Abstract
Background
There is growing concern on the residual risk of bacterial contamination of platelet concentrates in Germany, despite the reduction of the shelf-life of these concentrates and the introduction of bacterial screening. In this study, the applicability of the BactiFlow flow cytometric assay for bacterial screening of platelet concentrates on day 2 or 3 of their shelf-life was assessed in two German blood services. The results were used to evaluate currently implemented or newly discussed screening strategies.
Materials and methods
Two thousand and ten apheresis platelet concentrates were tested on day 2 or day 3 after donation using BactiFlow flow cytometry. Reactive samples were confirmed by the BacT/Alert culture system.
Results
Twenty-four of the 2,100 platelet concentrates tested were reactive in the first test by BactiFlow. Of these 24 platelet concentrates, 12 were false-positive and the other 12 were initially reactive. None of the microbiological cultures of the initially reactive samples was positive. Parallel examination of 1,026 platelet concentrates by culture revealed three positive platelet concentrates with bacteria detected only in the anaerobic culture bottle and identified as Staphylococcus species. Two platelet concentrates were confirmed positive for Staphylcoccus epidermidis by culture. Retrospective analysis of the growth kinetics of the bacteria indicated that the bacterial titres were most likely below the diagnostic sensitivity of the BactiFlow assay (<300 CFU/mL) and probably had no transfusion relevance.
Conclusions
The BactiFlow assay is very convenient for bacterial screening of platelet concentrates independently of the testing day and the screening strategy. Although the optimal screening strategy could not be defined, this study provides further data to help achieve this goal.
Keywords: bacterial contamination, flow cytometry, rapid method, BactiFlow, screening strategy
Introduction
The greatest risk of severe transfusion-transmitted infections in developed countries currently stems from bacterially contaminated platelet concentrates (PC), with the risk of such bacterial infections being approximately 50–250-fold higher than the combined risk of potential viral infections (HIV, HCV, HBV and HTLV-I/II1). Between 2005 and 2009 bacterial infection was the third leading cause of transfusion-related fatalities reported to the Food and Drug Administration2. PC storage conditions in gas-permeable bags with constant agitation at room temperature (20–24 °C) promote bacterial proliferation from small bacterial inocula immediately after donation to high titres during PC storage3,4. Bacterial screening of PC with culture methods, using an early-sampling strategy with the concept of release of negative-to-date PC5, was implemented in some transfusion facilities6–9 but was demonstrated to be unsatisfactory with regards to the risk of sampling error and time to results. Bacterial screening using BactiFlow (bioMérieux, Nürtingen, Germany), a flow cytometric-based technique, has been implemented on day 4 or 5 after production of the blood product10,11. Analysis of spiked PC with a rapid bacterial screening method (BactiFlow and 16S rDNA nucleic acid amplification technology [NAT]) on day 3, 4 or 5 has provided further perspectives12. The Pan Genera Detection assay is an immunological detection method that was introduced successfully as a day-of-issue screening method2 in a large study of 27,620 PC. This test had, however, already presented some shortcomings regarding the sensitivity of detection of Gram-negative bacteria13,14. In 2009, the shelf-life of PC was reduced in Germany from 5 to 4 days to minimise transfusion-associated bacterial sepsis because most platelet-related septic complications have been observed with older PC6,7,9,15–18. Nevertheless, in 2010 one fatal case with a bacterially contaminated PC transfused on day 4 still occurred, and consequently questions about the shelf-life of PC and the optimal screening strategy were raised again. The American Association of Blood Banks approved a new standard (5.1.5.1.1) in January 2011, resulting in the application of a point-of issue rapid assay for the screening of whole-blood-derived platelets19.
In the present study, we introduced the BactiFlow rapid flow cytometric assay10,11,20 in a routine test setting in two different transfusion facilities to screen for bacterial contamination of PC on day 2 or 3 of storage in order to provide data demonstrating the diverse possible applications of this method. Old and new thoughts regarding different screening strategies are discussed.
Materials and methods
Collection of platelet concentrates and study design
Apheresis-derived PC were analysed for the presence of bacteria on day 2 after production (sampling period: 12 months) by the Uni.Blutspendedienst OWL (ILTM, Bad Oeynhausen, Germany), or at the end of day 3 after production (sampling period: 4 months) by the German Red Cross Blood Transfusion Service West (GRCW, Central Laboratory, Hagen, Germany) using the BactiFlow flow cytometric assay. The ILTM performed parallel screening of all PC using the BacT/Alert culture system, whereas the GRCW confirmed BactiFlow reactive samples only. Pre-donation sampling was performed by collecting the first 30 to 40 mL of whole blood in a pre-donation sampling bag for blood tests and further laboratory examinations.
The ILTM prepared apheresis-derived single donor PC (APC) after standard processing with the Haemonetics MCS+ instrument (Haemonetics GmbH, Munich, Germany). Two APC were prepared by double apheresis from a single donor (donor-related PC, PC a/b). The donor’s arm was disinfected by spraying once with Kodan (Schuelke and Mayr, Norderstedt, Germany) and wiping with a sterile cotton swab. Extensive spraying was repeated once with a minimum liquid residence time of 90 seconds without further wiping. PC were stored in gas-permeable containers (LN994CF-CPP, Haemonetics GmbH) at 20–24 °C under constant agitation. The final product consisted of 2.0–4.0×1011 platelets/unit (205–295 mL) and 0.16–0.24 L/L ACD-A stabiliser and 0.76–0.84 L/L plasma per mL of preparation.
The GRCW also disinfected the donor’s arm with Kodan, spraying once and wiping with a sterile cotton swab, followed by a single, extensive spray and a minimum liquid residence of 30 seconds without further wiping. Routinely, one or two APC (PC a/b) were prepared from one donor after standard processing with different equipment -Trima Accel (formerly from Caridian BCT, now Terumo BCT, Belgium) and Haemonetics MCS+ (Haemonetics GmbH, München, Germany)- and stored in gas-permeable containers belonging to the respective machines at 20–24 °C under constant agitation. The final product consisted of 2.0–5.0×1011 platelets/unit (200–300 mL) and 0.11–0.22 L/L ACD-A stabiliser and 0.78–0.89 L/L plasma per mL preparation.
PC were analysed for the presence of bacteria on day 2 (ILTM) or day 3 (GRCW) after production. The criteria for evaluating the BactiFlow results were as follows: (a) BactiFlow initial testing <300 counts/mL (C/mL): negative, (b) BactiFlow initial testing ≥300 C/mL: reactive, (c) BactiFlow initial testing ≥300 C/mL, BactiFlow retesting (ILTM: duplicate, GRCW: single) <300 C/mL: initial reactive, (d) BactiFlow initial testing ≥300 C/mL, retesting ≥300 C/mL, negative culture result: false positive, (e) BactiFlow initial testing ≥300 C/mL, retesting ≥300 C/mL, positive culture result: positive. Records of patients receiving PC before the respective cultures became positive were reviewed for evidence of transfusion reactions, including inflammatory parameters (C-reactive protein, leucocyte count) and positive blood culture results.
Sterility testing of platelet concentrates and bacterial identification
The ILTM and GRWC collected samples (12–15 mL and 22–25 mL, respectively) of the PC into an additional sterile sampling bag using a flexible tube welding device (TSCD, Terumo Europe, Leuven, Belgium). Manually operated flow cytometric analysis was performed with 1 mL of PC sample as described previously20. Semiautomated flow cytometric analysis was performed with a minor modification from the previously published protocol11. The supernatant was discarded after the centrifiltration step, 1 mL buffer ChemSol B24 was added, then samples were placed on the BactiFlow ALS system (bioMérieux) and subsequently processed according the manufacturer’s instructions. For sterility testing at the ILTM, aliquots were split to inoculate aerobic and anaerobic culture bottles (BacT/Alert BPA/BPN, bioMérieux, Nürtingen, Germany), each with 5 mL of sample and 3 mL of PC were placed into an additional sterile tube for BactiFlow analysis. The GRCW inoculated aerobic and anaerobic culture bottles (BacT/Alert SA/SN, bioMérieux) with 10 mL of sample.
Culture bottles were incubated at 37 °C in the BacT/Alert automated culture system until a positive signal was detected, or for up to 7 days. Samples that did not react after 7 days of incubation were considered negative. Reactive culture bottles were subcultured on blood agar media (PVX, COS [bioMérieux]) and isolates were identified using conventional microbiological identification systems (API 20A, Vitek II [bioMérieux]), matrix assisted laser desorption time-of-flight (MALDI-ToF) mass spectrometry (Bruker Daltonics, Bremen, Germany) and molecular genetic identification by sequencing analysis of 16S ribosomal DNA21,22.
Spiking experiments with platelet concentrate isolates
For the determination of bacterial growth kinetics of potentially contaminating bacteria isolated from PC during this study, in each case two fresh PC units were inoculated with <1 colony-forming unit (CFU)/mL of Staphylococcus epidermidis (isolate 186 or 454), or Staphylococcus hominis isolate 4224. Bacterial titres of <1 CFU/mL were achieved by 10-fold serial dilution of stationary-grown overnight cultures in phosphate-buffered saline, followed by inoculation of the respective dilution as follows: (i) S. epidermidis isolate 186: 0.5 mL dilution 10−5 (15 CFU/mL), (ii) S. epidermidis isolate 454: 1 mL dilution 10−7 (6 CFU/mL) and (iii) S. hominis isolate 4224: 1 mL dilution 10−7 (15 CFU/mL). All PC used were sampled before bacterial inoculation to ensure baseline sterility.
In order to control bacterial inoculation of a PC unit, a sample was taken immediately after inoculation (0 h) and analysed with the BacT/Alert 3D continuous monitoring system. Sampling for BactiFlow analysis and cultivation was performed during storage at 22 °C with agitation on days 0, 2, 4, 5, and 7 after inoculation. Sampling and detection of bacteria by both methods were performed as described above. To monitor the growth kinetics of bacteria, samples were taken on days 1 to 5 and 7 after inoculation, and 100 μL aliquots of serial dilutions of PC samples were plated in triplicate onto tryptone soy agar and incubated at 37 °C for 48 hours. After incubation, the number of colonies was counted and the bacterial count per mL of sample was calculated.
Results
Bacterial screening
In a period of 12 months (ILTM) or 4 months (GRCW), 2100 PC were screened for bacterial contamination by BactiFlow flow cytometry on days 2 and 3 (Table I). In total, 24 PC were reactive in the first test (≥300 C/mL, Table I) by the BactiFlow assay (1.75% on day 2, 0.56% on day 3). Of these 24 PC, 12 were false-positive (0.57%), whereas the other 12 PC were only initially reactive, but retesting after an additional 2-hour incubation revealed negative results by BactiFlow and automated culture testing.
Table I.
Summary of screening results for bacterial contamination of apheresis-derived single donor platelet concentrates using BactiFlow flow cytometry.
| Total number | day 2 1,026 |
day 3 1,074 |
|
|---|---|---|---|
| negative (<300 C/mL) | 1,002 | 1,068 | |
| reactive | 18 | 6 | |
| BactiFlow | initially reactive* | 12 | 6 |
| false-positive** | 6 | 6 | |
| positive*** | - | - |
Initial testing ≥300 C/mL, retesting (ILTM: duplicate, GRCW: single) <300 C/mL;
Initial testing ≥300 C/mL, retesting ≥300 C/mL, negative culture result;
Initial testing ≥300 C/mL, retesting ≥300 C/mL, positive culture result.
Parallel screening by microbiological cultures performed by the ILTM revealed a positive signal for five PC samples (0.59%) whereas the BactiFlow assay remained negative (Table II). Only two donor-related PC (APC-1a/b, Table II) gave a confirmed positive culture result for S. epidermidis in both culture bottles during testing on day 2. A second sampling was not performed because both products had already been transfused at the time of the positive culture result. The remaining three culture-only positive samples exclusively gave a positive result in the anaerobic culture bottle (APC-2b, APC-3b: S. hominis;APC-4b: S. epidermidis, Table II). All PC had already been transfused without any transfusion reaction at the time of the positive culture result and no sample or predonation bag was available for confirmation of the positive result.
Table II.
Screening for bacterial contamination of apheresis-derived single donor platelet concentrates by BactiFlow flow cytometry and culture.
| Portion | Day of testing after donation (d0) | Time of transfusion after testinga | Aerobic culture detection (d)b | Anaerobic culture detection (d)c | Bacterial strains | BactiFlow results (C/mL) | |
|---|---|---|---|---|---|---|---|
| APC-1 | a | 2 | <24 h | Positive (1.24) | Positive (1.25) | S. epidermidis isolate 454 | Negative (30) |
| b | 2 | <24 h | Positive (1.23) | Positive (1.20) | S. epidermidis isolate 454 | Negative (0) | |
| APC-2 | a | 2 | <24 h | Negativec | Negativec | - | Negative (0) |
| b | 2 | <24 h | Negativec | Positive (1.02) | S. hominis | Negative (0) | |
| APC-3 | a | 2 | <12 h | Negativec | Negativec | - | Negative (61) |
| b | 2 | <24 h | Negativec | Positive (0.93) | S. hominis isolate 4224 | Negative (30) | |
| APC-4 | a | 2 | <12 h | Negativec | Negativec | - | Negative (0) |
| b | 2 | <12 h | Negativec | Positive (0.97) | S. epidermidis isolate 186 | Negative (0) |
Transfusion of PC based on the negative BactiFlow results and negative-to-date concept of BacT/alert culture results;
Detection time after sampling;
Negative, negative after 7 days of incubation;
Retesting was performed for available products immediately after receipt of a positive culture result.
Growth kinetics of bacteria
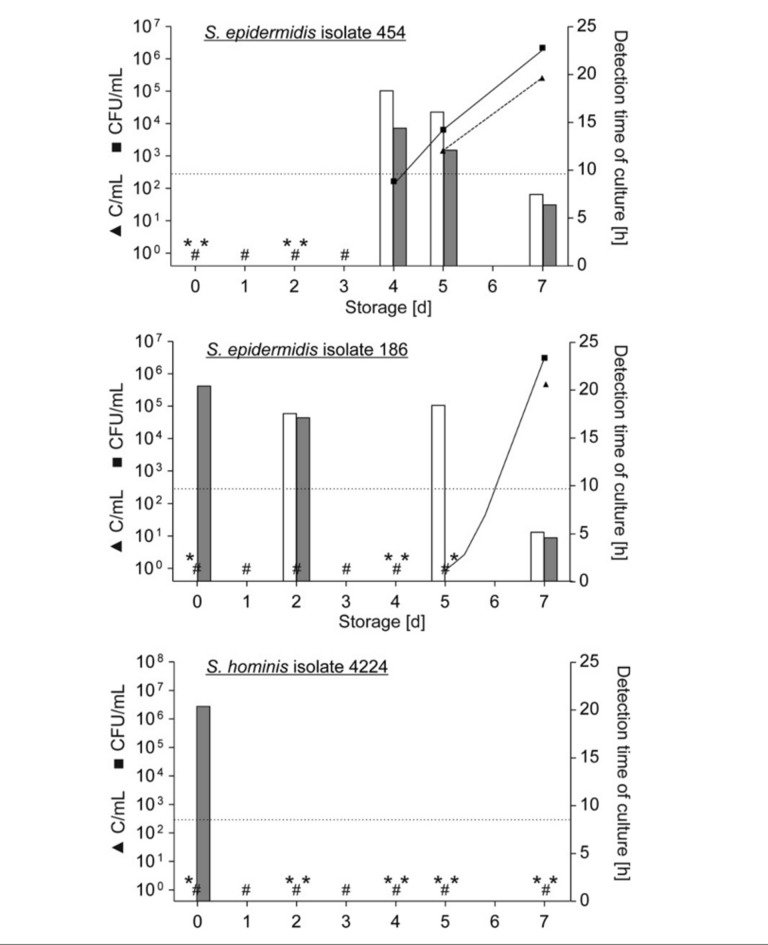
In order to analyse the probability of obtaining a positive BactiFlow result at the time of sampling, growth kinetics of three bacterial strains isolated from PC by the ILTM were evaluated. For each isolate two different fresh PC were inoculated immediately after production and analysed in parallel by standard cultures and BactiFlow assay four or five times during storage (Figure 1). Bacterial counts were determined by colony-forming assay on days 1, 2, 3, 4, 5 and 7 of storage. Bacterial proliferation was not detected prior to 72 hours of storage for both PC inoculated with the same strain. Subsequently only the results of the repeated proliferation experiment are presented in detail (Figure 1). S. hominis isolate 4224 did not proliferate in PC during storage for 7 days. However, PC contamination was proven by detection directly after inoculation in the anaerobic culture bottle, with a detection time of 20.40 hours. BactiFlow testing remained negative throughout the storage period of 7 days.
Figure 1.
Bacterial proliferation and detection by BactiFlow and cultures observed during the repeated proliferation experiment.
For each bacterium, a single APC was inoculated with <1 CFU/mL of S. epidermidis isolates 454 (6 CFU/PC) or 186 (7 CFU/PC) or S. hominis isolate 4224 (15 CFU/PC) respectively and stored at 22 °C with agitation. Samples for the plating assay were taken immediately (day 0) and on days 1, 2, 3, 4, 5 and 7 after inoculation; samples for flow cytometric analysis and microbial culture were taken on days 0, 2, 4, 5 and 7 after inoculation. Bacteria were enumerated by a colony-forming assay (■, solid lines) and the BactiFlow assay (▲, dashed lines), left scale. # represents negative plate count results. Negative BactiFlow results (<300 C/mL) were not displayed. The detection time of culture is displayed in bars (white bars: aerobic bottle, grey bars: anaerobic bottle), whereas * represents negative culture results, right scale. The dotted horizontal line represents the diagnostic sensitivity of the BactiFlow assay (300 CFU/mL).
S. epidermidis isolates 186 and 454 proliferated in PC: isolate 186 started to grow at around 5 days of incubation, whereas isolate 454 began to proliferate between 3 and 4 days of storage. For isolate 186, the BacT/Alert system gave positive results in the aerobic and/or anaerobic culture bottle before the onset of bacterial proliferation, with a detection time of 17.01–20.35 hours. The detection time of culture decreased considerably to 6.42 hours during bacterial proliferation and a corresponding titre of 106 CFU/mL on day 7. The BactiFlow assay gave the first positive results after 7 days of incubation, in accordance with the bacterial titre measured by the colony-forming assay. S. epidermidis isolate 454 was not detectable by the BacT/Alert automated culture system before the beginning of bacterial proliferation in PC. Subsequently, bacteria were detected with decreasing culture times from 14.43 hours to 6.42 hours according to the increasing bacterial titre. The first positive BactiFlow result appeared after 5 days of storage, consistent with the bacterial titre. The initiation of bacterial proliferation was already noted on day 4, but the bacterial titre determined by the plating assay was below the diagnostic sensitivity of 300 CFU/mL of the BactiFlow assay.
Look-back process
During the study period, we found five PC that were positive by culture for bacteria after transfusion. Positive PC were transfused within 12 hours (APC-4b, Table II) or 24 hours (APC-1a/b, APC-2b, APC-3b, Table II). The BacT/Alert culture system detected the positivity of these PC units between 1 and 7 days after sampling. We monitored the clinical characteristics of the recipients of these putatively contaminated PC units. Returned transfusion reports documented a transfusion without subsequent adverse reactions. All recipients had already been treated with antibiotics prior to transfusion of the putatively contaminated PC unit due to their underlying disease. Retrospective analysis of antibiosis related to the contaminating bacterial species revealed adequate therapy. An increase in inflammatory parameters was not observed.
Discussion
In recent years, there has been a unified effort to limit bacterial contamination of PC, including donor selection strategies, improved disinfection of the venipuncture site, diversion of the initial millilitres of blood23, collection guidelines, the use of detection methods after collection24 and pathogen inactivation methods25. In this study we show that screening for bacterial contamination using the BactiFlow assay can easily be adapted for each day of PC storage. The practicability, suitability and robustness of the BactiFlow assay in routine testing has been demonstrated previously in various studies10–14.
In the present study, bacterial screening on day 2 detected bacteria in five PC exclusively by culture methods (0.59%). The spectrum of bacteria detected exclusively included Gram-positive organisms, which were the most frequently isolated from PC in other studies26. These species are most often found as contaminants in clinical samples and symptomatic infections with these species are rare27. Only two PC were confirmed positive for S. epidermidis; confirmation of this positive result was based on the preparation of these two PC from the same donor. Proliferation studies of S. epidermidis isolate 454 supported the assumption of bacterial titres below the BactiFlow detection limit at this certain sampling time point because bacterial proliferation was not observed before 3 days of PC storage (Figure 1). Comparison of the BactiFlow results with those of the colony-forming assay show 5- to 10-fold higher numbers with corresponding bacterial titres of >6.5×103 CFU/mL. We previously observed a dose-dependent effect for S. epidermidis bacterial counts >5,000 CFU/mL, but this effect does not influence the assay validity, since the BactiFlow assay fulfils the imperative of correlation at low concentrations20. Higher deviations at high bacterial loads can be also explained by the BactiFlow counting strategy. The total sample volume is analysed with a maximum bacterial load of 105 CFU/mL, higher bacterial loads reduce the analysed sample volume to a tenth, and the count for the complete sample volume is calculated by the software.
All other culture-positive PC were only positive in anaerobic culture. The detection of bacteria only in one culture bottle suggested very low bacterial titres, basically resulting in a high risk of sampling errors. Furthermore, the sensitivity of the culture method with a stated detection limit of one CFU/mL6,8 is considerably higher than the diagnostic sensitivity of the BactiFlow assay (300 CFU/mL20). Alternatively, bacterial growth kinetics revealed bacterial titres below the diagnostic sensitivity of 300 CFU/mL of the BactiFlow assay. S. hominis isolate 4224 did not proliferate in PC. In contrast, both S. epidermidis isolates had the potential to proliferate in PC, but the bacterial titre on day 2 was most likely below the BactiFlow limit of detection. The examination of bacterial growth kinetics further supported the observation of positive results exclusively in the anaerobic culture bottles, most likely explained by very low bacterial titres connected to a high risk of sampling error. Of course, the low volume of 100 μL per plate used for the plating assay is also affected by this high risk of sampling error. However, in our opinion this will not result in a significant error regarding the determination of bacterial titres at the different sampling time points.
Furthermore, for the non-confirmed culture-positive PC, secondary contamination during the inoculation process of the culture bottles cannot be excluded. Review of the medical records of patients who received PC that initially tested positive showed that none of the recipients had symptoms or signs of febrile transfusion complications or evidence of an inflammatory event associated with the transfusion. Furthermore, the bacterial titre of potentially contaminated PC was below the level considered clinically significant (105 CFU/mL28,29).
Almost equal numbers of PC were tested on day 2 (manual sample preparation) and day 3 (semi-automated sample preparation), but the rate of reactive results was 3-fold higher for PC tested on day 2. This might be explained by the higher degree of automation and standardisation potential of the semi-automated method used for screening PC on day 3. Indeed, in the meantime, the manual protocol was standardised, based on experience from routine testing, and this resulted in a decreased false-positive rate of 0.13%10.
The subsequent reduction of the shelf-life of PC to 4 days challenges transfusion facilities to ensure adequate PC availability, especially around public holidays. Bacterial screening, concomitant with extension of the shelf-life, is necessary to overcome this problem. To our knowledge, only three different centres in Germany currently screen for bacterial contamination using rapid detection methods combined with a late sampling strategy in a routine setting, including testing on day 3 (GRCW and German Red Cross Frankfurt, personnel communication) or day 4 (German Red Cross Frankfurt, ILTM10,14). The GRCW and the ILTM use BactiFlow flow cytometry for bacterial screening on day 3 (GRCW) or day 4 (ILTM), whereas the German Red Cross Frankfurt use BactiFlow flow cytometry and 16S rDNA PCR for screening on day 3 or 4 (personnel communication Hourfar, KOLT meeting 2013). The possibilities for implementing screening strategies vary between blood service facilities with direct access to PC (e.g. ILTM) and blood services with distribution of products in different facilities (e.g. GRCW). The most critical point of different screening strategies is the sampling time point. Sampling errors and low rates of bacterial growth make it difficult to prevent transfusion of PC contaminated with slow-growing, most often Gram-positive organisms, sampled at early time-points (days 1 and 2). However, the risk of sampling errors decreases continuously during PC storage because micro-organisms have the potential to proliferate; sampling errors and bacterial growth kinetics become ever more negligible the later the sampling is performed.
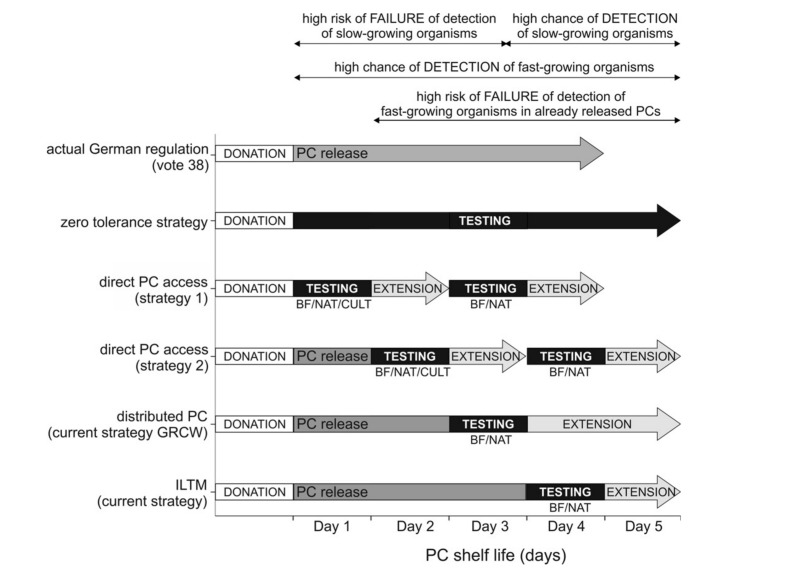
The different screening strategies under discussion with the German authority, the Paul-Ehrlich Institute, are shown in Figure 2. Currently, three routinely evaluated rapid methods are available: BactiFlow, NAT, and the Pan Genera Detection system (PGD)2,10–12,14,20,24,30,31. These methods differ significantly regarding their hands-on-time and time-to-result. The time-to-result is shorter for the PGD test and the BactiFlow assay (approximately 1.5 hours)20 than for NAT (3–4 hours)12. Sireis et al.12 also proposed the use of BacT/Alert as a possible rapid method for screening on day 3 or 4 after donation due to a maximum incubation time of <12 hours for the bacterial strains used. However, the bacterial panel used for spiking experiments in their study included bacteria with fast and stable growth characteristics32 achieving high bacterial titres at the first point of measurement; it would need to be confirmed that other isolates have these growth characteristics.
Figure 2.
Screening strategies.
Summary of the five main strategies discussed to improve the safety of PC, depending on the blood transfusion facility and methods used. BF: BactiFlow, NAT: nucleic acid amplification techniques, CULT: culture (BacT/Alert).
The screening strategy providing maximum safety and zero tolerance includes a bedside testing strategy immediately before transfusion, but this procedure is currently not an option, primarily due to the absence of an available screening method. Even the fast and easy-to-handle BactiFlow assay has limitations in overcoming these difficulties, primarily the laboratory environment needed for testing. The PGD test comes closest to a real point-of-care test, but this method has a poor sensitivity of >105 CFU/mL for the detection of some Gram-negative strains13,14. In our opinion, this test is not, therefore, suitable for preventing transfusion-associated infections. Secondly, point-of care testing causes great logistical complications for transfusion facilities, both with and without direct product access, including uncertain storage after release of the product and product loss accompanied by multiple screening approaches. Furthermore, the risk of secondary contamination increases with the frequency of sampling and originally negative PC samples can become positive due to product manipulation induced by sampling.
The strategies of testing on day 3 and/or 4 combined with the extension of PC storage to 5 days have two shortcomings: (i) fast-growing bacteria, such as Enterobacteriacae, contaminating products transfused before day 3 can be missed and (ii) slow-growing bacteria may not be detected because some PC isolates begin to show proliferation at a later stage10,13.
The clinical relevance of contamination with Gram-positive and Gram-negative bacteria is different. Review of the current German haemovigilance data revealed that most fatal outcomes occurred in cases of bacteria with high pathogenicity, e.g. S. pyogenes or S. aureus33. Unfortunately, the haemovigilance report did not provide information about the age of the contaminated PC and the bacterial load detected, which would aid the evaluation of the different screening strategies. Despite the fact that Gram-positive organisms are detected more frequently in PC, it is Gram-negative ones that account for the majority of transfusion fatalities26 as these show fast growth kinetics with bacterial titres of >106 CFU/mL within the first 24 to 48 hours13,20.
Screening on days 2 and 4 might possibly shorten the diagnostic window for fast-growing bacteria and increase the detection of slow-growing bacteria, but contaminated units transfused before day 2 are still invariably missed. Additionally, logistical problems occur because the number of PC requiring testing on day 2 is significantly higher than on day 3 or 4. In contrast, screening on days 1 and 3 further increases the detection of PC contaminated with fast-growing bacteria, but slow-growing bacteria may not be detected.
Application of BactiFlow flow cytometry or NAT as rapid testing methods showed the highest number of degrees of freedom. Certainly, for practical purposes, testing on more than one day implies a substantial effort, including more than doubling the number of analyses, significantly increased process times and greater costs for both methods. Although the currently established BactiFlow protocol is comparatively fast and easy to handle, it would benefit considerably from technical improvement through greater automation and reduction of sample set-up and analysis time. Realization of these aspects, accompanied by a significant reduction in test costs, will be necessary in order to implement a “2-or-more-testing-day strategy” in practice. So far, the data necessary to determine the optimal screening strategy, particularly regarding the best sampling time point, are incomplete. The results presented in this study provide additional data to help to resolve this issue; however, the number of PC tested PC was too small to reach definitive conclusions.
At present, screening strategies with a maintainable effort for both blood service facilities with direct and distributed PC access, include testing on day 3 and/or 4 to avoid transfusion of highly contaminated PC. These strategies may not avoid reactions to potentially low concentrations of bacterial endotoxins or exotoxins in the recipient, but it has been shown that the consequences are less severe28,34. The assessments of currently applied screening strategies presented in this study are only snapshots based on available data and require reconsideration for example in the case of septic complications from 1- or 2-day old PC.
In conclusion, the data presented in this study further support the proposal that the BactiFlow assay is very convenient for bacterial screening of PC, independently of the testing day and the screening strategy. Only NAT methods showed an almost equal applicability. None of the testing strategies discussed here can completely eliminate the risk of bacterial contamination of blood products. However, PC screening using the BactiFlow assay has some major advantages over other currently used strategies: (i) the diagnostic sensitivity is considerably higher than that of the PGD test, for example; and (ii) the later sampling day gives a higher probability of detecting highly contaminated PC compared to the negative-to-date concept.
Acknowledgements
The Authors thank Sarah Kirkby for her linguistic advice. Furthermore, we thank Anne-Kathrin Vollmer and Christoph Lichtenberg (ILTM), as well as Margarethe Rogowski and Malgorzata Krzos (GRCW) for their technical assistance.
Footnotes
The Authors declare no conflicts of interest.
References
- 1.Reading FC, Brecher ME. Transfusion-related bacterial sepsis. Curr Opin Hematol. 2001;8:380–6. doi: 10.1097/00062752-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs MR, Smith D, Heaton WA, et al. Detection of bacterial contamination in prestorage culture-negative apheresis platelets on day of issue with the Pan Genera Detection test. Transfusion. 2011;51:2573–82. doi: 10.1111/j.1537-2995.2011.03308.x. [DOI] [PubMed] [Google Scholar]
- 3.Montag T. Perspectives and limitations in the bacterial screening of platelet concentrates. J Lab Med. 2006;30:60–5. [Google Scholar]
- 4.Müller TH, Mohr H, Montag T. Methods for the detection of bacterial contamination in blood products. Clin Chem Lab Med. 2008;46:933–46. doi: 10.1515/CCLM.2008.154. [DOI] [PubMed] [Google Scholar]
- 5.de Korte D, Curvers J, de Kort WL, et al. Effects of skin disinfection method, deviation bag, and bacterial screening on clinical safety of platelet transfusions in the Netherlands. Transfusion. 2006;46:476–85. doi: 10.1111/j.1537-2995.2006.00746.x. [DOI] [PubMed] [Google Scholar]
- 6.Eder AF, Kennedy JM, Dy BA, et al. Bacterial screening of apheresis platelets and the residual risk of septic transfusion reactions: the American Red Cross experience (2004–2006) Transfusion. 2007;47:1134–42. doi: 10.1111/j.1537-2995.2007.01248.x. [DOI] [PubMed] [Google Scholar]
- 7.Larsen CP, Ezligini F, Hermansen NO, Kjeldsen-Kragh J. Six years’ experience of using the BacT/ALERT system to screen all platelet concentrates, and additional testing of outdated platelet concentrates to estimate the frequency of false-negative results. Vox Sang. 2005;88:93–7. doi: 10.1111/j.1423-0410.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- 8.McDonald CP, Pearce S, Wilkins K, et al. Pall eBDS: an enhanced bacterial detection system for screening platelet concentrates. Transfus Med. 2005;15:259–68. doi: 10.1111/j.0958-7578.2005.00587.x. [DOI] [PubMed] [Google Scholar]
- 9.Munksgaard L, Albjerg L, Lillevang ST, et al. Detection of bacterial contamination of platelet components: six years’ experience with the BacT/ALERT system. Transfusion. 2004;44:1166–73. doi: 10.1111/j.1537-2995.2004.04019.x. [DOI] [PubMed] [Google Scholar]
- 10.Vollmer T, Dreier J, Schottstedt V, et al. Detection of bacterial contamination in platelet concentrates by a sensitive flow cytometric assay (BactiFlow): a multicentre validation study. Transfus Med. 2012;22:262–71. doi: 10.1111/j.1365-3148.2012.01166.x. [DOI] [PubMed] [Google Scholar]
- 11.Vollmer T, Engemann J, Kleesiek K, Dreier J. Bacterial screening by flow cytometry offers potential for extension of platelet storage: results of 14 months of active surveillance. Transfus Med. 2011;21:175–82. doi: 10.1111/j.1365-3148.2011.01070.x. [DOI] [PubMed] [Google Scholar]
- 12.Sireis W, Ruster B, Daiss C, et al. Extension of platelet shelf life from 4 to 5 days by implementation of a new screening strategy in Germany. Vox Sang. 2011;101:191–9. doi: 10.1111/j.1423-0410.2011.01485.x. [DOI] [PubMed] [Google Scholar]
- 13.Vollmer T, Hinse D, Kleesiek K, Dreier J. The Pan Genera Detection immunoassay: a novel point-of-issue method for detection of bacterial contamination in platelet concentrates. J Clin Microbiol. 2010;48:3475–81. doi: 10.1128/JCM.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vollmer T, Hinse D, Schottstedt V, et al. Inter-laboratory comparison of different rapid methods for the detection of bacterial contamination in platelet concentrates. Vox Sang. 2012;103:1–9. doi: 10.1111/j.1423-0410.2011.01572.x. [DOI] [PubMed] [Google Scholar]
- 15.Fang CT, Chambers LA, Kennedy J, et al. Detection of bacterial contamination in apheresis platelet products: American Red Cross experience, 2004. Transfusion. 2005;45:1845–52. doi: 10.1111/j.1537-2995.2005.00650.x. [DOI] [PubMed] [Google Scholar]
- 16.Ramirez-Arcos S, Jenkins C, Dion J, et al. Canadian experience with detection of bacterial contamination in apheresis platelets. Transfusion. 2007;47:421–9. doi: 10.1111/j.1537-2995.2007.01131.x. [DOI] [PubMed] [Google Scholar]
- 17.Schrezenmeier H, Walther-Wenke G, Muller TH, et al. Bacterial contamination of platelet concentrates: results of a prospective multicenter study comparing pooled whole blood-derived platelets and apheresis platelets. Transfusion. 2007;47:644–52. doi: 10.1111/j.1537-2995.2007.01166.x. [DOI] [PubMed] [Google Scholar]
- 18.te Boekhorst PA, Beckers EA, Vos MC, et al. Clinical significance of bacteriologic screening in platelet concentrates. Transfusion. 2005;45:514–9. doi: 10.1111/j.0041-1132.2005.04270.x. [DOI] [PubMed] [Google Scholar]
- 19.Association Bulletin #10-02: Interim Standard 5.1.5.1.1. AABB. 2010;2010 [Google Scholar]
- 20.Dreier J, Vollmer T, Kleesiek K. Novel flow cytometry-based screening for bacterial contamination of donor platelet preparations compared with other rapid screening methods. Clin Chem. 2009;55:1492–502. doi: 10.1373/clinchem.2008.122515. [DOI] [PubMed] [Google Scholar]
- 21.Dreier J, Störmer M, Kleesiek K. Two novel real-time reverse transcriptase PCR assays for rapid detection of bacterial contamination in platelet concentrates. J Clin Microbiol. 2004;42:4759–64. doi: 10.1128/JCM.42.10.4759-4764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley BE, Linton CJ, Bennett DM, et al. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur J Clin Microbiol Infect Dis. 1998;17:247–53. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 23.Blut Arbeitskreis. Votum 27: Einführung des Predonation Samplings. Bundesgesundheitsblatt. 2002;45:756. [Google Scholar]
- 24.Palavecino EL, Yomtovian RA, Jacobs MR. Detecting bacterial contamination in platelet products. Clin Lab. 2006;52:443–56. [PubMed] [Google Scholar]
- 25.Seltsam A, Muller TH. Update on the use of pathogen-reduced human plasma and platelet concentrates. Br J Haematol. 2013;162:442–54. doi: 10.1111/bjh.12403. [DOI] [PubMed] [Google Scholar]
- 26.Brecher ME, Hay SN. Bacterial contamination of blood components. Clin Microbiol Rev. 2005;18:195–204. doi: 10.1128/CMR.18.1.195-204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatermann SG. Pathogenicity of bacteria contaminating blood products. Transfus Med Hemother. 2011;38:236–8. doi: 10.1159/000330425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacobs MR, Good CE, Lazarus HM, Yomtovian RA. Relationship between bacterial load, species virulence, and transfusion reaction with transfusion of bacterially contaminated platelets. Clin Infect Dis. 2008;46:1214–20. doi: 10.1086/529143. [DOI] [PubMed] [Google Scholar]
- 29.Muder RR, Yee YC, Rihs JD, Bunker M. Staphylococcus epidermidis bacteremia from transfusion of contaminated platelets: application of bacterial DNA analysis. Transfusion. 1992;32:771–4. doi: 10.1046/j.1537-2995.1992.32893032109.x. [DOI] [PubMed] [Google Scholar]
- 30.Harm SK, Delaney M, Charapata M, et al. Routine use of a rapid test to detect bacteria at the time of issue for nonleukoreduced, whole blood-derived platelets. Transfusion. 2013;53:843–50. doi: 10.1111/j.1537-2995.2012.03818.x. [DOI] [PubMed] [Google Scholar]
- 31.Prowse C. Zero tolerance. Transfusion. 2007;47:1106–9. doi: 10.1111/j.1537-2995.2007.01322.x. [DOI] [PubMed] [Google Scholar]
- 32.Störmer M, Arroyo A, Brachert J, et al. Establishment of the first International Repository for Transfusion-Relevant Bacteria Reference Strains: ISBT Working Party Transfusion-Transmitted Infectious Diseases (WP-TTID), Subgroup on Bacteria. Vox Sang. 2012;102:22–31. doi: 10.1111/j.1423-0410.2011.01510.x. [DOI] [PubMed] [Google Scholar]
- 33.Funk MB, Lohmann A, Guenay S, et al. Transfusion-transmitted bacterial infections - haemovigilance data of German Blood Establishments (1997–2010) Transfus Med Hemother. 2011;38:266–71. doi: 10.1159/000330372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montag T. Strategies of bacteria screening in cellular blood components. Clin Chem Lab Med. 2008;46:926–32. doi: 10.1515/CCLM.2008.176. [DOI] [PubMed] [Google Scholar]