Introduction
Discrepancy of results of ABO groups between red cell testing and serum testing is a significant issue in transfusion medicine1. One of its reported causes is blood chimerism, which shows mixed-field agglutination. However, mixed-field agglutination can be seen in various situations, including ABO-incompatible stem cell transplantation, recent transfusions of type-O red blood cells in a non-O recipient, and rare ABO subgroups such as B3 and chimera. Several molecular approaches have been employed to identify chimerism, namely ABO genotyping, human leucocyte antigen typing and DNA short tandem repeat assays2. These methods can typically detect three or more alleles of a chimera. However, some chimeras with a minor allele population might be overlooked because of preferential amplification of the major allele during ordinary polymerase chain reaction (PCR) analysis.
We report a case of blood chimerism in which B red cells accounted for less than 1% of the whole population. The B allele in peripheral blood mononuclear cells could not be identified by direct sequencing of ordinary PCR product involving exons 6 and 7 of the ABO gene, but was revealed using peptide nucleic acid (PNA)-mediated PCR clamping.
Case report with results
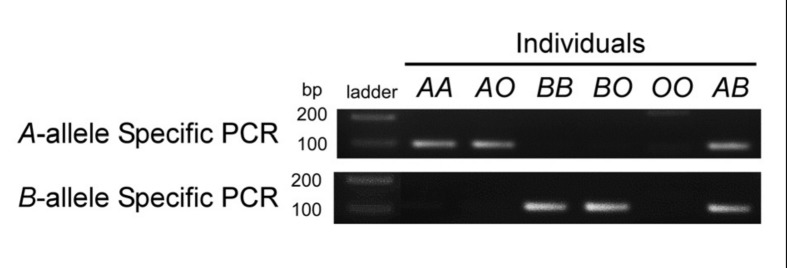
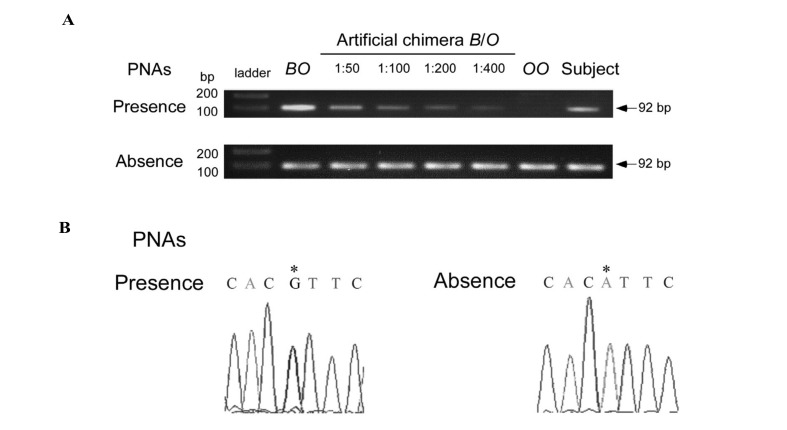
Serological tests conducted on a voluntary blood donor demonstrated a discrepancy between red cell testing and serum testing. The red blood cells were agglutinated by anti-H, but not macroscopically by anti-A and anti-B. However, a positive reaction was observed in the adsorption-elution test with anti-B. The serum contained antibodies that agglutinated A1 red cells, but lacked antibodies that agglutinated B red cells. The individual’s saliva contained equivalent quantities of B and H substances. In addition, the degree of B-transferase activity in serum was significantly lower than that in common B individuals. Since the results of these tests seemed to be consistent with the characteristics of a chimera rather than phenotypic subgroups such as B3 and Bm, flow cytometric analysis of ABH antigens on the erythrocytes was carried out and this revealed that the red cells comprised 0.3% B cells and 99.7% O cells. To verify the B allele in blood, we carried out ordinary ABO genotyping using direct sequencing of the PCR products including exons 6 and 7 of the ABO gene3,4 and this demonstrated only the O01 allele. The individual was not a twin and did not have any history of transfusion, bone marrow transplantation or foeto-maternal haemorrhages. The inconsistency between the antigen analysis and the ABO genotyping could have been attributable to the very low population of B red cells, because the sensitivity of direct sequencing depends on the percentage of the mutant allele in the sample analysed. To detect the minor allele, we employed allele-specific PNA-mediated PCR clamping, as this is more sensitive than direct sequencing, having the ability to detect mutations in samples containing less than 1% mutant alleles5,6. For PCR, 100 ng of genomic DNA prepared from peripheral blood mononuclear cells of the subject, as well as from control individuals with the genotypes AA, AO, BB, BO, OO and AB, were mixed in a 50-μL final volume containing 1×PCR buffer with TaKaRa Taq polymerase (TaKaRa, Shiga, Japan), 0.2 mmol/L dNTP, 0.2 μmol/L of each primer, and 1.6 μmol PNA. The primers used were ABO+17674S and ABO+17765AS, which have the sequences 5′–CAGTAGGAAGGATGTCCTCGTG–3′ and 5′–TGAACTGCTCGTTGAGGATG–3′, respectively (Figure 1). A allele-specific PCR was performed using both PNA ABOc253CLΔ and ABOc278CLG, which have the sequences N–CTCGTGGTACCCCTTG–C and N–ATGTTGAACGTGCCCTCC–C, respectively. B allele-specific PCR was performed using both PNA ABOc253CLΔ and ABOc278CLA, the sequence of which is N–ATGTTGAATGTGCCCTCC–C. PNA ABOc253CLΔ corresponds to deletion of G at nucleotide 261 of the cDNA and is used in order to amplify the DNA region exclusively from intron 5 to exon 6 on genomic DNA of the common A and B alleles. The nucleotides underlined in PNA ABOc278CLA and ABOc278CLG are complementary to nucleotide A or G at position 297 on the cDNA which is associated with the A or B allele, and each suppresses amplification of the A or B allele, respectively. The conditions used for amplification were 94 ºC for 3 minutes, 40 cycles of 98 ºC for 10 seconds, 58 ºC for 30 seconds, and 72 ºC for 1 minute. The A-specific PCR showed an amplified 92-bp product in the individuals with genotypes AA, AO and AB, but not in BB, BO and OO individuals (Figure 2). Subsequently, direct sequencing of these PCR products verified the nucleotides of exon 6 from A allele. Similarly, the B-specific PCR demonstrated the amplification in individuals with BB, BO and AB genotypes, but not in individuals with AA, AO and OO genotypes. Subsequent direct sequencing of these PCR products verified the sequences of exon 6 from the B allele. To examine the sensitivity of the B-specific PCR, genomic DNA from an individual with the BO genotype was mixed with that of an individual with the OO genotype at ratios of 1:10, 1:50, 1:100, and 1:200, followed by PCR amplification with the PNA. The intensity of the 92-bp band was proportional to the ratios of the B allele in all of the diluted preparations, but not in the OO DNA sample. In contrast, similar bands appeared in all samples, irrespective of the ratios of the B allele in a control PCR without those PNA (Figure 3A). Because the B-specific PCR seemed to be sufficiently sensitive to detect the minor B allele at a B and O allele copy ratio of 1:400, we carried out PCR with the genomic DNA of the subject and observed that the band had lower intensity, demonstrating the presence of the B allele in the subject’s blood cells. Subsequently, direct sequencing of the B-specific PCR product showed G at position 297 on the cDNA associated with the B allele (Figure 3B). In contrast, the non-clamping PCR product demonstrated A at the same position associated with the O allele. Thus, B allele-specific PNA-mediated PCR clamping is likely to be valid for detection of blood chimerism with a minor allele.
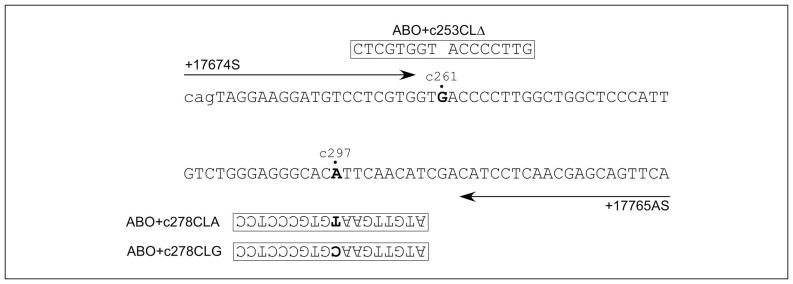
Figure 1.
Schematic illustration of primers and PNA used for allele-specific PNA-mediated PCR clamping.
Two common nucleotide primers, ABO+17674S and +17765AS, used for PCR amplification, are indicated by arrows, while the PNA ABO+c253CLΔ, ABO+c278CLA and ABO+c278CLG are represented by squares with the corresponding sequences. The genomic sequence between positions +17674 and +17765 relative to the translation start site of the human ABO gene is shown. The sequence was from genomic DNA from the B allele (GenBank accession number AJ920329.1). The nucleotides in exon 6 are denoted by the upper letters, while those in intron 5 are denoted by the lower letters. c261 shows the location of nucleotide G, which is absent from the O alleles including O01 and O02. c297 indicates the location of nucleotide A or G at position 297 on the cDNA which is associated with the A/O or B allele, respectively.
Figure 2.
Representative PCR products obtained from PNA-mediated PCR clamping using genomic DNA from individuals with genotypes AA, AO, BB, BO, OO and AB.
A or B allele-specific PNA-mediated PCR clamping was performed, followed by 2% agarose gel electrophoresis and staining with ethidium bromide. TrackIt™ 1kb PLUS DNA Ladder (Life Technologies) was used as a molecular size marker.
Figure 3.
Sensitivity and specificity of the B allele-specific PNA-mediated PCR clamping.
(A) Sensitivity of the B-specific PCR. Genomic DNA of the BO genotype was mixed with that of the OO genotype at ratios of 1:10, 1:50, 1:100, and 1:200, followed by B-specific PCR with the PNA ABOc253CLΔ and ABOc278CLA or a control PCR without these PNA. The numbers indicate copy ratios between the B and O alleles. DNA from the subject was also examined using B-specific PCR or non-clamping PCR. TrackIt™ 1kb PLUS DNA Ladder was used as a molecular size marker. (B) Specificity of the B-specific PCR. Direct sequence analyses showed a difference in the nucleotides at position 297 between the B-specific PCR product and the non-clamping PCR product. The sequences are demonstrated between positions 294 and 300 of the cDNA which were obtained from the B-specific PCR or the non-clamping PCR. The change in the nucleotide to G and A at position 297 indicated by an asterisk is associated with the B and A/O allele, respectively.
Discussion
In this study, serological tests for the case reported highlighted a discrepancy between the results of red cell testing and serum testing, and subsequent flow cytometric analysis of ABH antigens on erythrocytes demonstrated a chimera comprising 99.7% O red cells and 0.3% B red cells. Consistently, PNA-mediated PCR clamping was able to detect the B allele in genomic DNA prepared from the subject’s blood mononuclear cells. PNA-mediated PCR clamping is, therefore, a valid technique for the delineation of chimerism.
ABO genotyping has been performed using PCR-restriction fragment length polymorphism, single-strand conformation polymorphism, PCR with sequence-specific primers and direct sequencing of the PCR product7–10. Allele-specific primers are available for ABO genotyping, although most 3′ mismatches between a primer and its template do not significantly impair the PCR reaction11. Although direct sequencing of a PCR product is a popular method for genotyping, some chimeras with a minor allele population might be overlooked because of preferential amplification of the major allele. For detection of any minor allele in a chimera, Won et al. developed an amplification refractory mutation system, which was able to reveal artificial chimerism comprising 5% A red cells and 95% O red cells12. Recently, ABO chimerism was determined by real-time PCR analysis, the sensitivity of which was 0.1% to 1%13. PNA-mediated PCR clamping is another sensitive method for detecting a minor allele. PNA oligomers suppress amplification of the complementary sequence confined by a pair of DNA oliginucleotide primers because PNA is not a substrate for DNA polymerase. The PNA-clamped probe assay is more sensitive than direct sequencing, having the ability to detect mutations in samples containing less than 1% mutant alleles. This method is reportedly available for detection of c-Kit point mutations and K-Ras mutations in various tumour samples14–16. The A or B allele-specific PNA-mediated PCR clamping technique could, therefore, be suitable for the detection of a minor allele in an ABO blood group chimera as well as a nucleotide substitution in ABO subgroups.
Footnotes
The Authors declare no conflicts of interest.
Sources of support
This work was supported in part by a Grant-in-Aid from Japan Society for the Promotion of Science (21590732 to TN, 25860486 to YT).
References
- 1.Bluth MH, Reid ME, Manny N. Chimerism in the immunohematology laboratory in the molecular biology era. Transfus Med Rev. 2007;21:134–46. doi: 10.1016/j.tmrv.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Cho D, Lee JS, Yazer MH, et al. Chimerism and mosaicism are important causes of ABO phenotype and genotype discrepancies. Immunohematology. 2006;22:183–7. [PubMed] [Google Scholar]
- 3.Shima M, Nishimura T, Yoshioka A, et al. The ABO blood group genotype in Japanese determined by polymerase chain reaction (PCR) using genomic DNA. Jpn J Transfus Med. 1992;38:542–7. [Google Scholar]
- 4.Sano R, Nakajima T, Takahashi K, et al. Expression of ABO blood-group genes is dependent upon an erythroid cell-specific regulatory element that is deleted in persons with the B(m) phenotype. Blood. 2012;119:5301–10. doi: 10.1182/blood-2011-10-387167. [DOI] [PubMed] [Google Scholar]
- 5.Nagai Y, Miyazawa H, Huqun, et al. Genetic heterogeneity of the epidermal growth factor receptor in non-small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–82. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Ladanyi M. Epidermal growth factor receptor mutation testing in lung cancer: searching for the ideal method. Clin Cancer Res. 2007;13:4954–5. doi: 10.1158/1078-0432.CCR-07-1387. [DOI] [PubMed] [Google Scholar]
- 7.Lee JC, Chang JG. ABO genotyping by polymerase chain reaction. J Foresic Sci. 1992;37:1269–75. [PubMed] [Google Scholar]
- 8.Yip SP. Single-tube multiplex PCR-SSCP analysis distinguishable 7 common ABO alleles and readily identifies new alleles. Blood. 2000;95:1487–92. [PubMed] [Google Scholar]
- 9.Gassner C, Schmarda A, Nussbaumer W, et al. ABO glycosyltransferase genotyping by polymerase chain reaction using sequence-specific primers. Blood. 1996;88:1852–6. [PubMed] [Google Scholar]
- 10.Muro T, Fujihara J, Imamura S, et al. Determination of ABO genotypes by real-time PCR using allele-specific primers. Leg Med (Tokyo) 2012;14:47–50. doi: 10.1016/j.legalmed.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Kwok S, Kellogg DE, McKinney N, et al. Effects of primer-template mismatches on the polymerase chain reaction: human immunodeficiency virus type 1 model studies. Nucl Acid Res. 1990;18:999–1005. doi: 10.1093/nar/18.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Won EJ, Park HR, Park TS, et al. Amplification refractory mutation system-PCR is essential for the detection of chimaeras with a minor allele population: a case report. J Clin Pathol. 2013;66:446–8. doi: 10.1136/jclinpath-2012-201355. [DOI] [PubMed] [Google Scholar]
- 13.Liu F, Li G, Mao X, Hu L. ABO chimerism determined by real-time polymerase chain reaction analysis after ABO-incompatible haematopoietic stem cell transplantation. Blood Transfus. 2013;11:43–52. doi: 10.2450/2012.0013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sotlar K, Escribano L, Landt O, et al. One-step detection of c-kit point mutations using peptide nucleic acid-mediated polymerase chain reaction clamping and hybridization probes. Am J Pathol. 2003;162:737–46. doi: 10.1016/S0002-9440(10)63870-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Hung K, Wu L, et al. Detection of tumor mutations in the presence of excess amounts of normal DNA. Nat Biotechnol. 2002;20:186–9. doi: 10.1038/nbt0202-186. [DOI] [PubMed] [Google Scholar]
- 16.Beau-Faller M, Legrain M, Voegeli AC, et al. Detection of K-Ras mutations in tumour samples of patients with non-small cell lung cancer using PNA-mediated PCR clamping. Br J Cancer. 2009;100:985–92. doi: 10.1038/sj.bjc.6604925. [DOI] [PMC free article] [PubMed] [Google Scholar]