffee in drinking water ad libitum in order to select a dose not affecting body weight; mice were then administrated with 3% w/v of coffee for 2 wk. Mice were sacrificed, and tissues were recovered and immediately frozen in liquid nitrogen 24, 48, or 72 h, 1 wk and 2 wk after treatment. For short-term coffee experiments, mice were administered with 3% w/v dose of regular and decaffeinated coffee by gavage and sacrificed after 1, 2, 4, and 6 h, when tissues were immediately frozen in liquid nitrogen. After extraction, tissues were homogenized during 2 cycles for 20 s at 5500 rpm using a Precellys 24 tissue homogenator (Bertin Technologies) in a 20 mM Tris buffer (pH 7.4) containing 150 mM NaCl, 1% Triton X-100, 10 mM EDTA, and Complete® protease inhibitor cocktail (Roche Applied Science). Tissue extracts were then centrifuged at 12 000 g at 4 °C, and supernatants were collected. Protein concentration in the supernatants was evaluated by the bicinchoninic acid technique (BCA protein assay kit, Pierce Biotechnology).
Quantitative analysis of GFP-LC3 dots in mouse tissue sections and immunofluorescence
To avoid postmortem autophagy induction, dead mice were immediately perfused with 4% paraformaldehyde (w:v in PBS, pH 7.4). Tissues were then harvested and further fixed with the same solution for at least 4 h, followed by treatment with 15% sucrose (w:v in PBS) for 4 h and with 30% sucrose (w:v in PBS) overnight. Tissue samples were embedded in Tissue-Tek OCT compound (Sakura Finetechnical Co, Ltd) and stored at −80 °C. Five micrometer (5 μm) thick tissue sections were prepared with a CM3050 S cryostat (Leica Microsystems), air-dried for 1 h, washed in PBS for 5 min, dried at RT for 30 min, and mounted with VECTASHIELD anti-fading medium. For acetylation staining, 5-μm-thick tissue sections were prepared. Tissues were permeabilized with 0.1% Triton and blocked with 5% BSA, then incubated overnight at 4 °C with primary antibody in 2% BSA. After 1 h RT incubation with secondary-HRP conjugated antibody, tissues were mounted as described above. For each organ, approximately 10 pictures of 5 independent visual fields from at least 3 mice were acquired using an Axio Observer inverted fluorescence microscope equipped with Apotome confocal-like system (Carl Zeiss). GFP-LC3 dots per area of cells and acetylation intensity were quantified by means of Metamorph (Molecular Devices) software.
Fluorescence microscopy
For the analysis of GFP-LC3 mice tissue sections, a Leica APO 63× NA 1.15 immersion objective was employed. For acetylation analysis, a Leica APO 40× NA 1.15 immersion objective was used. Zeiss Immersol® immersion oil was used for all microscopic analyses. Images were acquired with a Leica DFC 350 Fx camera (version 1.8.0) using Leica LAS AF software and processed with Adobe Photoshop (version CS5) software.
Immunoblotting
For immunoblotting, 25 μg of proteins were separated on 4–12% bis-tris acrylamide (Invitrogen) or 12% tris-glycine SDS-PAGE precast gels (Biorad) and electrotransferred to Immobilon™ membranes (Millipore Corporation). Membranes were then sliced horizontally in different parts according to the molecular weight of the protein of interest to allow simultaneous detection of different antigens within the same experiment.27,28 Unspecific binding sites were saturated by incubating membranes for 1 h in 0.05% Tween 20 (v:v in TBS) supplemented with 5% non-fat powdered milk (w:v in TBS), followed by an overnight incubation with primary antibodies specific for acetylated-lysine, LC3B, phospho-AMPK (Thr172), AMPK, phospho-ribosomal protein S6 kinase (Thr421/Ser424), ribosomal protein S6 kinase, (Cell Signaling Technology), or STQM/p62 (Santa Cruz Biotechnology). Development was performed with appropriate horseradish peroxidase (HRP)-labeled secondary antibodies (Southern Biotech) plus the SuperSignal West Pico chemoluminescent substrate (Thermo Scientific-Pierce). An anti-glyceraldehyde-3-phosphate dehydrogenase antibody (Chemicon International) was used to control equal loading of lanes. Immunoblotting quantifications were performed through densitometric analysis by means of ImageJ software in 3 independent experiments.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Noboru Mizushima for the generous gift of GFP-LC3 transgenic mice. G.K. is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR); Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Fondation Bettencourt-Schueller; Fondation de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the European Research Council (ERC); the LabEx Immuno-Oncology; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI). F.M. is supported by FWF grants LIPOTOX, P23490-B12, I1000, and P24381-B20.
References
- 1.Kempf K, Herder C, Erlund I, Kolb H, Martin S, Carstensen M, Koenig W, Sundvall J, Bidel S, Kuha S, et al. Effects of coffee consumption on subclinical inflammation and other risk factors for type 2 diabetes: a clinical trial. Am J Clin Nutr. 2010;91:950–7. doi: 10.3945/ajcn.2009.28548. [DOI] [PubMed] [Google Scholar]
- 2.Vinson JA, Burnham BR, Nagendran MV. Randomized, double-blind, placebo-controlled, linear dose, crossover study to evaluate the efficacy and safety of a green coffee bean extract in overweight subjects. Diabetes Metab Syndr Obes. 2012;5:21–7. doi: 10.2147/DMSO.S27665. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Greenberg JA, Boozer CN, Geliebter A. Coffee, diabetes, and weight control. Am J Clin Nutr. 2006;84:682–93. doi: 10.1093/ajcn/84.4.682. [DOI] [PubMed] [Google Scholar]
- 4.Freedman ND, Park Y, Abnet CC, Hollenbeck AR, Sinha R. Association of coffee drinking with total and cause-specific mortality. N Engl J Med. 2012;366:1891–904. doi: 10.1056/NEJMoa1112010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malerba S, Turati F, Galeone C, Pelucchi C, Verga F, La Vecchia C, Tavani A. A meta-analysis of prospective studies of coffee consumption and mortality for all causes, cancers and cardiovascular diseases. Eur J Epidemiol. 2013;28:527–39. doi: 10.1007/s10654-013-9834-7. [DOI] [PubMed] [Google Scholar]
- 6.Terry P, Vainio H, Wolk A, Weiderpass E. Dietary factors in relation to endometrial cancer: a nationwide case-control study in Sweden. Nutr Cancer. 2002;42:25–32. doi: 10.1207/S15327914NC421_4. [DOI] [PubMed] [Google Scholar]
- 7.Gunter MJ, Schaub JA, Xue X, Freedman ND, Gaudet MM, Rohan TE, Hollenbeck AR, Sinha R. A prospective investigation of coffee drinking and endometrial cancer incidence. Int J Cancer. 2012;131:E530–6. doi: 10.1002/ijc.26482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotsopoulos J, Ghadirian P, El-Sohemy A, Lynch HT, Snyder C, Daly M, Domchek S, Randall S, Karlan B, Zhang P, et al. The CYP1A2 genotype modifies the association between coffee consumption and breast cancer risk among BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2007;16:912–6. doi: 10.1158/1055-9965.EPI-06-1074. [DOI] [PubMed] [Google Scholar]
- 9.Bravi F, Bosetti C, Tavani A, Bagnardi V, Gallus S, Negri E, Franceschi S, La Vecchia C. Coffee drinking and hepatocellular carcinoma risk: a meta-analysis. Hepatology. 2007;46:430–5. doi: 10.1002/hep.21708. [DOI] [PubMed] [Google Scholar]
- 10.Sinha R, Cross AJ, Daniel CR, Graubard BI, Wu JW, Hollenbeck AR, Gunter MJ, Park Y, Freedman ND. Caffeinated and decaffeinated coffee and tea intakes and risk of colorectal cancer in a large prospective study. Am J Clin Nutr. 2012;96:374–81. doi: 10.3945/ajcn.111.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Zhai L, Zeng J, Peng Q, Wang J, Deng Y, Xie L, Mo C, Yang S, Li S, et al. Coffee consumption and prostate cancer risk: an updated meta-analysis. Cancer Causes Control. 2014;25:591–604. doi: 10.1007/s10552-014-0364-8. [DOI] [PubMed] [Google Scholar]
- 12.Blagosklonny MV. Answering the ultimate question “what is the proximal cause of aging?”. Aging (Albany NY) 2012;4:861–77. doi: 10.18632/aging.100525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blagosklonny MV. Hypoxia, MTOR and autophagy: converging on senescence or quiescence. Autophagy. 2013;9:260–2. doi: 10.4161/auto.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blagosklonny MV. Rapamycin extends life- and health span because it slows aging. Aging (Albany NY) 2013;5:592–8. doi: 10.18632/aging.100591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–37. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–5. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–95. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 19.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martins I, Galluzzi L, Kroemer G. Hormesis, cell death and aging. Aging (Albany NY) 2011;3:821–8. doi: 10.18632/aging.100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 22.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–6. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 23.Kroemer G, Jäättelä M. Lysosomes and autophagy in cell death control. Nat Rev Cancer. 2005;5:886–97. doi: 10.1038/nrc1738. [DOI] [PubMed] [Google Scholar]
- 24.Morselli E, Galluzzi L, Kepp O, Mariño G, Michaud M, Vitale I, Maiuri MC, Kroemer G. Oncosuppressive functions of autophagy. Antioxid Redox Signal. 2011;14:2251–69. doi: 10.1089/ars.2010.3478. [DOI] [PubMed] [Google Scholar]
- 25.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma Y, Galluzzi L, Zitvogel L, Kroemer G. Autophagy and cellular immune responses. Immunity. 2013;39:211–27. doi: 10.1016/j.immuni.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Criollo A, Niso-Santano M, Malik SA, Michaud M, Morselli E, Mariño G, Lachkar S, Arkhipenko AV, Harper F, Pierron G, et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 2011;30:4908–20. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen S, Niso-Santano M, Adjemian S, Takehara T, Malik SA, Minoux H, Souquere S, Mariño G, Lachkar S, Senovilla L, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012;48:667–80. doi: 10.1016/j.molcel.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Sahani MH, Itakura E, Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–41. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashimoto D, Ohmuraya M, Hirota M, Yamamoto A, Suyama K, Ida S, Okumura Y, Takahashi E, Kido H, Araki K, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181:1065–72. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Ho CT. Polyphenolic chemistry of tea and coffee: a century of progress. J Agric Food Chem. 2009;57:8109–14. doi: 10.1021/jf804025c. [DOI] [PubMed] [Google Scholar]
- 32.Lecour S, Lamont KT. Natural polyphenols and cardioprotection. Mini Rev Med Chem. 2011;11:1191–9. doi: 10.2174/13895575111091191. [DOI] [PubMed] [Google Scholar]
- 33.Pietrocola F, Mariño G, Lissa D, Vacchelli E, Malik SA, Niso-Santano M, Zamzami N, Galluzzi L, Maiuri MC, Kroemer G. Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell Cycle. 2012;11:3851–60. doi: 10.4161/cc.22027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sinha RA, Farah BL, Singh BK, Siddique MM, Li Y, Wu Y, Ilkayeva OR, Gooding J, Ching J, Zhou J, et al. Caffeine stimulates hepatic lipid metabolism by the autophagy-lysosomal pathway in mice. Hepatology. 2014;59:1366–80. doi: 10.1002/hep.26667. [DOI] [PubMed] [Google Scholar]
- 35.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–22. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morselli E, Mariño G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192:615–29. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennetzen MV, Mariño G, Pultz D, Morselli E, Færgeman NJ, Kroemer G, Andersen JS. Phosphoproteomic analysis of cells treated with longevity-related autophagy inducers. Cell Cycle. 2012;11:1827–40. doi: 10.4161/cc.20233. [DOI] [PubMed] [Google Scholar]
- 39.Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, et al. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–25. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Küttner V, Bhukel A, Mariño G, Pietrocola F, Harger A, Zimmermann A, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–44. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mariño G, Morselli E, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Bénit P, Rustin P, Criollo A, et al. Longevity-relevant regulation of autophagy at the level of the acetylproteome. Autophagy. 2011;7:647–9. doi: 10.4161/auto.7.6.15191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.