Abstract
Multiple MHC loci encoding human leukocyte antigens (HLA) have allelic variants unequivocally associated with differential immune control of HIV-1 infection. Fine mapping based on single nucleotide polymorphisms (SNPs) in the extended MHC (xMHC) region is expected to reveal causal or novel factors and to justify a search for functional mechanisms. We have tested the utility of a custom fine-mapping platform (the ImmunoChip) for 172 HIV-1 seroconverters (SCs) and 449 seroprevalent individuals (SPs) from Lusaka, Zambia, with a focus on more than 6,400 informative xMHC SNPs. When conditioned on HLA and non-genetic factors previously associated with HIV-1 viral load (VL) in the study cohort, penalized approaches (HyperLasso models) identified an intergenic SNP (rs3094626 between RPP21 and HLA-E) and an intronic SNP (rs3134931 in NOTCH4) as novel correlates of early set-point VL in SCs. The minor allele of rs2857114 (downstream from HLA-DOB) was an unfavorable factor in SPs. Joint models based on demographic features, HLA alleles and the newly identified SNP variants could explain 29% and 15% of VL variance in SCs and SPs, respectively. These findings and bioinformatics strongly suggest that both classic and non-classic MHC genes deserve further investigation, especially in Africans with relatively short haplotype blocks.
Keywords: HIV-1, HLA, human MHC, SNP, viral load
Introduction
In the era of genome-wide association studies (GWAS), the search for definitive host genetic factors underlying effective immune control of HIV-1 infection has yet to yield convincing results beyond those already obtained through hypothesis-driven approaches.1 To date, the most consistent observations center on the importance of human leukocyte antigen class I (HLA-I) genes within the human major histocompatibility complex (MHC) region on chromosome 6p21, although other candidates in the extended MHC (xMHC) may also play some role.2, 3 From the nine HIV-1-related GWAS4–12, only two single nucleotide polymorphisms (SNPs) within the xMHC have consistently shown favorable impact on set-point viral load (VL): rs2395029 within the HCP5 gene and rs9264942 located 35 kb upstream of HLA-C. Follow-up studies have noted strong linkage disequilibrium (LD) between rs2395029 and HLA-B*57:01, an established allele for favorable outcomes after HIV-1 infection.10, 13, 14
The majority of reported GWAS have been performed in populations of European ancestry, while two have scanned African-American populations, without detecting any robust novel associations with disease progression.10, 11 The overall lack of consensus in various findings may reflect the evolving viral diversity, as well as the complexity of human population heterogeneity in genetic architecture.15, 16 Fine-mapping of implicated genetic regions in diverse racial groups is an important step in follow-up studies, as variable LD patterns can help resolve specific haplotype blocks for in-depth interrogation of causal variants or primary association signals that are not fully captured by genome-wide scans.17 In sub-Saharan African populations with the greatest burden of HIV/AIDS and profound genetic diversity, fine-mapping of genetic factors related to immune control of HIV-1 infection may help steer future translational research, especially if dense coverage of candidate loci can effectively rule out the uncertainty with local and extended haplotypes.
Previous studies based on a large Zambian cohort with longitudinal data have identified several host genetic factors that have a cumulative impact on VL.18–22 These studies have also highlighted the importance of viral evolution and adaptation.21–24 With a focus on both early VL in recent HIV-1 seroconverters (SCs) and chronic VL in seroprevalent individuals (SPs), we have tested the utility of a custom genotyping platform (the ImmunoChip) for fine-mapping of host genetic factors in the xMHC region.
RESULTS
Characteristics of seroconverted and seroprevalent individuals included for final analysis
A total of 172 eligible SCs and 449 SPs had successful HLA and ImmunoChip genotyping results that met all inclusion criteria and passed several layers of quality control procedures. In comparisons of overall characteristics of individuals within the two subgroups (Table 1), there were more women in the SC group than in the SP group, with a male-to-female sex ratio of 0.6 and 1.1, respectively. On average, SCs were younger than SPs. Previously established HLA factors had similar distribution in SCs and SPs, except that HLA-B*57 (a favorable factor) was more common in SPs (12.9%) than in SCs (7.6%). Mean log10-transformed VL was slightly lower in SCs (4.6 ± 0.7) than in SPs (4.8 ± 0.7).
Table 1.
Overall characteristics of 172 HIV-1 seroconverters and 449 seroprevalent subjects with HLA and SNP genotyping results.
| Characteristica | SCs (N = 172) | SPs (N = 449) | P |
|---|---|---|---|
| Sex ratio, M/F (no. of individuals) | 0.6 (66/106) | 1.1 (237/212) | 0.001 |
| Age: mean ± SD (year)b | 30.5 ± 7.5 | 32.7 ± 7.9 | 0.002 |
| Enrollment dates | 0.387 | ||
| Earliest | Apr 1995 | Mar 1995 | |
| Latest | Feb 2008 | Sept 2008 | |
| Estimated dates of infection (EDI) | |||
| Earliest | Dec 1995 | - | NA |
| Latest | May 2009 | - | NA |
| Duration of follow-up (month): median (IQR) | - | 17.9 (8.0–31.6) | NA |
| Viral load (VL) outcome | |||
| Mean ± SD (log10)c | 4.6 ± 0.7 | 4.8 ± 0.7 | <0.001 |
| Categories: n (%) | 0.004 | ||
| <10,000 copies/mL | 38 (22.1) | 76 (16.9) | 0.137 |
| 10,000–100,000 copies/mL | 83 (48.3) | 174 (38.8) | 0.031 |
| >100,000 copies/mL | 51 (29.6) | 199 (44.3) | 0.001 |
| Key HLA variants of interest: n (%) | |||
| A*36 | 27 (15.7) | 47 (10.5) | 0.072 |
| A*74 | 22 (12.8) | 62 (13.8) | 0.740 |
| B*13 | 4 (2.3) | 17 (3.8) | 0.368 |
| B*57 | 13 (7.6) | 58 (12.9) | 0.060 |
| B*81 | 11 (6.4) | 39 (8.7) | 0.348 |
| DRB1*01:02 | 22 (12.8) | 34 (7.6) | 0.042d |
SCs, HIV-1 seroconverters; SPs, seroprevalent individuals; M, male; F, female; SD, standard deviation; IQR, interquartile range; NA, not applicable.
At EDI for SCs; at viral load sampling date for SPs.
Geometric mean of all VL measurements in the 3–12 months interval after the EDI in SCs versus first available VL in the SPs.
q = 0.144 (for comparing six HLA variants of interest).
Haplotype blocks defined by informative xMHC SNPs
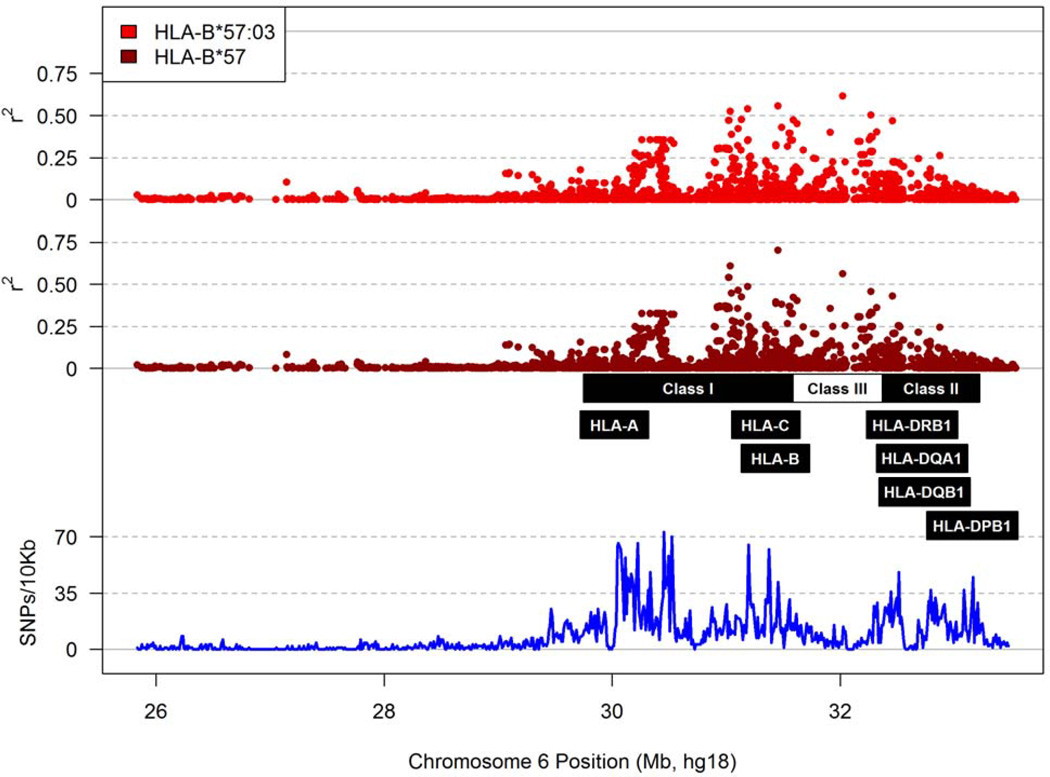
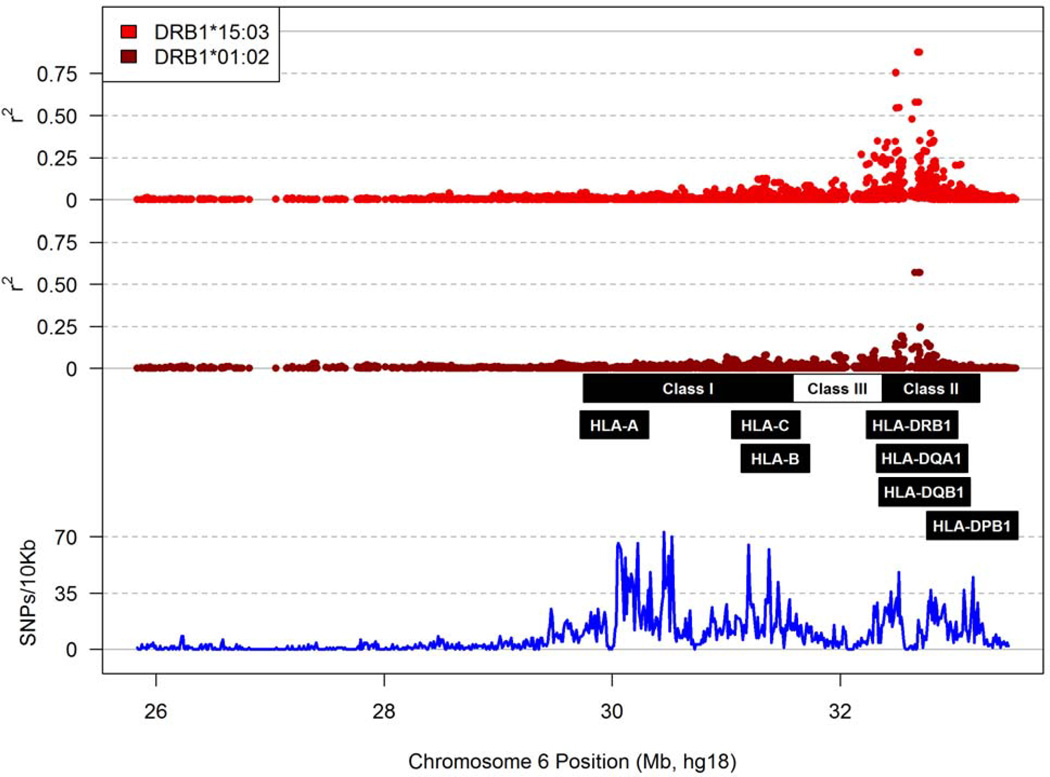
Following the removal of 591 MHC SNPs (~7% of the total) known to be duplicates on the ImmunoChip (Table S1 in Supplemental Materials), 6,417 SNPs in SCs and 6,708 in SPs passed several genotyping quality control procedures and showed minor allele frequencies (MAF) over the analysis thresholds (≥0.025 in SCs and ≥0.015 in SPs, for a minimum of 10 observations in each group) (Tables S2 and S3 in Supplemental Materials). These informative SNPs divided the xMHC region into 410 haplotype blocks of various sizes (Tables S2 and S3) in Supplemental Materials), but they rarely tagged HLA class I or class II alleles (i.e., pairwise r2 >0.80). Accordingly, major HLA factors pertinent to the study population (Figure 1 and Figure 2) were mostly independent of the SNP genotypes.
Figure 1.
Patterns of linkage disequilibrium between individual MHC SNPs and HLA-B*57. The r2 values for B*57:03 and B*57 are shown separately. The relative location of major HLA loci of interest is indicated, so is the density of SNP coverage along the MHC region.
Figure 2.
Patterns of linkage disequilibrium between MHC SNPs and two HLA-DRB1 alleles of interest. The r2 values for DRB1*01:02 and DRB1*15:03 are plotted using a strategy described in Figure 1.
xMHC SNP variants and set-point VL in HIV-1 seroconverters
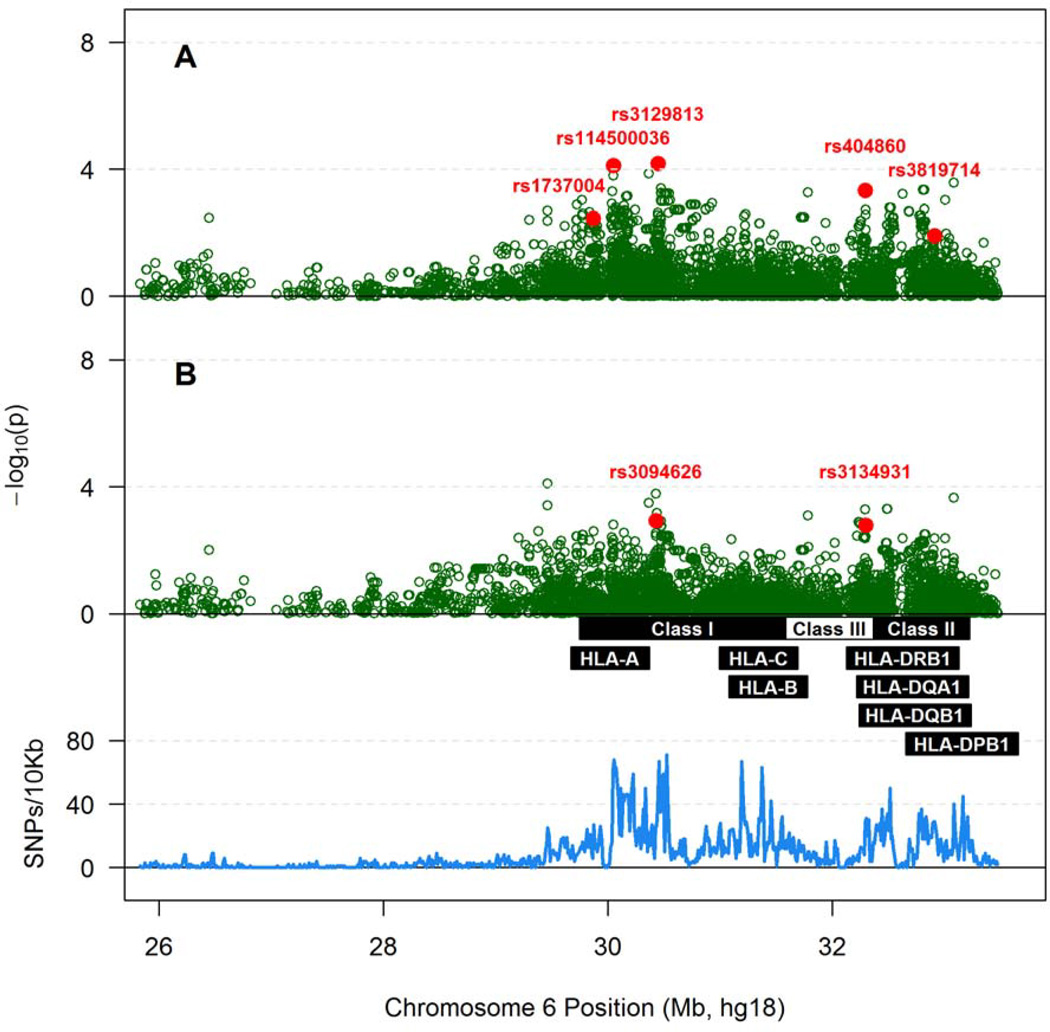
Conventional approach to association analyses did not identify any xMHC SNP with an adjusted p value less than 2.8×10−5, the threshold for xMHC-wide statistical significance after correction for 1,800 independent tests. Several trends (p <0.001) were noted for multiple SNPs mapped to NOTCH4 and to an intergenic region between RPP21 and HLA-E (Table S4 in Supplemental Materials). On the other hand, high-dimensional HyperLasso model (penalized regression) indicated that three SNPs in the MHC class I region and two others in class II and class III might contribute to VL variability in SCs (Figure 3A). These five SNPs, along with sex and age, explained 34.4% of the overall VL variance (p <0.0001) (Table 2). When conditioned further on three prominent HLA factors (A*74, B*13 and B*57), only rs3094626 in the MHC class I region (intergenic between RPP21 and HLA-E) and rs3134931 in class III (intronic within NOTCH4) showed independent contributions to VL variability (Figure 3B). In the reduced multivariable model (Table 2), sex, age, rs3094626 (allele C), rs3134931 (allele G) and the three known HLA factors collectively explained 28.9% of the overall VL variance (p <0.0001). The two MHC SNPs were also among the top hits in single-variant models, with the 11th and 16th ranked p-values, respectively, after statistical adjustment for demographic features and HLA factors (Table S4 in Supplemental Materials).
Figure 3.
Associations of SNPs within the extended MHC with Box-Cox transformed log10 viral load in 172 seroconverters. A) Results adjusted for sex and age at time of HIV-1 infection; B) Results adjusted for sex, age at time of infection and three prominent HLA factors (HLA-A*74, B*13 and B*57). As in Figure 1, the density of SNP coverage along the MHC region is also shown. One of the SNPs, rs114500036, is also known as rs2394250 (see Table 2)
Table 2.
Summary of HyperLasso results from analyses of Box-Cox-transformed log10 viral load in HIV-1 seroconverters.a
| SNP ID | Chr 6 position | Gene | Class | MAF | Allele | Haplotype block numberb |
Relation to VL |
|---|---|---|---|---|---|---|---|
| 1st analysisc | |||||||
| rs1737004 | 29,869,190 | HCG4|HLA-G | upstream | 0.439 | C | 93 | Low |
| rs2394250 | 30,051,635 | HCG9 | ncRNA | 0.282 | T | NA | Low |
| rs3129813 | 30,446,226 | RPP21|HLA-E | intergenic | 0.436 | G | NA | High |
| rs404860 | 32,292,323 | NOTCH4 | intron | 0.334 | G | 285 | High |
| rs3819714 | 32,912,195 | TAP2 | intron | 0.422 | G | 351 | Low |
| 2nd analysisd | |||||||
| rs3094626 | 30,431,602 | RPP21|HLA-E | intergenic | 0.430 | C | 133 | Low |
| rs3134931 | 32,298,598 | NOTCH4 | intron | 0.517 | G | NA | High |
Testing additive effects for the minor alleles (nucleotides) of single nucleotide polymorphism (SNPs); Chr, chromosome; MAF, minor allele frequency; VL, viral load
Not all eligible SNPs can be assigned to haplotype blocks (details are shown in Supplemental Materials). NA, not applicable.
First analysis has age and sex as covariates; overall R2=34.4% for the model.
Second analysis has age, sex, HLA-A*74, B*13 and B*57 as covariates; overall R2=28.9 for the model.
xMHC SNP variants and chronic VL in seroprevalent individuals
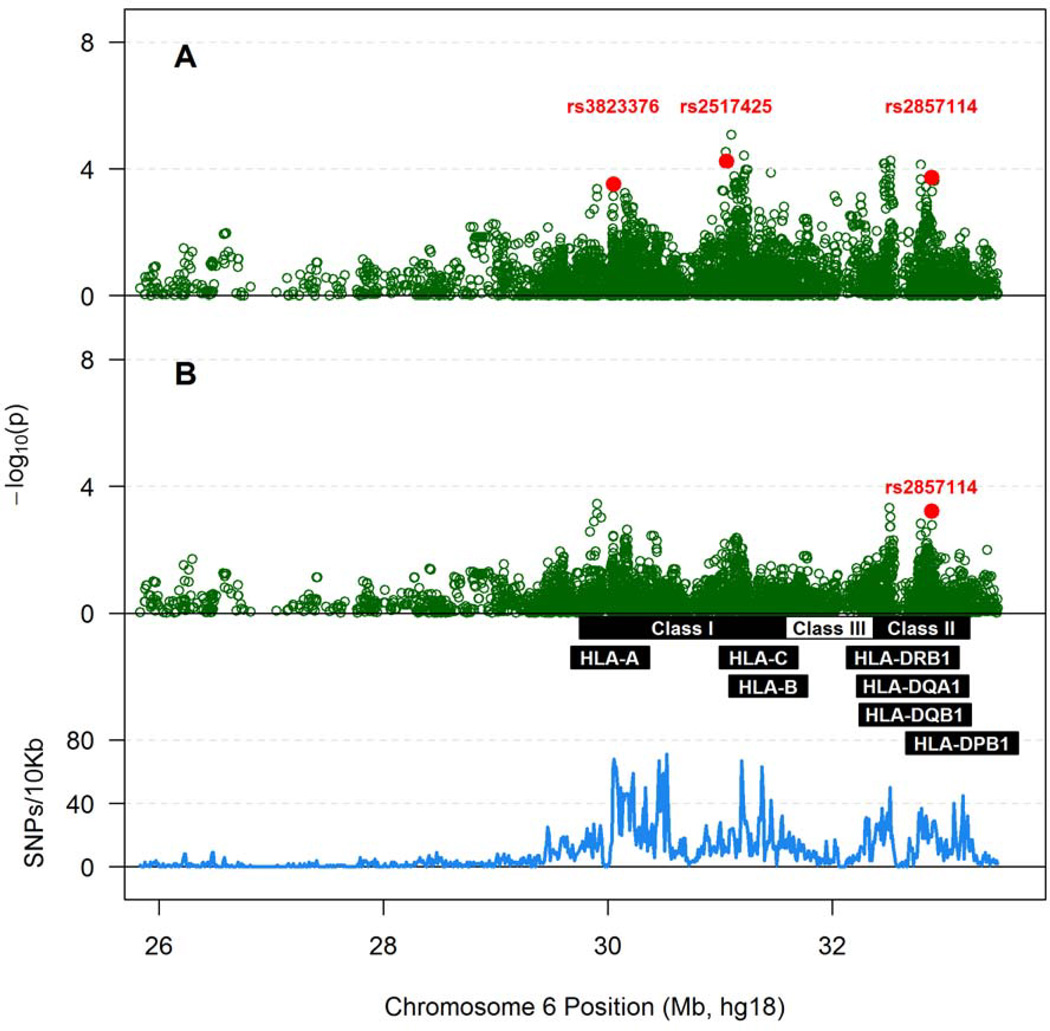
When tested individually, no single SNPs reached the Bonferroni-corrected, statistical significance threshold for potential association with chronic VL in SPs (Table S5 in Supplemental Materials). HyperLasso model initially identified two SNPs in the MHC class I region (rs3823376 and rs2517425) and another in class II (rs2875114) as apparent contributors to VL variability in SPs (Figure 4A). These three SNPs, along with sex and age, could explain 14.8% of the overall VL variance (p <0.0001) (Table 3). Indeed, the allele C for rs2857114, a SNP downstream from HLA-DOB in the MHC class II region, remained as an independent marker associated with unfavorable VL outcome after additional adjustment for previously identified HLA alleles (Figure 4B and Table 3). The final model explained 14.9% of the overall VL variance (p <0.0001) (Table 3).
Figure 4.
Associations of SNPs within the extended MHC with Box-Cox transformed log10 viral load (VL) in 449 Zambians with seroprevalent HIV-1 infection. A) Results adjusted for sex and age at the time of VL measurement; B) Results adjusted for sex, age at the time of VL measurement and five known HLA factors (HLA-A*36, A*74, B*57, B*81 and DRB1*01:02). As in Figure 1, the density of SNP coverage along the MHC region is also shown.
Table 3.
Summary of HyperLasso results from analysis of Box-Cox transformed log10 viral load (VL) in seroprevalent individuals.a
| SNP ID | Chr 6 position | Gene | Class | MAF | Allele | Haplotype block numberb |
Relation to VL |
|---|---|---|---|---|---|---|---|
| 1st Analysisc | |||||||
| rs3823376 | 30,052,163 | HCG9 | ncRNA | 0.404 | G | NA | Low |
| rs2517425 | 31,057,649 | MUC21 | intergenic | 0.280 | C | 170 | Low |
| rs2857114 | 32,887,974 | HLA-DQB2|HLA-DOB | downstream | 0.457 | C | 346 | High |
| 2nd analysisd | |||||||
| rs2857114 | 32,887,974 | HLA-DQB2|HLA-DOB | downstream | 0.457 | C | 346 | High |
Testing additive effects for the minor alleles of single nucleotide polymorphisms (SNPs); MAF, minor allele frequency.
Not all eligible SNPs can be assigned to haplotype blocks (details are shown in Supplemental Materials). NA, not applicable.
First analysis has adjusted for sex and age at VL measurement; overall R2=14.8% for the model.
Second analysis has adjusted for sex, age at VL measurement, HLA-A*36, A*74, B*57, B*81 and DRB1*01:02; Overall R2=14.9% for the model.
Findings from bioinformatics
A search in the HaploReg database (http://www.broadinstitute.org/mammals/haploreg/haploreg.php) revealed that the three independent associations (rs3094626, rs3134931, and rs2857114) identified by HyperLasso models were not complicated by tagging SNPs (r2 ≥0.80) reported in African populations (Figure S1 in Supplemental Materials). First, neither rs3094626 nor rs3134931 can tag other SNPs. Second, the only SNP (rs2621331 in HLA-DOB) tagged by rs2857114 (r2 = 1.0) has less functional attributes than rs2857114. In terms of previously reported associations captured by the NCBI Global Cross-database, rs2857114 has been implicated as an expression QTL for TAP2 and HLA-DOB in B-cells and monocytes.25
In populations of European ancestry, however, two of the three SNPs of major interest have quite different LD patterns (Figure S2). In particular, rs2857114 can effectively tag 70 other SNPs at the HLA-DOB locus, including two (rs2187684 and rs2257789) that may govern promoter functions; one of these (rs2187684) has been associated with Juvenile idiopathic arthritis (JIA).26 No other clues can point to the causal variants.
DISCUSSION
Guided by strong epidemiologic evidence from hypothesis-driven approaches,21, 22 our study here revealed several xMHC SNPs as novel correlates of differential immune control of HIV-1 infection in an African cohort. Of note, when results obtained from three analytical approaches (linear regression, penalized regression and ordinal regression) are compared side by side for seroconverters (recent HIV-1 infection) and seroprevalent subjects (unknown but longer duration of infection), there is a striking contrast for the list of independent markers at the gene and SNP levels (e.g., Figure 3 versus Figure 4). In light of the various pathways for viral adaptation and immune escape mutations in individuals and populations,24, 27, 28 it is now evident that seroconverters and seroprevalent subjects should be treated as distinct subgroups for certain epidemiologic analysis even when they are derived from the same location, because lumping them together (for the sake of statistical power) can obscure the search for early correlates and mechanisms of durable immune control.21, 29
Our work has also demonstrated that penalized regression can be advantageous when a large number of individuals SNPs with modest effects are analyzed. Penalized regression, such as HyperLasso, focuses on prediction and not on hypothesis testing, which requires moving away from the usual and overwhelming reliance on p values in genetic studies.30 Although the models can be calibrated to offer some control of overall false discovery rates, regularization also induces biased coefficient estimates and complicates the calculations of standard errors for variables selected into the model. Methods such as HyperLasso typically include the best SNPs out of a pool of closely related variants for characterizing the association.31 Thus, the resulting models are not necessarily unique, i.e., multiple competing models may provide virtually identical predictive accuracy. To this end, it is often necessary to emphasize the specific genes or haplotype blocks rather than the individual SNPs. Surveys based on various databases, including The 1000 Genomes Project,32 ENCODE33, 34, and the NCBI Global Cross-database, can help narrow the search for consensus findings or causal variants, especially when functional attributes in various cell lineages are known.25
The novel MHC SNPs identified by two series of HyperLasso models can explain a good proportion of the variability in both early and chronic VL (Tables 2 and 3), clearly beyond those already attributed to classical HLA alleles in the Zambian cohort.22 In particular, the association of allele G for rs3134931 (NOTCH4) with set-point VL has some consistency with reported literature, as two previous studies on HIV-1 have highlighted a nonsynonymous SNP (rs8192591) within exon 9 of NOTCH4, corresponding to Gly534Ser.8, 35 Similar to our findings on rs3134931-G and high set-point VL, an unfavorable relationship has been reported for rs8192591 and disease progression.35 The conservative amino acid substitution introduced by rs8192591 is not expected to alter the protein function.8 The low minor allele frequency for rs8192591 in our study cohort also rules out the possibility of a major impact. In contrast, the allelic variants of rs3134931 (a SNP mapped to an enhancer region) can have distinct binding affinity for PU1 in fibroblasts and TH1 cells (Figure S1). While rs3134931-G may well be a causal variant of immunological importance, recognition of NOTCH4 (http://www.ncbi.nlm.nih.gov/gene/4855) as a hot spot for autoimmune disorders and infectious diseases8, 35–38 still justifies an in-depth evaluation of neighboring variants in follow-up studies.
The biological relevance of other xMHC SNPs associated with set-point and chronic VL is less obvious, even when LD patterns and other datasets are considered (Figure S1). The association signal for rs2857114 may point to the potential role of HLA-DOB (http://www.ncbi.nlm.nih.gov/gene/3112), a non-classic HLA gene encoding the DOβ chain that forms a heterodimer with HLA-DOα. The DOαβ complex is typically stored in lysosomes to regulate HLA-DM-mediated loading of antigenic peptides to MHC class II molecules.39–41 In the absence of strong LD with known SNPs beyond the HLA-DOB locus, further analysis of rs2857114 in African populations can be highly advantageous because there is little complication by neighboring variants (Figure S1). Conversely, presence of conserved extended haplotypes in populations of European ancestry42 can obscure the search for causal variants within cis- and trans-acting elements.25
Several studies have noted the benefits of the popular ImmunoChip for detecting independent association signals for coding variants at many loci.43, 44 By design, the ImmunoChip may allow for fine-mapping in populations of European ancestry, with clear improvement for SNP coverage when compared with existing GWAS arrays.45, 46 However, as far as the xMHC region is concerned, the first-generation ImmunoChip still has a major caveat in its lack of dense coverage (e.g., Figure 1), especially for coding SNPs (Table S2 and Table S3). It is also apparent that multiple MHC SNPs (i.e. rs2395029 and rs9264942) highly relevant to HIV/AIDS and other infectious diseases are not covered at all by the ImmunoChip. Our statistical models should continue to improve when fine-mapping based on sequencing can adequately capture variants in both classic and non-classic MHC loci. Further analyses of rare variants in large cohort should be informative as well. Future generations of the ImmunoChip may need to meet these needs and minimize uncertainties about coverage for the xMHC region.
PATIENTS AND METHODS
Study population
Subjects available for this study came from the Zambia-Emory HIV-1 Research Project (ZEHRP). Based on HIV-1 infection status at the time of enrollment, subjects were classified into two subgroups: 242 seroconverters (SCs) who became infected during quarterly follow-up visits and 482 seroprevalent individuals (SPs) who were already infected at baseline. The overall study design and follow-up strategies have been described in detail elsewhere for this cohort.47–49
Ethical aspects
The research outlined in this study was approved by the institutional review boards at clinical sites in Lusaka, Zambia and at two collaborating institutions (Emory University and University of Alabama at Birmingham) in the United States. All subjects gave written informed consent for screening and participation.
Assessment of virologic outcomes
Plasma VL was measured as HIV-1 RNA concentration (copies/mL) using the Roche Amplicor 1.0 assay (Roche Diagnostics Systems Inc., Branchburg, NJ) in a laboratory certified by the virology quality assurance program of the AIDS Clinical Trial Group. Results after the initiation of antiretroviral therapy (ART) were censored. For SCs, set-point VL corresponded to the log10-transformed geometric mean of all available VL measurements during the 3–12 months interval after the estimated date of infection (EDI). For SPs, the first log10-transformed VL measurement available was used for analysis. Subjects (49 SCs and 5 SPs) who had missing VL data were excluded first, so were those (5 SCs and 13 SPs) who had VL below the lower limit of detection (i.e., <400 copies/ml). The remaining SCs (n=188) and SPs (n=464) were retained for further analyses.
Genotyping
Allelic variants for three classic HLA class I genes (HLA-A, HLA-B and HLA-C) and two HLA class II loci (HLA-DRB1 and HLA-DQB1) were resolved to the first four digits (distinct protein sequences) using a combination of PCR-based methods, including PCR with sequence-specific primers (SSP) (Dynal/Invirtrogen, Brown Deer, WI), automated sequence-specific oligonucleotide (SSO) probe hybridization (Innogenetics, Alpharetta, GA), and sequencing-based typing (SBT) (Abbott Molecular, Inc., Des Plaines, IL). SNP genotyping with the Illumina ImmunoChip46 was processed at a genomics core facility (University of Alabama at Birmingham); SNP alleles were inferred using the joint calling and haplotype phasing algorithm implemented in BEAGLECALL.50 We completed a series of data cleaning and quality control procedures for SNPs in the xMHC region, excluding SNPs based on the following criteria: (i) duplication (Table S1), (ii) missingness (call rate <98.5%), (iii) minor allele frequency (MAF) <0.025 in SCs and <0.015 in SPs, and (iv) deviation from Hardy-Weinberg equilibrium (HWE) (p <10−6). In the end, 6,417 informative SNPs in SCs (Tables S3) and 6,708 SNPs in SPs (Table S3) were mapped to the xMHC region (~7.5 Mb between rs498548 and rs2772390).51
Data cleaning based on ImmunoChip results and population stratification
Subjects with ImmunoChip data were further evaluated against three additional (and occasionally overlapping) quality control criteria for final exclusion: (i) overall call rates <95% (8 SCs and 3 SPs), (ii) failing sex determination (n=0); (iii) up to third degree familial relationships (4 SCs and 5 SPs) according to kinship coefficients (the KING software package).52, 53 Population stratification was assessed using multidimensional scaling (MDS) implemented in KING.53, 54 We gathered SNP data for 1,184 unrelated individuals from the eleven study populations in Phase 3 of the International HapMap Project, defined by the Illumina 1M and Affymetrix 6.0 genome-wide SNP arrays. For the MDS analysis, we used a subset of SNPs (~30,000) that were (i) annotated with reference sequence (rs) IDs on the ImmunoChip, (ii) outside of regions of known extended LD in European populations, and (iii) reliably aligned with the HapMap 3 data (i.e., removing SNPs with ambiguous A/T and C/G variants).55 The MDS data revealed four outliers (1 SC and 3 SPs, possibly immigrants) that were not suitable for final analysis.
Analysis of general features and population genetics
The overall characteristics of SCs and SPs were compared using Student’s t test (for continuous variables), χ2 test (for categorical variables), and the Wilcoxon-Mann-Whitney test (for non-parametric variables) (Table 1). HLA alleles and xMHC SNP genotypes in SCs and SPs were also analyzed separately for patterns of LD, with SNPs assigned to individual haplotype blocks using HaploView56 whenever possible (Tables S2 and S3). SNPs that can effectively tag individual HLA alleles (pairwise r2 >0.80) received special attention, especially in terms of their physical location (relative to the HLA alleles and other MHC genes) and potential function (transcription and translation).
Strategies for association analysis
To effectively identify novel SNPs that are independently associated with VL outcomes in SCs and SPs, we adopted a high-dimension method known as penalized regression (HyperLasso),31 which tests all eligible xMHC SNPs at once and minimizes the risk of false discovery from random testing. The HyperLasso model uses a hierarchical Normal-Exponential-Gamma (NEG) regression coefficient, which is parameterized in terms of shape and scale. Based on the method reported by Vignal et al.,57 the shape parameter was set to 1, and 100 null permutations were analyzed for both the SC and SP datasets (for a given set of non-SNP covariates, i.e. non-genetic factors and previously identified HLA alleles) over a grid of scale parameter values. The null datasets were created by randomly pairing phenotypes/non-genetic covariates with genotypes (thus maintaining the relationship between the non-genetic covariates and VLs). We then selected a value for the scale parameter that allowed no more than one spurious SNP signal in the final regression model. The minor alleles of individual SNPs were tested for additive effect, and they were included only if the minor allele was observed in at least 10 subjects (either heterozygote or homozygote) in a given dataset. For analyses of set-point VL in SCs, the HyperLasso model was adjusted for potential effects of sex, age at EDI and three known HLA factors (A*74, B*13, and B*57).21 Analyses of chronic VL in SPs were conditioned on sex, age at VL measurement and five known HLA factors (HLA-A*36, A*74, B*57, B*81, and DRB1*01:02) as reported earlier.21
To facilitate comparison with findings from multiple GWAS, we also applied a standard approach to testing individual candidate SNPs and ranked them based on their adjusted p values in generalized linear models (GLMs) (SAS version 9.2). To account for the widespread LD within the xMHC region, SimpleM was used to calculate the number of independent tests for Bonferroni correction of p values.58, 59 For both SCs and SPs, the SimpleM estimate for the number of independent tests was nearly 1,800. Thus, a p value of 2.8×10−5 was considered statistically significant for the MHC-wide random testing for individual SNPs.
For all linear regression models, the assumption of a normal VL data distribution was assessed using the Kolmogorov-Smirnov test. The standard log10-transformation of VL helped with data normalization, but Box-Cox transformation of log10 VL was generally applicable to separate analyses of SCs and SPs. We further obtained summary statistics (proportion odds ratios, 95% confidence intervals and p values) from ordinal logistic regression models for three VL categories: (i) <10,000 copies/mL (low), (ii) 10,000–100,000 copies/mL (medium), and (iii) >100,000 copies/mL (high). This alternative strategy is expected to alleviate concerns about minor VL fluctuations within individuals and inter-assay variability in VL data. More importantly, these VL categories have strong and persistent implications for rates of heterosexual HIV-1 transmission.47, 60
Bioinformatics for individual SNPs of interest
To gather evidence for functional relevance, SNPs associated with HIV-1 VL in HyperLasso models were first queried in HaploReg61 for known LD with SNPs documented by the 1000 Genomes Project (not necessary covered by the ImmunoChip) or annotated by the ENCODE project.33, 34 MHC SNPs implicated for other diseases (dbGaP) and/or gene expression QTL (eQTL) in various cell lines25 were also surveyed (NCBI Global Cross-database, http://www.ncbi.nlm.nih.gov/), with a focus on functional attributes in cells that mediate immune pathways. These procedures further took into account of (i) cis- and trans-acting elements, (ii) local haplotypes resolved by HaploView,56 (iii) conserved extended haplotypes known for various HLA alleles.42
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by multiple grants, including R01 AI071906 (to R.A.K. and J.T.) and R01 AI064060 (to E.H.) from the National Institute of Allergy and Infectious Diseases (NIAID). We are grateful to several associates, especially Ilene Brill, Paul Farmer, Travis Porter and Wei Song, for their contribution to data management and genotyping.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interests.
REFERENCES
- 1.Prentice HA, Tang J. HIV-1 dynamics: a reappraisal of host and viral factors, as well as methodological issues. Viruses. 2012;4(10):2080–2096. doi: 10.3390/v4102080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guergnon J, Theodorou I. What did we learn on host's genetics by studying large cohorts of HIV-1-infected patients in the genome-wide association era? Curr Opin HIV AIDS. 2011;6(4):290–296. doi: 10.1097/COH.0b013e3283478449. [DOI] [PubMed] [Google Scholar]
- 3.Aouizerat BE, Pearce CL, Miaskowski C. The search for host genetic factors of HIV/AIDS pathogenesis in the post-genome era: progress to date and new avenues for discovery. Curr HIV/AIDS Rep. 2011;8(1):38–44. doi: 10.1007/s11904-010-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalmasso C, Carpentier W, Meyer L, Rouzioux C, Goujard C, Chaix ML, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PLOS ONE. 2008;3(12):e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limou S, Le Clerc S, Coulonges C, Carpentier W, Dina C, Delaneau O, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199(3):419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 7.Le Clerc S, Limou S, Coulonges C, Carpentier W, Dina C, Taing L, et al. Genomewide association study of a rapid progression cohort identifies new susceptibility alleles for AIDS (ANRS Genomewide Association Study 03) J Infect Dis. 2009;200(8):1194–1201. doi: 10.1086/605892. [DOI] [PubMed] [Google Scholar]
- 8.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5(12):e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbeck JT, Gottlieb GS, Winkler CA, Nelson GW, An P, Maust BS, et al. Multistage genomewide association study identifies a locus at 1q41 associated with rate of HIV-1 disease progression to clinical AIDS. J Infect Dis. 2010;201(4):618–626. doi: 10.1086/649842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelak K, Goldstein DB, Walley NM, Fellay J, Ge D, Shianna KV, et al. Host determinants of HIV-1 control in African Americans. J Infect Dis. 2010;201(8):1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330(6010):1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartha I, Carlson JM, Brumme CJ, McLaren PJ, Brumme ZL, John M, et al. A genome-to-genome analysis of associations between human genetic variation, HIV-1 sequence diversity, and viral control. Elife. 2013;2:e01123. doi: 10.7554/eLife.01123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catano G, Kulkarni H, He W, Marconi VC, Agan BK, Landrum M, et al. HIV-1 disease-influencing effects associated with ZNRD1, HCP5 and HLA-C alleles are attributable mainly to either HLA-A10 or HLA-B*57 alleles. PLOS ONE. 2008;3(11):e3636. doi: 10.1371/journal.pone.0003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrestha S, Aissani B, Song W, Wilson CM, Kaslow RA, Tang J. Host genetics and HIV-1 viral load set-point in African-Americans. AIDS. 2009;23(6):673–677. doi: 10.1097/QAD.0b013e328325d414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman SJ, Hill AV. Human genetic susceptibility to infectious disease. Nat Rev Genet. 2012;13(3):175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- 16.Donfack J, Buchinsky FJ, Post JC, Ehrlich GD. Human susceptibility to viral infection: the search for HIV-protective alleles among Africans by means of genome-wide studies. AIDS Res Hum Retroviruses. 2006;22(10):925–930. doi: 10.1089/aid.2006.22.925. [DOI] [PubMed] [Google Scholar]
- 17.McLaren PJ, Ripke S, Pelak K, Weintrob AC, Patsopoulos NA, Jia X, et al. Fine-mapping classical HLA variation associated with durable host control of HIV-1 infection in African Americans. Hum Mol Genet. 2012;21(19):4334–4347. doi: 10.1093/hmg/dds226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G, et al. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol. 2002;76(16):8276–8284. doi: 10.1128/JVI.76.16.8276-8284.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang J, Tang S, Lobashevsky E, Zulu I, Aldrovandi G, Allen S, et al. HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res Hum Retroviruses. 2004;20(1):19–25. doi: 10.1089/088922204322749468. [DOI] [PubMed] [Google Scholar]
- 20.Lazaryan A, Lobashevsky E, Mulenga J, Karita E, Allen S, Tang J, et al. Human leukocyte antigen B58 supertype and human immunodeficiency virus type 1 infection in native Africans. J Virol. 2006;80(12):6056–6060. doi: 10.1128/JVI.02119-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang J, Malhotra R, Song W, Brill I, Hu L, Farmer PK, et al. Human leukocyte antigens and HIV type 1 viral load in early and chronic infection: predominance of evolving relationships. PLOS ONE. 2010;5(3):e9629. doi: 10.1371/journal.pone.0009629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yue L, Prentice HA, Farmer P, Song W, He D, Lakhi S, et al. Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J Virol. 2012;87(2):708–715. doi: 10.1128/JVI.02118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J, Cormier E, Gilmour J, Price MA, Prentice HA, Song W, et al. Human leukocyte antigen variants B*44 and B*57 are consistently favorable during two distinct phases of primary HIV-1 infection in sub-Saharan Africans with several viral subtypes. J Virol. 2011;85(17):8894–8902. doi: 10.1128/JVI.00439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crawford H, Matthews PC, Schaefer M, Carlson JM, Leslie A, Kilembe W, et al. The hypervariable HIV-1 capsid protein residues comprise HLA-driven CD8+ T-cell escape mutations and covarying HLA-independent polymorphisms. J Virol. 2011;85(3):1384–1390. doi: 10.1128/JVI.01879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44(5):502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinks A, Barton A, Shephard N, Eyre S, Bowes J, Cargill M, et al. Identification of a novel susceptibility locus for juvenile idiopathic arthritis by genome-wide association analysis. Arthritis Rheumat. 2009;60(1):258–263. doi: 10.1002/art.24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal S. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296(5572):1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 28.Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458(7238):641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prentice HA, Porter TR, Price MA, Cormier E, He D, Farmer PK, et al. HLA-B*57 versus HLA-B*81 in HIV-1 infection: slow and steady wins the race? J Virol. 2013;87(7):4043–4051. doi: 10.1128/JVI.03302-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayers KL, Cordell HJ. SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genet Epidemiol. 2010;34(8):879–891. doi: 10.1002/gepi.20543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoggart CJ, Whittaker JC, De Iorio M, Balding DJ. Simultaneous analysis of all SNPs in genome-wide and re-sequencing association studies. PLoS Genet. 2008;4(7):e1000130. doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Via M, Gignoux C, Burchard EG. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2(1):3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38(Database issue):D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Clerc S, Coulonges C, Delaneau O, Van Manen D, Herbeck JT, Limou S, et al. Screening low-frequency SNPS from genome-wide association study reveals a new risk allele for progression to AIDS. J Acquir Immune Defic Syndr. 2011;56(3):279–284. doi: 10.1097/QAI.0b013e318204982b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, Hamon S, Li D, Barral-Rodriguez S, Ott J, Diabetes Genetics C. MHC fine mapping of human type 1 diabetes using the T1DGC data. Diabetes, Obesity & Metabol. 2009;11(Suppl 1):53–59. doi: 10.1111/j.1463-1326.2008.01003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdes AM, Thomson G Type 1 Diabetes Genetics C. Several loci in the HLA class III region are associated with T1D risk after adjusting for DRB1-DQB1. Diabetes, Obesity & Metabol. 2009;11(Suppl 1):46–52. doi: 10.1111/j.1463-1326.2008.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morris DL, Taylor KE, Fernando MM, Nititham J, Alarcon-Riquelme ME, Barcellos LF, et al. Unraveling multiple MHC gene associations with systemic lupus erythematosus: model choice indicates a role for HLA alleles and non-HLA genes in Europeans. Am J Hum Genet. 2012;91(5):778–793. doi: 10.1016/j.ajhg.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santin I, Castellanos-Rubio A, Aransay AM, Gutierrez G, Gaztambide S, Rica I, et al. Exploring the diabetogenicity of the HLA-B18-DR3 CEH: independent association with T1D genetic risk close to HLA-DOA. Genes Immun. 2009;10(6):596–600. doi: 10.1038/gene.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Souwer Y, Chamuleau ME, van de Loosdrecht AA, Tolosa E, Jorritsma T, Muris JJ, et al. Detection of aberrant transcription of major histocompatibility complex class II antigen presentation genes in chronic lymphocytic leukaemia identifies HLA-DOA mRNA as a prognostic factor for survival. Br J Haematol. 2009;145(3):334–343. doi: 10.1111/j.1365-2141.2009.07625.x. [DOI] [PubMed] [Google Scholar]
- 41.Xiu F, Côté MH, Bourgeois-Daigneault MC, Brunet A, Gauvreau MÉ, Shaw A, et al. Cutting edge: HLA-DO impairs the incorporation of HLA-DM into exosomes. J Immunol. 2011;187(4):1547–1551. doi: 10.4049/jimmunol.1100199. [DOI] [PubMed] [Google Scholar]
- 42.Dorak MT, Shao W, Machulla HK, Lobashevsky ES, Tang J, Park MH, et al. Conserved extended haplotypes of the major histocompatibility complex: further characterization. Genes Immun. 2006;7(6):450–467. doi: 10.1038/sj.gene.6364315. [DOI] [PubMed] [Google Scholar]
- 43.Polychronakos C. Fine points in mapping autoimmunity. Nat Genet. 2011;43(12):1173–1174. doi: 10.1038/ng.1015. [DOI] [PubMed] [Google Scholar]
- 44.Trynka G, Hunt KA, Bockett NA, Romanos J, Mistry V, Szperl A, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43(12):1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikula T, West A, Katajamaa M, Lönnberg T, Sara R, Aittokallio T, et al. A human ImmunoChip cDNA microarray provides a comprehensive tool to study immune responses. J Immunol Methods. 2005;303(1–2):122–134. doi: 10.1016/j.jim.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13(1):101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, et al. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17(10):901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, et al. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76(1):397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kempf MC, Allen S, Zulu I, Kancheya N, Stephenson R, Brill I, et al. Enrollment and retention of HIV discordant couples in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2008;47(1):116–125. doi: 10.1097/QAI.0b013e31815d2f3f. [DOI] [PubMed] [Google Scholar]
- 50.Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85(6):847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, et al. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet. 2012;8(4):e1002640. doi: 10.1371/journal.pgen.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratification. Am J Hum Genet. 2008;82(2):352–365. doi: 10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 57.Vignal CM, Bansal AT, Balding DJ. Using penalised logistic regression to fine map HLA variants for rheumatoid arthritis. Ann Hum Genet. 2011;75(6):655–664. doi: 10.1111/j.1469-1809.2011.00670.x. [DOI] [PubMed] [Google Scholar]
- 58.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32(4):361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 59.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34(1):100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, et al. Human leukocyte antigen class I genotyes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008;181(4):2626–2635. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.