Abstract
Metoprolol is a selective β-1 adrenergic receptor blocker that undergoes extensive metabolism by the polymorphic enzyme, CYP2D6. Our objective was to investigate the influence of CYP2D6 polymorphisms on efficacy and tolerability of metoprolol tartrate. 281 study participants with uncomplicated hypertension received 50 mg of metoprolol twice daily followed by response guided titration to 100 mg twice daily. Phenotypes were assigned based on results of CYP2D6 genotyping and copy number variation assays. Clinical response to metoprolol and adverse effect rates were analyzed in relation to CYP2D6 phenotypes by using appropriate statistical tests. Heart rate response differed significantly by CYP2D6 phenotype (p-value <0.0001) with poor metabolizers & intermediate metabolizers showing greater HR reduction. However, blood pressure response and adverse effect rates were not significantly different by CYP2D6 phenotype. Other than a significant difference in heart rate response, CYP2D6 polymorphisms were not a determinant of the variability in response or tolerability to metoprolol.
Keywords: CYP2D6, metoprolol, genotype, phenotype, copy number variation, clinical efficacy, tolerability
Introduction
Metoprolol is a cardioselective β-1 adrenergic receptor blocker that is considered a cornerstone therapy (1–4) for the treatment of various cardiovascular diseases owing to the mortality benefits demonstrated in the setting of coronary artery diseases, heart failure, and hypertension (5, 6). With an estimated 72 million prescriptions filled annually, metoprolol (as tartrate and succinate) ranks 4th among the top 10 most prescribed medications, making it the most commonly used β-1 blocker in clinical practice (7). However, response to metoprolol therapy is highly variable particularly in hypertension (HTN) and heart failure (HF) (8, 9). Despite its wide spread use, much remains to be learned about the determinants of variability in metroprolol response.
Like other lipophilic β-1 blockers, metoprolol undergoes extensive metabolism in the liver via three distinct oxidative pathways: O-demethylation, N-dealkylation and alpha hydroxylation (10–12). The first comprises the principle route of metabolism, contributing to about 65% of the overall metabolism, whereas the other two pathways are considered minor, each accounting for 10% (13, 14). Although several cytochrome enzymes are involved in its metabolism, studies have shown that CYP2D6 is responsible for metabolizing about 60% of the oral metoprolol dose (15).
Understanding the genetics of CYP2D6 is of utmost importance from a clinical standpoint since the enzyme is responsible for metabolizing more than 30% of drugs belonging to different therapeutic classes. Unlike other cytochromes, polymorphisms in CYP2D6 result from not only single nucleotide polymorphism (SNPs), but also from insertion/deletions of nucleotide bases (indels), as well as whole gene deletions, duplications and multiplications. As a consequence, more than 100 variant alleles have been identified thus far (http://www.imm.ki.se/CYPalleles) giving rise to gene products or enzymes with various activities. The CYP2D6 activity score system (16), a relatively new and simple method, assigns a score to each variant allele based on its predicted function, and subsequently it allows rapid classification of an individual’s CYP2D6 metabolizer phenotype into one of the four predicted phenotypes: poor metabolizers (PMs), intermediate metabolizers (IMs), extensive metabolizers (EMs), and ultrarapid metabolizers (UMs). The prevalence of each phenotype varies among racial groups with the PM phenotype having the highest frequency among Caucasians (5–10%) compared with the other races (17).
There is ample evidence in the literature suggesting that the CYP2D6 polymorphisms impact the pharmacokinetics of metoprolol as well as other β blockers (18–22). Whether these differences in metoprolol pharmacokinetics translate into variability in response is a subject of ongoing debate (23–25). Hence, the main objective of our study was to assess the influence of CYP2D6 polymorphisms on the clinical efficacy and tolerability of metoprolol when used for the treatment of uncomplicated hypertension.
Results
Baseline characteristics for the 218 study participants are summarized in Table 1. Although not shown here, it is important to note that they were well balanced across the four CYP2D6 phenotypes. All of the observed allele frequencies were in Hardy Weinberg Equilibrium, and the most prevalent genotypes were *1/*1 and *1/*2 which collectively accounted for 30% of all the genotypes. As for CYP2D6 phenotype distribution, almost 84% (184) of the study participants were EMs (Table 2). For three samples carrying variations in the CYP2D6 gene copy number, inferring the CYP2D6 metabolizer phenotype was only achievable after performing the Pyrosequencing-based method for allele quantification (28). Further information on CYP2D6 phenotype distribution by race is shown in supplement.
Table 1.
Summary statistics of demographic and baseline characteristics of the study population, and breakdown of demographic and baseline characteristics by race
| N=218 | Nonblack (n=134) | Black (n=84) | |
|---|---|---|---|
|
| |||
| Age (years) | |||
| Mean (SD) | 50.8 (9.0) | 50.7 (9.1) | 50.9 (9.0) |
| Gender | |||
| Female, n (%) | 52.2 | 47.4 | 65.5 |
| Race, n (%) | |||
| Asian | 0.5 | ||
| Black-African American (%) | 38.5 | ||
| White-European American (%) | 57.5 | ||
| Other (%) | 3.5 | ||
| Baseline SBP (mm Hg) | |||
| Mean (SD) | 147.4 (11.1) | 147.5 (11.0) | 147.3 (11.0) |
| Baseline DBP (mm Hg) | |||
| Mean (SD) | 94.5 (5.8) | 94.2 (5.5) | 95.0 (6.2) |
| Baseline HR (beats/min) | |||
| Mean (SD) | 79.0 (9.5) | 78.0 (9.4) | 80.8 (9.3) |
| Duration of hypertension (years) | |||
| Mean (SD) | 7.3 (6.6) | 7.5 (7.3) | 7.0 (5.3) |
| Family history of hypertension, n (%) | 76.6 | 73.3 | 81.0 |
| Never taken antihypertensive medication, n (%) | 7.0 | 5.2 | 8.3 |
| Taking antihypertensive medication at entry, n (%) | 77.2 | 81.5 | 70.2 |
| Smoking status, n (%) | |||
| - Current smoker | 17.6 | 11.1 | 27.4 |
| - Former smoker | 26.0 | 30.3 | 17.8 |
| - Ever smoker | 43.7 | 42.4 | 45.7 |
| BMI (kg/m2) | |||
| Mean (SD) | 30.6 (5.0) | 30.7 (5.0) | 30.4 (5.1) |
| Waist circumference (inches) | |||
| Mean (SD) | 38.6 (4.8) | 39.5 (4.9) | 37.3 (4.4) |
| Hip circumference (inches) | |||
| Mean (SD) | 43.3 (4.4) | 43.5 (4.3) | 43.1 (4.7) |
Table 2.
Distribution of the inferred CYP2D6 metabolizer phenotypes in study participants (n= 218):
| Genotype | n | Activity score | Phenotype | Phenotype frequency |
|---|---|---|---|---|
|
| ||||
| *3/*4 | 2 | 0 | PM | 5.0% (11) |
| *4/*4 | 7 | |||
| *4/*5 | 1 | |||
| *4/*6 | 1 | |||
|
| ||||
| *3/*17 | 1 | 0.5 | IM | 6.8% (15) |
| *4/*10 | 2 | |||
| *4/*17 | 1 | |||
| *4/*41 | 7 | |||
| *5/*10 | 1 | |||
| *5/*17 | 3 | |||
|
| ||||
| *1/*1 | 30 | 2 | EM | 84% (184) |
| *1/*2 | 41 | 2 | ||
| *1/*3 | 5 | 1 | ||
| *1/*4 | 16 | 1 | ||
| *1/*5 | 7 | 1 | ||
| *1/*6 | 2 | 1 | ||
| *1/*10 | 6 | 1.5 | ||
| *1/*17 | 6 | 1.5 | ||
| *1/*41 | 8 | 1.5 | ||
| *1xN/*4 | 2 | 2 | ||
| *1/*10xN† | 1 | 2 | ||
| *1/*17xN† | 1 | 2 | ||
| *2/*2 | 11 | 2 | ||
| *2/*4 | 18 | 1 | ||
| *2/*4xN | 1 | 1 | ||
| *2/*5 | 6 | 1 | ||
| *2/*10 | 4 | 1.5 | ||
| *2/*17 | 5 | 1.5 | ||
| *2/*41 | 5 | 1.5 | ||
| *10/*17 | 2 | 1 | ||
| *10xN/*41 | 1 | 1.5 | ||
| *17/*17 | 3 | 1 | ||
| *17/*41 | 2 | 1 | ||
| *41/*41 | 1 | 1 | ||
|
| ||||
| *1/*1xN | 2 | 3 | UM | 3.6% (8) |
| *1xN/*2 | 2 | 3 | ||
| *1/*2xN | 2 | 3 | ||
| *2xN/*10† | 1 | 2.5 | ||
| *2xN/*2 | 1 | 3 | ||
Pyrosequencing based method was used to identify duplicated allele and predict CYP2D6 metabolizer phenotype
There was no statistically significant difference between the four groups in terms of the mean daily dose of metoprolol, 200 mg in PMs, 193.74 mg (± 12.5) in IMs, 195 mg (± 10.9) in EMs & 200 mg in UMs (p-value=0.77). In ten study participants (5%), metoprolol was not titrated to the maximum recommended dosage, of whom nine were EMs and one was an IM.
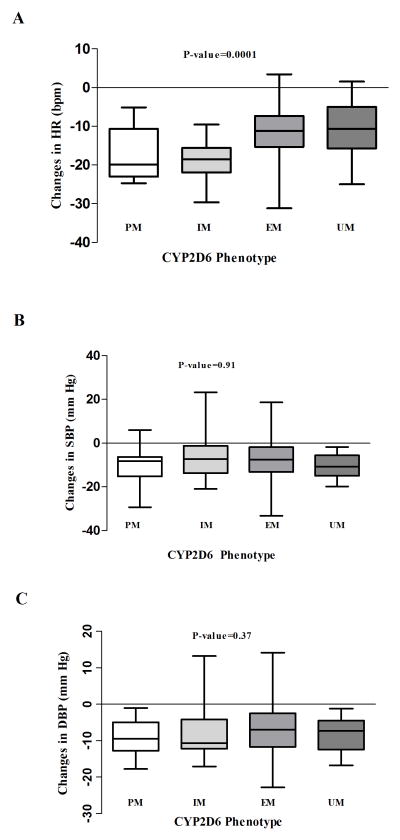
At the end of the study period, reduction in systolic (SBP) and diastolic blood pressure (DBP) did not vary significantly between the four CYP2D6 phenotypes (Table 3). However, the decline in heart rate (HR) was significantly greater in PMs & IMs compared with EMs & UMs (Table 3 and figure 1).
Table 3.
Changes from baseline in heart rate (HR), systolic blood pressure (SBP), & diastolic blood pressure (DBP) in poor (PM), intermediate (IM), extensive (EM) and ultrarapid (UM) metabolizers of CYP2D6 treated with metoprolol
| Parameter | PM | IM | EM | UM | P |
|---|---|---|---|---|---|
|
| |||||
| Change in HR (beats/min) | −16.6± 6.9 | − 18.6± 5.1 | −11.4± 6.6 | −11.2± 8.2 | 0.0001 |
| Change in SBP (mm Hg) | −9.4± 9.0 | −6.5± 11.1 | −7.1± 9.8 | −9.4± 5.1 | 0.91 |
| Change in DBP (mm Hg) | −9.3± 5.1 | −8.3± 7.5 | −7.0± 6.6 | −7.5± 4.8 | 0.37 |
Figure 1. Clinical response to metoprolol therapy by CYP2D6 phenotype.
Figure 1A. Comparison of the changes in heart rate (HR) from baseline by CYP2D6 phenotype. The bottom and top of the box represent 25th and 75th percentiles, the band in the middle represent the median (50th percentile), The lower whisker represent the minimum value of the data while the upper whisker represents the maximum value of the data (ANOVA p-value)
Figure 1B. Comparison of the changes in systolic blood pressure (SBP) from baseline by CYP2D6 phenotype. The bottom and top of the box represent 25th and 75th percentiles, the band in the middle represent the median (50th percentile), The lower whisker represent the minimum value of the data while the upper whisker represents the maximum value of the data (ANOVA p-value)
Figure 1C. Comparison of the changes in diastolic blood pressure (DBP) from baseline by CYP2D6 phenotype. The bottom and top of the box represent 25th and 75th percentiles, the band in the middle represent the median (50th percentile), The lower whisker represent the minimum value of the data while the upper whisker represents the maximum value of the data (ANOVA p-value)
The Analysis of Covariance (ANCOVA) test showed a statistically significant difference in HR change from baseline by CYP2D6 phenotype, but not in SBP or DBP change. In addition to age, baseline HR, and race, CYP2D6 phenotype was among the most significant predictors of the variability in HR response to metoprolol (table 4). The overall incidence of clinical bradycardia, defined as a resting HR less than 60 beats/minute, was observed in 25% of the patients (n=55). Stratification of the incidence of clinical bradycardia was not significantly different by CYP2D6 phenotype: 27% in PMs, 25% in IMs, 25% in EMs and 11% in UMs (p= 0.86). Of the 55 cases of clinical bradycardia, only four patients experienced symptomatic bradycardia. One of them had a PM phenotype while the others were EMs.
Table 4.
Results of Analysis of Covariance (ANCOVA) test showing CYP2D6 phenotype as a significant predictor of variability in heart rate changes only with metoprolol.
| Variable | Heart rate | Systolic blood pressure | Diastolic blood pressure | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| beta | SE* | 95% CI ** | p | beta | SE* | 95% CI** | p | beta | SE* | 95% CI ** | p | |
| CYP2D6 phenotype (PM) | 2.10 | 0.70 | 0.73,3.47 | 0.004 | − 0.03 | 1.6 | −3.17,3.11 | 0.97 | 0.50 | 0.7 | −2.8,3.8 | 0.50 |
| Baseline heart rate (bpm) | − 0.34 | 0.04 | −0.42, −0.26 | < 0.0001 | ||||||||
| Baseline SBP (mm Hg) | − 0.17 | 0.06 | −0.30, −0.05 | 0.003 | ||||||||
| Baseline DBP (mm Hg) | − 0.21 | 0.07 | −0.35, −0.07 | 0.0026 | ||||||||
| Age (years) | − 0.16 | 0.04 | −0.08, −0.24 | 0.0002 | 0.04 | 0.07 | −0.10,0.18 | 0.52 | − 0.01 | 0.04 | −0.09,0.07 | 0.74 |
| Race (African American) | − 2.24 | 0.80 | −0.67, −3.8 | 0.007 | − 6.61 | 1.30 | −9.16, −4.06 | < 0.0001 | − 5.01 | 0.86 | −3.33, −7.70 | < 0.0001 |
| Gender (Female) | 0.55 | 0.80 | −1.02,2.12 | 0.49 | 4.57 | 1.25 | 2.12,7.02 | 0.0003 | 3.48 | 0.82 | 1.88,5.36 | < 0.0001 |
SE: standard error,
CI: confidence interval
A total of 67 side effects (30.7%) were reported during the study period (table 5). However the rates of these side effects did not differ significantly between the CYP2D6 metabolizer phenotypes. Side effect rates were 36.4% (4), 26.7% (4), 31.0% (57), and 25% (2), in PMs, IMs, EMs and UMs, respectively (p=0.92). Further grouping of the PMs and IMs into one category and the EMs and UMs into the other, suggested no significant difference in the rates of adverse effects. Additionally, almost all of the reported side effects occurred following dose titration to 100 mg twice daily except for two cases of fatigue/tiredness which occurred at 50 mg twice daily. However, the genotype derived CYP2D6 phenotypes for these two study participants were EMs. This is suggestive that metoprolol was well tolerated even by study participants with impaired CYP2D6 function.
Table 5.
Types of adverse event rates in poor (PM), intermediate (IM), extensive (EM), & ultrarapid (UM) metabolizers
| Phenotype (n) | Type of Adverse Event* | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None | Fatigue/tiredness | Depression | Symptomatic bradycardia | Dizziness | Nausea | Diarrhea | Hypokalemia | Headache | Others† | |
| PMs (11) | 64% (7) | 0 | 0 | 9% (1) | 18% (2) | 0 | 0 | 0 | 0 | 9% (1) |
| IMs (15) | 73.3% (11) | 0 | 0 | 0 | 13.3% (2) | 0 | 6.7% (1) | 0 | 6.7% (1) | 0 |
| EMs (184) | 69.0% (127) | 11.4% (21) | 2.1% (4) | 1.6%(3) | 5.4% (10) | 0.6% (1) | 0.6% (1) | 0.6% (1) | 2.7% (5) | 6% (11) |
| UMs (8) | 75% (6) | 0 | 0 | 0 | 12.5% (1) | 0 | 0 | 0 | 0 | 12.5% (1) |
P-value= 0.41,
side effects reported by study participants: back pain, cough, muscle pain, bronchitis
Discussion
This study was designed to determine whether polymorphisms in CYP2D6 have an impact on the variability in efficacy and adverse responses observed among individuals treated with metoprolol. Although our study enrolled patients with uncomplicated hypertension, this should not preclude the generalizability of our results to other patient populations where metoprolol is considered the standard of care. After eight weeks of metoprolol therapy, our findings suggest that there is no relationship between CYP2D6 polymorphism and variability in the clinical efficacy of metoprolol as it pertains to SBP & DBP responses. Study participants with the PM and IM phenotype exhibited a greater reduction in HR, but not in SBP and DBP despite the fact that these phenotypes are linked to impaired metoprolol metabolism. Furthermore, higher doses of metoprolol were well tolerated by those with the PM and IM phenotype given the comparable rates of adverse effects that occurred in each CYP2D6 phenotype group.
Because of the consistent findings from published trials (18–22) demonstrating that the pharmacokinetic disposition of metoprolol is significantly altered among individuals with PM or IM phenotype compared with those with the EM or UM phenotype, we opted not to perform any pharmacokinetic study. The results from these studies were further substantiated in the recent meta-analysis conducted by Blacke et al (26) which included 13 studies with an overall number of 264 participants. In the meta-analysis, the IM and PM phenotypes had a 3–5 fold increase in all the pharmacokinetic parameters of metoprolol (AUC, Cmax, and terminal half-life) compared with the EM and UM phenotypes.
If the response to metoprolol is proportional to its plasma concentration, then the profound differences in drug exposure should translate into differences in either its clinical efficacy parameters or adverse effect rates depending on CYP2D6 phenotype. This is the rationale for the hypothesis that individuals with low or high CYP2D6 activity are predisposed to either undesired side effects or to therapeutic failure. Several clinical trials sought to further investigate this concept i.e. whether the interindividual differences in metoprolol plasma levels, driven primarily by polymorphisms in CYP2D6 gene, translate into a variability in drug response. However, the reported results were conflicting. While some studies showed that the variation in metoprolol response could be explained by polymorphisms in the CYP2D6 gene (23–25), others did not. This might be attributed to the small sample size of the PM phenotype in the studies that did not find an association for any phenotype (18–20).
Given this controversy, we conducted this clinical trial to determine whether CYP2D6 polymorphisms are responsible for the variation in metoprolol response. Hence, it is important to highlight the strengths of our study. Its prospective design enabled us to assess the recorded home and office HR and BP measurements as well as adverse effect rates throughout the trial period, and most importantly during the metoprolol dose titration phase, so that we can accurately examine the relationship between CYP2D6 polymorphisms and the variation in clinical efficacy and adverse effect responses to metoprolol.
We genotyped the CYP2D6 gene for the most studied variants as recommended by the CPIC guidelines (28) mainly because of the highly polymorphic nature that the gene exhibits, and the differences in the frequencies of the alleles among different ethnic groups. Given the diverse ethnic background of our study participants, it was of paramount importance that our genotyping methods targeted all the variant alleles. Because CYP2D6 polymorphisms result from whole gene duplication or deletion in addition to single nucleotide substitution, we performed Taqman® copy number assay to estimate the number of CYP2D6 gene copies carried in each sample. We also utilized the novel Pyrosequencing-based assay for allele quantification, recently described by Langaee et al (28) to define the duplicated/multiplied allele in samples with problematic genotypes due to copy number variations. As a consequence, our genotyping approach is very comprehensive compared to some studies in the literature, which studied only the *4 loss of function allele. Additionally, our study is unique in terms of utilizing the activity score method endorsed by the CPIC guidelines to infer CYP2D6 phenotype from the genotype and copy number variation information. Hence the CYP2D6 phenotypes of our study participants comprised the CYP2D6 activity spectrum, and not just the PM and EM phenotypes (24, 25). Subsequently, our findings provide more complete data on the influence of CYP2D6 polymorphisms on response to metoprolol. As suggested by our results, CYP2D6 polymorphisms impacted HR response, but not DBP or SBP responses to metoprolol. Of note, our sample size was large enough to detect the difference in DBP response (−4 mm Hg) between CYP2D6 phenotypes as reported by some studies (24), indicating clinically meaningful differences in BP response by CYP2D6 genotype do not exist.
It has been suggested that the differences in response to metoprolol between PMs and non PMs occur during the dose titration phase of the drug only, when the dose-response relationship is linear, and these differences tend to subside or become insignificant with higher doses (e.g. 200 mg/day), when the sigmoid portion of the response curve is reached. This notion could explain the discrepancies between our findings and what was reported in the literature as it pertains to DBP changes since higher doses of metoprolol were used in the current study as opposed to 50 mg of metoprolol XL in the study by Rau et al. (25). Although we do not have data on 50 mg daily, our participants were initiated on 50 mg twice daily (which is also likely to be on the linear portion of the dose-response curve), and the data at that dose were in line with our findings at 100 mg twice daily. Thus, it remains unclear if differences in the doses studied account for the differences in relationship with the BP response by CYP2D6 phenotype.
The adverse effect rates observed in this study are comparable to those reported in other trials (30%) despite the stringent inclusion criteria that were put in place in this study. It is worth noting that only one patient withdrew from the study due to symptomatic bradycardia (47 beats/min), which occurred following administration of the second dose of metoprolol 50 mg. Contrary to what was anticipated, his genotype-predicted phenotype was EM.
In conclusion, we observed a significant relationship between CYP2D6 genotype-derived phenotype and HR response to metoprolol, but no relationship with BP or adverse effects. Metoprolol was well tolerated by all study participants, and having the PM or IM phenotype did not preclude titrating metoprolol up to the maximum protocol dose of 100 mg twice daily. Furthermore, BP response did not differ by CYP2D6 phenotype. The only response we observed to be influenced by CYP2D6 phenotype was HR, which is readily measured in the clinical setting. In this well-powered, carefully genotyped study, there is no evidence for the clinical utility of CYP2D6 genotyping to guide metoprolol therapy.
Methods
Patient selection
The Pharmacogenomic Evaluation of Antihypertensive Responses-2 (PEAR-2) study is a prospective, multicenter, open label, sequential monotherapy clinical trial (clinical trial.gov identifier: NCT01203852). Participants enrolled in this study were recruited at three sites: the University of Florida (Gainesville, FL), Emory University (Atlanta, GA), and Mayo Clinic (Rochester, MN). The study protocol was approved by the Institutional Review Board of each participating institution, and all study participants provided written informed consent for screening prior to active enrollment.
The criteria for inclusion into the study were: males or females, 18–65 years of age, of any race or ethnicity, with mild to moderate newly diagnosed, untreated or known uncomplicated hypertension. Patients were excluded if they had any of the following exclusion criteria: secondary forms of hypertension, treated office systolic blood pressure > 170 mm Hg, isolated systolic hypertension, other diseases requiring treatment with blood pressure lowering medications, known cardiovascular diseases, diabetes mellitus, primary renal disease, renal insufficiency (serum creatinine > 1.5 in men or 1.4 in women), liver abnormalities (liver enzymes > 2.5 upper limits of normal), pregnancy or lactation, chronic use of medications that could potentially affect the blood pressure. Additionally, exclusion criteria specific for β-1 blocker therapy were: HR < 55 beats/min (in the absence of a β-1 blocker), presence of a cardiac pacemaker, history of Raynaud’s phenomenon, as well as asthma or COPD.
BP screening for inclusion
Study participants, not meeting any of the aforementioned exclusion criteria underwent further screening for inclusion in the study based upon their home blood pressure (HBP) and office blood pressure (OBP) measurements. Hence, they undertook an extensive training on how to use the HBP monitors by Microlife, model #s BP3AC1-PC and BP3MC1-PC (Dunedin, FL), provided to them for use at home with appropriately sized cuffs. Of note, the accuracy of these monitors was inspected by comparing the BP readings recorded by these monitors against those performed manually by trained study coordinators prior to the study participants taking them home for use. Initially, the study participants underwent a 4 week wash-out period, during which they were instructed to measure their HBP in triplicate twice daily (in the morning and evening).
Following completion of the washout period, participants returned to the clinic with their HBP monitors for assessment of the recorded HBP measurements. HBP readings were accepted if at least 5 morning and 5 evening BP measurements were recorded in the previous 7 days. Study participants with insufficient data were asked to return to the clinic when they had sufficient HBP readings. During the clinic visit, OBP measurements were also obtained using the same HBP monitor assigned to each subject so as to avoid device related discrepancies in BP measurements. Participants were deemed eligible to proceed with the trial if their BP readings met the following criteria: average home (HSBP) and average office systolic blood pressures (OSBP) less than 180 mm Hg, average home diastolic blood pressure (HDBP) was greater than 85 mm Hg, but less than 110 mm Hg, and average office diastolic blood pressure (ODBP) greater than 90 mm Hg, but less than 110 mm Hg. Accordingly, study participants were excluded if, by either BP measurement method, DBP was greater than 110 mm Hg or SBP greater than 180 mm Hg. For those who met the BP inclusion criteria, baseline studies (collection of HBP & OBP readings as well as collection of biological samples for laboratory, DNA and RNA analysis) were initiated.
Intervention
eligible study participants were started on metoprolol tartrate at an initial dose of 50 mg twice daily. The drug was provided to all patients in blister packs as means to improve and assess adherence to therapy. After two weeks of therapy, patients returned to the clinic for drug response assessment. Those with average HBP or OBP readings > 120/70 mm Hg and HR > 55 beats per minute (bpm) had their metoprolol dose titrated to 100 mg twice daily for an additional 6 week period. During the final visit, data collection similar to baseline was also performed. Afterwards, study participants had their metoprolol dose tapered in increments of 50 mg every 2 days until it was completely stopped, and they entered the second washout period of the study with subsequent treatment with chlorthalidone. During each visit, study participants were asked open ended questions to assess whether they were any experiencing adverse effect that might have occurred following the initiation of metoprolol. Side effects of interest that were potentially β-1 blocker related included, but were not limited to fatigue, tiredness, depression, bradycardia, and dizziness.
Genotyping
Genomic DNA was isolated from lymphocytes in whole blood using the FlexiGene DNA kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Genotyping for CYP2D6*2,*3,*4,*6,*10,*17 and *41 variant alleles was performed by polymerase chain reaction (PCR) followed by pyrosequencing using a PSQ HS96A SNP reagent kit (Qiagen, Valencia, CA), (27)and method paper). The variant alleles that were genotyped were those recommended by the recent Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy, and these include:*2, *3, *4,*6,*10,*17 and*41 (28). If no sequence variation was detected, then the allele was by default given the wild type name,*1. Of note, CYP2D6 genotyping was performed after the study participants finished the metoprolol phase of the trial so as to avoid any potential for bias.
CYP2D6 copy number assay
Copy number variations in the CYP2D6 gene were estimated by TaqMan® Copy Number Assay (Life Technologies, CA) and the novel Pyrosequencing-based method for allele quantification (methods paper ref). In the TaqMan® method RNase P served as the internal control for copy number analysis. The primers used in this method were selected to target a specific sequence on exon 9. All samples were run in quadruplicate along with DNA samples with known copy number i.e. carrying deletions, or two, three and four gene copies, used as positive controls. Relative quantification of CYP2D6 gene copy number was performed by using CopyCaller Software (Applied Biosystems, Inc).
The newly described Pyrosequencing based approach for allele quantification (28) was performed only on samples with CYP2D6 copy number variations where inferring the CYP2D6 metabolizer phenotype from the genotypic information may be problematic i.e. varies depending on which of the two inherited CYP2D6 alleles is duplicated or multiplied. Briefly, the software for Pyrosequencing analysis measures at the end of the pyrosequencing reaction, the percent distribution of each allelic base at the polymorphic region. Any deviation from a percent distribution of 50% for either allele is suggestive of variations in its copy number. Accordingly, the allele with a higher percent distribution is the duplicated/multiplied allele.
Phenotype derivation
The activity score method was used to predict the CYP2D6 metabolizer phenotype from the genotype data, and is shown in Supplemental table 1(15, 26). Each allele is given a value ranging from 0 to 1 depending on the functional status of the CYP2D6 enzyme. Alleles encoding a fully functional enzyme are given a value of 1, whereas those encoding an enzyme with null or partial function are given a value of either 0 or 0.5. If there are two or more copies of a particular allele, then the value is multiplied by the copy number of the allele. The final activity score is the sum of the values assigned to each of the alleles constituting a genotype. Thus, four genotype derived phenotypes arise: the PM, IM, EM, and UM phenotypes (Supplemental table 2).
Statistical Analysis
Only study participants who completed the metoprolol portion of the study at the time this work was done, were included in the final analysis (n=218). All demographic numerical data were expressed as mean values with standard deviations whereas categorical data were expressed as percentages. Departure from Hardy Weinberg Equilibrium (HWE) was tested for each CYP2D6 variant allele by race/ethnic group using chi-squared test with one degree of freedom. Differences in the mean daily dose of metoprolol between PMs, IMs, EMs, & UMs were compared by Kruskal Wallis test. The ANOVA test was used to compare the means of the HR and BP (SBP & DBP) changes from baseline between the four CYP2D6 phenotypes. To control for the effects of baseline characteristics (age, gender, race, baseline HR and baseline BP) on metoprolol response, the Analysis of Covariance (ANCOVA) test was also performed to evaluate the differences in BP and HR changes with CYP2D6 phenotype as the explanatory variable. Post-hoc analysis was performed with Tukey’s test to find the means that were significantly different from the others. P values less than 0.05 were considered statistically significant. The study had a power of 89% to detect an effect size of at least 0.6 for HR and BP changes from baseline. To compare the rates of adverse effects between the four phenotypes, the Fisher’s exact test was used. All statistical analyses were performed with SAS (version 9.3, SAS Institute Inc., Cary, NC).
Supplementary Material
Study highlights.
What is the current knowledge on the topic?
The impact of CYP2D6 polymorphisms on response to metoprolol is controversial due to the conflicting results from various studies with different study designs.
What question this study addressed?
This study was designed to investigate whether polymorphisms in CYP2D6 gene impact the clinical efficacy and tolerability of metoprolol in patients with uncomplicated hypertension.
What this study adds to our knowledge?
This study further substantiates the findings from other clinical trials that functional polymorphisms in the CYP2D6 gene do not affect BP or adverse responses to metoprolol. The HR response in affected by CYP2D6 polymorphisms.
How might this change clinical pharmacology and therapeutics?
These data suggest limited clinical utility of CYP2D6 genotypic information to guide use or dosing of metoprolol.
Acknowledgments
The PEAR-2 study was supported by the National Institute of Health Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic). PEAR-2 was also supported by funds from the Mayo Foundation.
Footnotes
Conflict of Interest:
No conflicts to be disclosed
Author contributions:
I.S.H. wrote the manuscript. A.B.C., J.G.G., S.T.T., R.M.C.D. and J.A.J. designed the research. I.S.H., T.Y.L., R.D., S.G. and B.M.B. performed the research. I.S.H. and Y.G. analyzed the data. T.Y.L. Contributed New Reagents/Analytical Tools
References
- 1.Yancy CW, et al. 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013 doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 2.Chobanian AV, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289:2560–72. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 3.O’Gara PT, et al. 2013 ACCF/AHA guideline for the management of ST-elevation myocardial infarction: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2013;61:e78–140. doi: 10.1016/j.jacc.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Jneid H, et al. 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/non-ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2012;60:645–81. doi: 10.1016/j.jacc.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Gottlieb SS, et al. Tolerability of beta-blocker initiation and titration in the Metoprolol CR/XL Randomized Intervention Trial in Congestive Heart Failure (MERIT-HF) Circulation. 2002;105:1182–8. doi: 10.1161/hc1002.105180. [DOI] [PubMed] [Google Scholar]
- 6.Wikstrand J, et al. Metoprolol versus thiazide diuretics in hypertension. Morbidity results from the MAPHY Study. Hypertension. 1991;17:579–88. doi: 10.1161/01.hyp.17.4.579. [DOI] [PubMed] [Google Scholar]
- 7.informatics, I.i.f.h. The use of medications in the United States: Review of 2010. Apr, 2011. (April, 2011) [Google Scholar]
- 8.Materson BJ, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med. 1993;328:914–21. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 9.Deedwania PC, Gottlieb S, Ghali JK, Waagstein F, Wikstrand JC. Efficacy, safety and tolerability of beta-adrenergic blockade with metoprolol CR/XL in elderly patients with heart failure. Eur Heart J. 2004;25:1300–9. doi: 10.1016/j.ehj.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Morris TH, Kaumann AJ. Different steric characteristics of beta 1- and beta 2-adrenoceptors. Naunyn Schmiedebergs Arch Pharmacol. 1984;327:176–9. doi: 10.1007/BF00500913. [DOI] [PubMed] [Google Scholar]
- 11.El Tayar N, Testa B, van de Waterbeemd H, Carrupt PA, Kaumann AJ. Influence of lipophilicity and chirality on the selectivity of ligands for beta 1- and beta 2-adrenoceptors. J Pharm Pharmacol. 1988;40:609–12. doi: 10.1111/j.2042-7158.1988.tb05319.x. [DOI] [PubMed] [Google Scholar]
- 12.Toda N, Hayashi S, Hatano Y, Okunishi H, Miyazaki M. Selectivity and steric effects of metoprolol isomers on isolated rabbit atria, arteries and tracheal muscles. J Pharmacol Exp Ther. 1978;207:311–9. [PubMed] [Google Scholar]
- 13.Johnson JA, Burlew BS. Metoprolol metabolism via cytochrome P4502D6 in ethnic populations. Drug Metab Dispos. 1996;24:350–5. [PubMed] [Google Scholar]
- 14.Jonkers RE, Koopmans RP, Portier EJ, van Boxtel CJ. Debrisoquine phenotype and the pharmacokinetics and beta-2 receptor pharmacodynamics of metoprolol and its enantiomers. J Pharmacol Exp Ther. 1991;256:959–66. [PubMed] [Google Scholar]
- 15.Lennard MS, Silas JH, Freestone S, Ramsay LE, Tucker GT, Woods HF. Oxidation phenotype--a major determinant of metoprolol metabolism and response. N Engl J Med. 1982;307:1558–60. doi: 10.1056/NEJM198212163072505. [DOI] [PubMed] [Google Scholar]
- 16.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–42. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 17.Griese EU, et al. Assessment of the predictive power of genotypes for the in-vivo catalytic function of CYP2D6 in a German population. Pharmacogenetics. 1998;8:15–26. doi: 10.1097/00008571-199802000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Zineh I, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther. 2004;76:536–44. doi: 10.1016/j.clpt.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 19.Fux R, et al. Impact of CYP2D6 genotype on adverse effects during treatment with metoprolol: a prospective clinical study. Clin Pharmacol Ther. 2005;78:378–87. doi: 10.1016/j.clpt.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Lefebvre J, Poirier L, Poirier P, Turgeon J, Lacourciere Y. The influence of CYP2D6 phenotype on the clinical response of nebivolol in patients with essential hypertension. Br J Clin Pharmacol. 2007;63:575–82. doi: 10.1111/j.1365-2125.2006.02796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozawa T, et al. Influence of CYP2D6 genotype on metoprolol plasma concentration and beta-adrenergic inhibition during long-term treatment: a comparison with bisoprolol. J Cardiovasc Pharmacol. 2005;46:713–20. doi: 10.1097/01.fjc.0000184117.76188.68. [DOI] [PubMed] [Google Scholar]
- 22.Rau T, et al. Effect of the CYP2D6 genotype on metoprolol metabolism persists during long-term treatment. Pharmacogenetics. 2002;12:465–72. doi: 10.1097/00008571-200208000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Wuttke H, et al. Increased frequency of cytochrome P450 2D6 poor metabolizers among patients with metoprolol-associated adverse effects. Clin Pharmacol Ther. 2002;72:429–37. doi: 10.1067/mcp.2002.127111. [DOI] [PubMed] [Google Scholar]
- 24.Bijl MJ, et al. Genetic variation in the CYP2D6 gene is associated with a lower heart rate and blood pressure in beta-blocker users. Clin Pharmacol Ther. 2009;85:45–50. doi: 10.1038/clpt.2008.172. [DOI] [PubMed] [Google Scholar]
- 25.Rau T, et al. Impact of the CYP2D6 genotype on the clinical effects of metoprolol: a prospective longitudinal study. Clin Pharmacol Ther. 2009;85:269–72. doi: 10.1038/clpt.2008.218. [DOI] [PubMed] [Google Scholar]
- 26.Blake CM, Kharasch ED, Schwab M, Nagele P. A Meta-Analysis of CYP2D6 Metabolizer Phenotype and Metoprolol Pharmacokinetics. Clin Pharmacol Ther. 2013;94:394–9. doi: 10.1038/clpt.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91:321–6. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langaee T, Hamadeh I, Chapman AB, Gums JG, Johnson JA. A novel simple method for determining CYP2D6 copy number and identify allele(s) with duplication/multiplication. Manuscript is submitted for publication. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.