Abstract
Growth control scales morphological attributes and, therefore, provides a critical contribution to the evolution of adaptive traits. Yet, the genetic mechanisms underlying growth in the context of specific ecological adaptations are poorly understood. In water striders, adaptation to locomotion on the water surface is associated with allometric and functional changes in thoracic appendages, such that T2-legs, used as propelling oars, are longer than T3-legs, used as steering rudders. The Hox gene Ubx establishes this derived morphology by elongating T2-legs but shortening T3-legs. Using gene expression assays, RNAi knockdown, and comparative transcriptomics, we demonstrate that the evolution of water surface rowing as a novel means of locomotion is associated with the evolution of a dose-dependent promoting-repressing effect of Ubx on leg growth. In the water strider Limnoporus dissortis, T3-legs express six to seven times higher levels of Ubx compared to T2-legs. Ubx RNAi shortens T2-legs and the severity of this phenotype increases with increased depletion of Ubx protein. Conversely, Ubx RNAi lengthens T3-legs but this phenotype is partially rescued when Ubx protein is further depleted. This dose-dependent effect of Ubx on leg growth is absent in non-rowing relatives that retain the ancestral relative leg length. We also show that the spatial patterns of expression of dpp, wg, hh, egfr, dll, exd, hth, and dac are unchanged in Ubx RNAi treatments. This indicates that the dose-dependent opposite effect of Ubx on T2- and T3-legs operates without any apparent effect on the spatial expression of major leg patterning genes. Our data suggest that scaling of adaptive allometries can evolve through changes in the levels of expression of Hox proteins early during ontogeny, and in the sensitivity of the tissues that express them, without any major effects on pattern formation.
Keywords: Hox genes, Adaptation, Allometry, Morphological evolution, RNA interference
Highlights
-
•
Ubx is generally expressed at higher levels in T3- relative to T2-legs in semi-aquatic insects.
-
•
It is only in the derived Gerridae where the high levels of Ubx result in reduced T3-leg length.
-
•
In the Gerridae, the response of leg tissues to Ubx levels is bimodal.
-
•
Changes in Ubx regulation and function have evolved in Limnoporus without disrupting patterning hierarchies.
-
•
Changes in Hox protein levels and emergence of tissue sensitivity to these levels can shape adaptive morphological traits.
Introduction
Growth control properly scales different body parts and is therefore a critical process in organismal development and evolution (Abzhanov et al., 2004; Bronikowski, 2000; Emlen et al., 2012; Nijhout and Grunert, 2010; O’Farrell, 2003; Stern and Emlen, 1999). A major challenge for understanding how growth can shape animal diversity is to decipher the mechanisms of growth control in the context of the particular ecological environment underlying species-specific life histories (Abzhanov et al., 2004; Emlen et al., 2012; Moczek and Rose, 2009; Stern and Emlen, 1999; Stillwell et al., 2011). Growth is intricately tied to pattern formation and biological systems must integrate inputs from both processes (Baena-Lopez et al., 2012). Organisms have been facing this challenge throughout evolution, and a variety of routes to reconciling growth and pattern formation were taken depending on the trait. For example in many systems, modulation of the growth of some traits is delayed to near the end of ontogeny, after all other essential body parts are specified and their identity acquired. It is possible that such late fine-tuning strategy circumvents the constraints imposed by the tight and pleiotropic interactions of genetic processes during development (Carroll, 2008; Duboule et al., 2007; Gibson, 1996; Wilkins, 2002). However, this case is restricted to some traits especially those required only during adulthood and have no or minimal function during juvenile stages. Examples include the development and evolution of a variety of exaggerated secondary sexual traits in beetles and water striders (Emlen et al., 2012; Khila et al., 2012), or appendage differentiation in holometabolous insects (Loehlin and Werren, 2012; Nijhout and Grunert, 2010). A greater challenge facing organisms during development and evolution is when structures must be specified and the scaling of their allometry fine-tuned during early development. This is generally required for attributes that are essential for both juveniles and adults; such is the case of locomotory appendages in hemimetabolous non-metamorphosing systems.
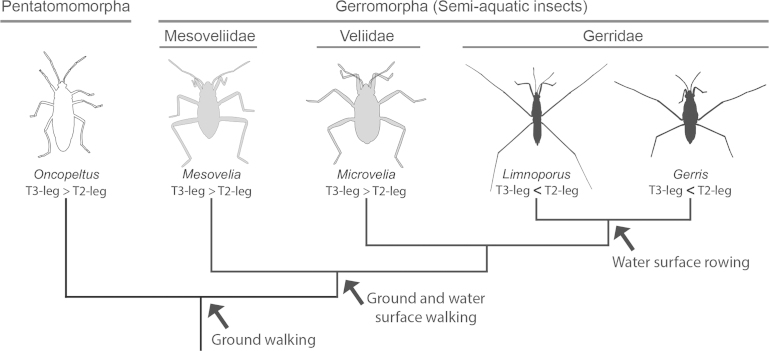
The case of water striders—a monophyletic group of semi-aquatic heteropterans—is a striking example where animals must pattern, dramatically grow, and fine-tune the final allometry of their appendages before the end of embryonic development, to adapt to the challenging requirements of their habitat (Fig. 1) (Andersen, 1982; Hu and Bush, 2010). Throughout the evolution of the semi-aquatic insects, the transition to life on water surface seems to be associated with a gradual increase in leg length relative to body length. Basally branching semi-aquatic insects can walk both on water and land, and share a common relative leg length plan, where T3-legs are longer than T2-legs, with their close relatives that are exclusively terrestrial (Fig. 1). Conversely derived lineages, which have specialized in water-surface locomotion through rowing, have evolved a novel adaptive morphology where the length of locomotory appendages has been reversed such that T2-legs are now longer than T3-legs (Fig. 1) (Andersen, 1982; Damgaard et al., 2005; Tseng and Rowe, 1999). Water striders use their T2-legs as propelling oars and their T3-legs as steering rudders (Andersen, 1982; Hu and Bush, 2010). T1-legs are much shorter and function primarily for stability, grasping the female during mating, and for handling preys (Andersen, 1982). These changes in the allometry of posterior thoracic appendages facilitated their functional adaptation and enabled the animals to generate efficient propulsion on the water–air interface (Andersen, 1976; Hu et al., 2003). As such, water striders have diversified to dominate a range of water surface niches, including ponds, streams, lakes, and even oceans (Andersen, 1982).
Fig. 1.
Leg morphology and mode of locomotion across the semi-aquatic insects and the closely related terrestrial outgroup Oncopeltus. Both terrestrial insects (highlighted in white) and most semi-aquatic insects (highlighted in gray) retain the ancestral relative leg length where T2-legs are shorter than T3-legs. Derived lineages (highlighted in black), which specialize in open water rowing, have a reversed relative leg length where T2-legs are now longer than T3-legs.
We are beginning to understand some of the genetic mechanisms responsible for these adaptive changes in leg allometry in water striders (Khila et al., 2009). Hox genes are conserved transcription factors that impart segment identity along the body axis across animals, through establishing a set of characters that distinguish segments that express them from those that do not (Akam, 1998a). Hox mutations result in the loss of the distinguishing characters and cause homeotic transformation of the affected segment to the likeness of another (Struhl, 1982). The Hox gene Ultrabithorax (Ubx), in particular, manages a multitude of small-scale differences between the second and third thoracic segments across a variety of arthropod species (Akam, 1998c; Stern, 1998, 2003). Among these phenotypic differences between segments, Ubx protein regulates the growth of the third thoracic appendages in a number of species, including holometabolous and hemimetabolous insects (Mahfooz et al., 2007; Stern, 2003). In holometabolous insects, there is a temporal decoupling of segmentation along the anterior–posterior axis of the embryo and appendage differentiation during post-embryonic stages (Baker, 1988). Late during Drosophila pupal development, Ubx modulates the shape and size of T3-legs, and establishes inter-species differences in trichome patterns (Stern, 1998, 2003). High levels of Ubx protein repress trichome development on T2 femur, and variation in Ubx regulation underlies variation in trichome patterns across species (Stern, 1998). During wing development, T2 imaginal discs, which do not express Ubx, differentiate into functional adult wings. However T3 imaginal discs, which do express Ubx, differentiate into the much smaller halteres (Roch and Akam, 2000). Ubx controls haltere morphogenesis through regulation of a large number of genetic factors at distinct developmental stages, including signaling molecules, transcription factors, and growth pathways (Castelli-Gair and Akam, 1995; Crickmore and Mann, 2006, 2008; Pavlopoulos and Akam, 2011; Roch and Akam, 2000). This indicates that in flies, Ubx can regulate specific morphogenetic processes during post-embryonic stages. In the hemimetabolous water striders however, the dramatic growth of the legs and the requirement for Ubx in fine-tuning their allometry coincide with the requirement for patterning both the appendages and the segments along the anterior–posterior axis of the embryo. In the embryo of water striders, Ubx protein is expressed in both T2- and T3-legs, and functions to lengthen T2- but to shorten T3-legs (Khila et al., 2009). This fine-tuning of relative leg length by Ubx is driven by changes in the spatial expression and function of the protein in the second and third thoracic segments (Khila et al., 2009). The novel deployment of Ubx in T2-legs distinguishes water striders from close terrestrial relatives, such as the milkweed bug Oncopeltus fasciatus where Ubx expression is restricted to T3-legs (Mahfooz et al., 2007). Similarly, the reversed function of Ubx to shorten T3-legs in water striders distinguishes them from sister basally branching semi-aquatic insects taxa (Khila et al., 2014). In the water strider Gerris buenoi, Ubx seems to be expressed at higher levels in T3-legs compared to T2-legs (Khila et al., 2009). However whether these differential levels of expression have a relevant role in regulating leg allometry remains to be tested. Here we investigate a possible role of Ubx levels in shaping the morphological differences that distinguish the second from the third thoracic segments during the evolution of the semi-aquatic insects. Although Ubx controls multiple morphological attributes in these segments, we focus on the morphology of the legs in the context of adaptation to life on the water surface. We use parental RNAi, which depletes Ubx from the start of embryogenesis and across all tissues expressing the gene. Although we obtain partial transformation of the third thoracic segment to the likeness of the second thoracic segment, our conclusions throughout the manuscript focus primarily on the response of the legs in each segment to Ubx RNAi. By manipulating the strength of RNAi knockdown and by analyzing the association between the responses of T2- and T3-legs to Ubx RNAi, we describe the evolution of tissue sensitivity to Ubx levels across a selection of semi-aquatic insects representing both basal and derived taxa. We also examine whether or not major leg patterning hierarchies are affected by the changes in Ubx regulation that led to the specialization of water striders in surface rowing.
Methods
Animal collection and rearing
Adult individuals of the water strider Limnoporus dissortis were collected from ‘Rivière de l׳Acadie,’ and Microvelia americana from ‘Rivière du Nord’ at the vicinity of Montréal, Québec, Canada. Mesovelia furcata was collected in Vilette d׳Anthon, ‘plans d׳eau de Salonique’ lake in Lyon, France. Animals were kept in aquaria at 25 °C with a 14-h light/10-h dark cycle, and were fed on live crickets. Pieces of floating Styrofoam were regularly supplied for the females to lay eggs.
Gene cloning
Limnoporus total RNA was extracted from different embryonic and nymphal stages. First strand cDNA synthesis was then performed, using total RNA as a template, according to Invitrogen manual instructions. To clone Limnoporus dll, hh, and wg, degenerate primers were designed based on sequence alignments of Tribolium, Acyrthosiphon, and Pediculus. Specific primers for dac, dpp, egfr, hth, and Ubx were designed based on sequences obtained from a whole transcriptome of Limnoporus. Primer sequences and Genbank accession numbers are presented in Table S1.
Embryo collection and dissection
Embryos were collected, treated with 25% bleach, and then washed with PTW 0.05% (1X PBS; 0.05% Tween-20). For image acquisition, late embryos were dissected out of the egg, fixed in 4% Formaldehyde, and their images captured using a Zeiss Discovery 12 scope. For staining, embryos of various early stages were dissected, cleaned from yolk, and kept briefly in PTW 0.05% on ice until fixation with the appropriate method, based on the type of subsequent staining (see below).
in situ hybridization
Dissected embryos were fixed in 200 µl 4% Paraformaldehyde (PFA)+20 µl Dimethyl Sulfoxide (DMSO), and 600 µl heptane for 20 min at room temperature with shaking. Embryos were then washed several times in cold methanol and rehydrated in decreasing concentrations of methanol in PTW 0.05%. These embryos were washed three times in PTW 0.05%, three times in PBT 0.3% (1X PBS; 0.3% Triton X100), and twice with PBT 1% (1X PBS; 1% Triton X100). Following these washes, embryos were transferred to 1:1 PBT 1%/hybridization solution (50% Formamide; 5% dextran sulfate; 100 µg/ml yeast tRNA; 1X salts). Embryos were pre-hybridized for 1 h at 60 °C, followed by addition of a Dig-labeled RNA probe overnight at 60 °C. Embryos were then transferred gradually from hybridization solution to PBT 0.3% through consecutive washes with 3:1, 1:1, 1:3 pre-warmed hybridization solution: PBT 0.3% gradient. A blocking step was performed with PAT (1X PBS; 1% Triton X100; 1% BSA) at room temperature followed by incubation with anti-DIG antibody coupled with alkaline phosphatase for 2 h at room temperature, or at 4 °C overnight. Embryos were washed several times in PBT 0.3% then in PTW 0.05% before color reaction is conducted with NBT/BCIP in alkaline phosphatase buffer (0.1 M Tris pH 9.5; 0.05 M MgCl2; 0.1 M NaCl; 0.1% Tween-20).
Antibody staining
Dissected embryos were fixed in 4% Formaldehyde for 10–15 min at room temperature, washed three times in PTW 0.05% then three times in PBT 0.3%. Embryos were then incubated in a blocking solution (1X PBS; 0.1% Triton X100; 0.1% BSA; 10% NGS) for 1 h at room temperature, then incubated with a 1:5 dilution of the primary antibody (the FP6.87 mouse anti-UbdA or anti-Exd from Developmental Studies Hybridoma Bank (DSHB), rabbit anti-Mef2, and Alexa488-coupled anti-horseradish peroxidase) overnight at 4 °C. Both FP6.87 aliquots obtained from R. H. White and from DSHB cross-react in the semi-aquatic insects. Embryos were then washed four times in PTW 0.05%, three times in blocking solution and incubated in blocking solution for 30 min. Embryos were then incubated with the anti-mouse secondary antibody coupled with horseradish peroxidase at 1:1000 dilution for 2 h at room temperature. Several washes in PBT 0.3% and PTW 0.05% were performed before color reaction was conducted in DAB; 0.064% NiCl2; 0.009% H2O2 for a black color, or without NiCl2 for a brown color.
Quantification of expression using comparative transcriptomics
Limnoporus T2- and T3-legs from wild type and a sample of pooled Ubx RNAi treatments with a ~2 μg/μl ds-Ubx concentration, spanning mid- and late stages, were dissected and used for total RNA extraction. The RNA from these legs was used by ProfilExperts (Lyon-France) to conduct deep sequencing using Illumina technology. A total of ~50 million reads were generated per sample. These reads were aligned against a pre-existing full Limnoporus transcriptome, using BLASTn, to quantify Ubx transcript levels by determining the number of Reads Per Kilobase per Million mapped reads (RPKM) in each sample (Mortazavi et al., 2008).
Parental RNAi
Gene knockdown using parental RNAi was conducted following the protocol in (Khila et al., 2009). Control RNAi was conducted by injecting either ds-yfp (double stranded RNA against yellow fluorescent protein) or injection buffer. Template for in vitro transcription to produce ds-RNA for each gene was prepared using T7-tagged primers in Table S2. Embryos were collected from the Styrofoam stripes, treated with 25% bleach, and washed in 0.05% PTW. Embryos were routinely screened for phenotypes.
Leg measurements
A sample of 10 embryos (N=10) is used for each RNAi group in Fig. 3E and a total of 92 embryos in Fig. 4A–D. Two sets of measurements were recorded: egg length and the length of the tarsus, tibia, and femur individually. Total leg length was calculated as the sum of the length of the three leg segments. After measuring egg length, late embryos prior to hatching were dissected and mounted on slides in Hoyer׳s medium. Measurements for each leg segment of each pair of legs were recorded on a Zeiss microscope using the Zen software.
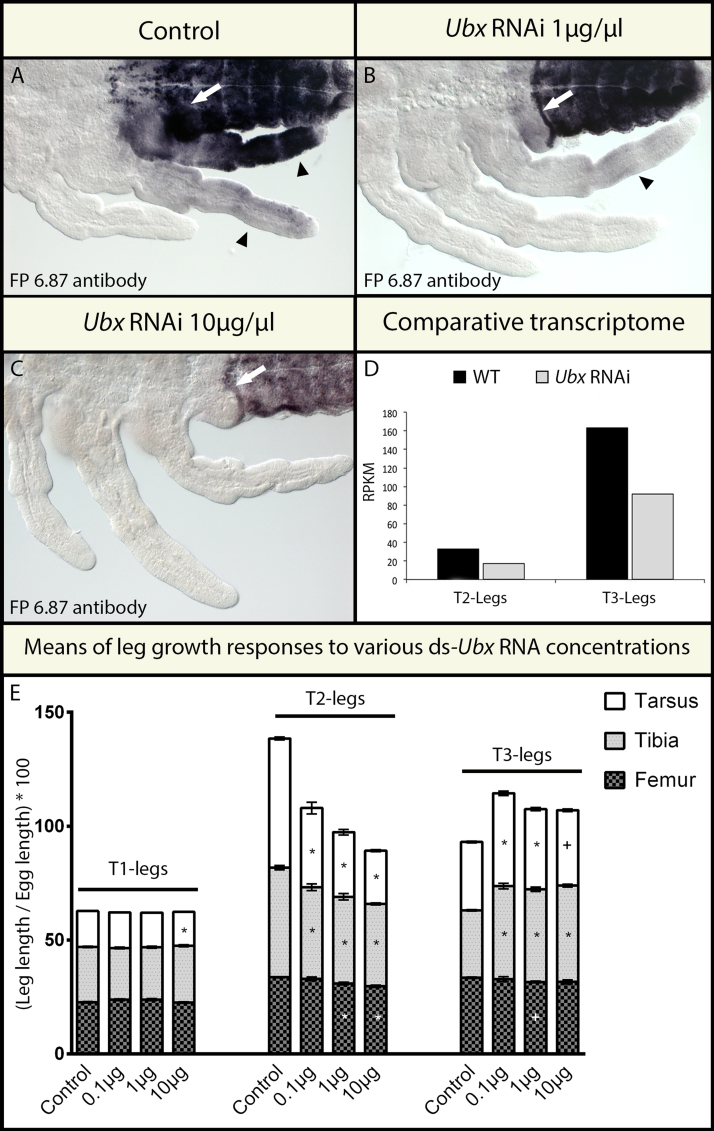
Fig. 3.
Effect of RNAi on Ubx levels and on leg length in Limnoporus embryos. (A) Anti-UbdA staining is unaffected in control embryos and shows faint Ubx expression in T2-legs and stronger expression in T3-legs. In the abdomen, the anterior-most boundary of Abd-A is masked by the strong expression of Ubx (white arrow). (B) When a 1 μg/μl concentration of ds-Ubx was injected, Ubx is now undetectable in T2- and becomes faint in T3-legs. Note that the anterior-most boundary of Abd-A has now become sharp due to faint Ubx in abdominal segment A1 (white arrow). (C) When the even higher 10 μg/μl concentration of ds-Ubx was injected, Ubx is now undetectable in T2-legs, T3-legs, or in the abdomen. The anterior-most boundary of Abd-A remains intact (white arrow). Because Ubx is also expressed in abdominal segments and contributes to the strong signal there (Fig. 2A), the strong depletion of Ubx results in a lower signal in the abdomen, even when the reaction is left to develop longer. (D) Quantification, using deep sequencing, of the efficiency of Ubx transcript depletion in response to Ubx RNAi (pool of embryos from a ~2 μg/μl treatment). Ubx is depleted approximately by half and this depletion is uniform in T2- and T3-legs, such that the difference in Ubx levels remains between the two legs. (E) Growth response of T2- and T3-legs to the injection of increasing concentrations of ds-Ubx. * indicates statistical significance at P≤0.01 and + at P≤0.05. N=10 in all samples.
Fig. 4.
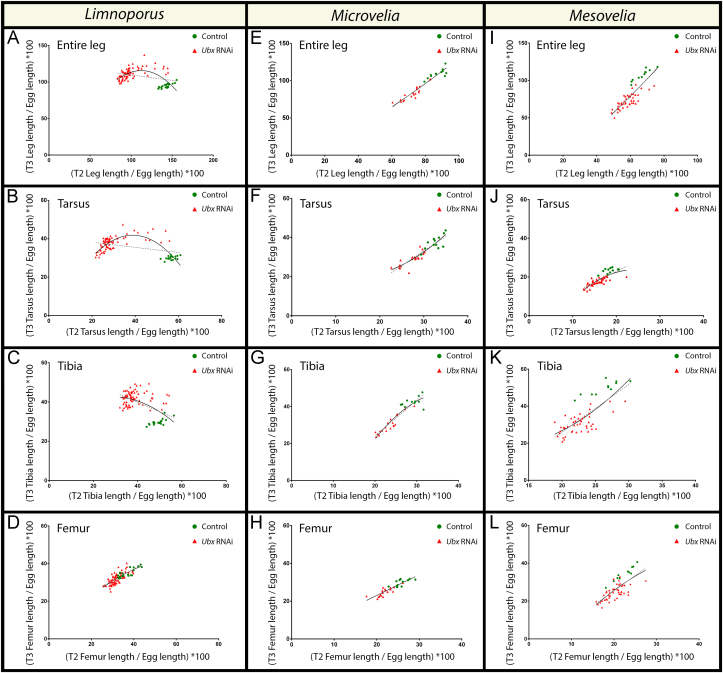
Correlation between the responses of T2- and T3-legs to Ubx RNAi in the three semi-aquatic insects. Plotted values of leg length were corrected to egg length. Two fit models were plotted on the data for comparison: a centered second order polynomial (quadratic) curve shown as a solid black curve, and a linear regression fit shown as a dashed line. Each data point represents an individual; ds-Ubx treated individuals are represented by red triangles and control individuals are represented by green dots. (A) Limnoporus total T3-leg length relative to T2-leg length showing that the data points follow the curved model (R²=0.3457) rather than the linear model (R²=0.1197); N=92. (B–D) Individual leg segments of the T3-leg were plotted against their T2-leg counterparts to analyze the contribution of each to the global effect on the total curve. (B) Limnoporus T3-leg tarsus; (C) Limnoporus T3-leg tibia; and (D) Limnoporus T3-leg femur. The same analyses were conducted on Microvelia (E–H), N=31; and Mesovelia (I–L), N=56. In these two species, the centered second order polynomial follows the same curve as the linear model. Value of R² showing best fit for these models can be found in Table S3.
Statistical analyses
Statistical significance in leg length between each RNAi group and controls was determined by performing an analysis of variance (ANOVA) with the mean value of each pair of leg segments corrected to the egg length to account for embryo size variations. Statistical analyses were performed using the SPSS (Statistical Package for the Social Sciences) software package (IBM Corporation). In the scatter plots in Fig. 4, the centered second order polynomial (quadratic) model, and the linear regression fit model were plotted using GraphPad Prism (version 6.01). R² values for the models are presented in Table S3.
Results
Ubx is expressed at higher levels in T3-legs relative to T2-legs across the semi-aquatic insects
To examine the evolution of differential levels of Ubx expression across the semi-aquatic insects we chose representatives from basally branching and derived lineages, some of which walk and others row on water (Fig. 1). In the surface rowing water strider Limnoporus, in situ hybridization revealed that the expression of Ubx mRNA is much stronger in T3- compared to T2-legs (Fig. 2A). Ubx expression is also detectable in abdominal segments, in addition to the thorax (Fig. 2A). To further confirm this difference in the levels of expression between the two legs, we detected Ubx protein using the FP6.87 antibody known to recognize both Ubx and Abdominal-A (Abd-A) proteins (Kelsh et al., 1994; White and Wilcox, 1984). Consistent with in situ hybridization, antibody staining is much stronger in T3-legs (Fig. 2B), supporting the conclusion that both Ubx mRNA and Ubx protein are expressed at higher levels in T3- relative to T2-legs in Limnoporus embryos. Interestingly, these clear differences in the levels of Ubx in the legs are not seen in a set of neurons located on T2 and T3 segments (Fig. S1). To verify that FP6.87 antibody reacts with Ubx protein only in the thorax, we stained embryos with Abd-A mRNA probe. Abd-A mRNA is restricted to abdominal segments A2–A8 and no staining was observed in the thorax (contrast Fig. 2A–C). Therefore, the strong signal of FP6.87 staining in T3-legs corresponds to Ubx alone, whereas the strong signal in the abdominal segments is a result of co-expression of Ubx and Abd-A there. Finally, to determine the differences in the levels of Ubx expression between T2- and T3-legs, we performed a comparative transcriptome between Limnoporus T2- and T3-legs using deep sequencing. This method indicated that the levels of Ubx transcript are over six to seven times higher in T3-legs compared to T2-legs, during both mid and late embryogenesis (Fig. 2D).
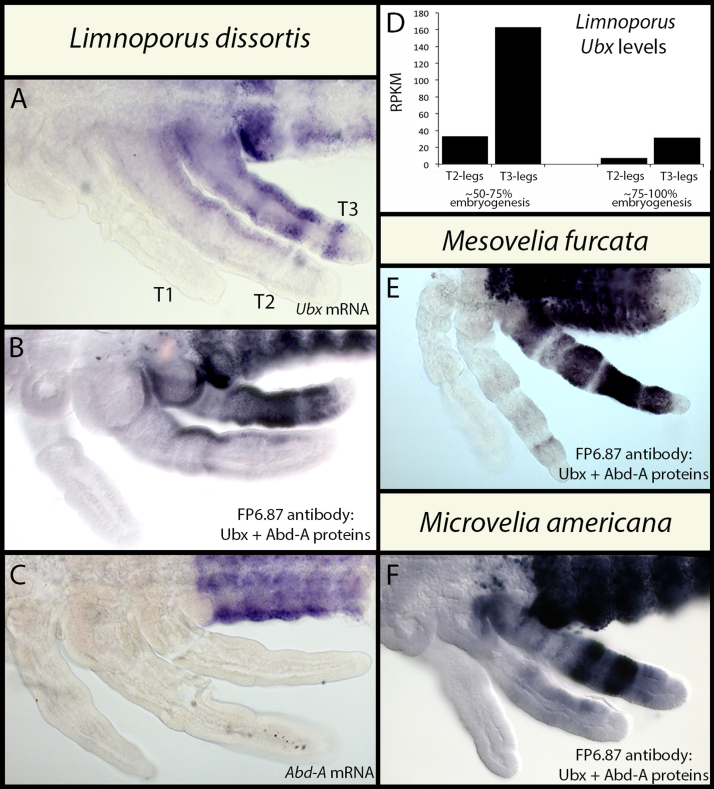
Fig. 2.
Levels of Ubx mRNA and Ubx protein expression in T2- and T3-legs of three species of semi-aquatic insects. (A) Detection of Limnoporus Ubx mRNA using a Digoxygenin labeled probe. (B) Detection of both Ubx and Abd-A proteins using the FP6.87 anti-UbdA antibody in Limnoporus embryo. (C) Detection of Limnoporus Abd-A mRNA using a Digoxygenin labeled probe. Comparison between A, B, and C shows that Ubx is expressed in T2 and T3 thoracic as well as abdominal segments, whereas Abd-A is absent from thoracic segments. (D) Quantification, using deep sequencing, of the differences of Ubx transcript levels between T2- and T3-legs at mid- and late embryogenesis in Limnoporus. Ubx levels are consistently 6–7 times higher in T3- compared to T2-legs. (E and F) Detection of Ubx and Abd-A proteins using the FP6.87 anti-UbdA antibody in Mesovelia (E) and Microvelia (F) embryos. RPKM: Reads Per Kilobase per Million. T1–3: thoracic appendages 1–3.
Apart from the Gerridae, most other semi-aquatic insects retain the ancestral leg allometry where T2-legs are shorter than T3-legs (Fig. 1) (Andersen, 1982; Damgaard et al., 2005). Therefore, we wanted to know whether this difference in Ubx levels between T2- and T3-legs is characteristic of lineages where relative leg length is reversed or whether it is a general feature of semi-aquatic insects regardless of relative leg length. We therefore chose M. furcata, a representative of the most basally branching Mesoveliidae, and M. americana, a representative of the derived Veliidae (Andersen, 1982; Damgaard et al., 2005). In contrast to Limnoporus, both Mesovelia and Microvelia retain ancestral leg length plan and can walk on water and land (Fig. 1) (Andersen, 1982). Surprisingly in both species, FP6.87 staining revealed a much stronger signal in T3- relative to T2-legs (Fig. 2E and F), just like in Limnoporus. These results suggest that across the semi-aquatic insects, Ubx is expressed at higher levels in T3- compared to T2-legs, regardless of phylogenetic position, relative leg length, or whether the legs are used for walking or rowing as a mode of water surface locomotion.
Stronger Ubx depletion results in stronger length phenotype in Limnoporus T2- but not T3-legs
By the end of embryo development, Limnoporus T2-legs reach a much longer size relative to T3-legs (compare control T2-leg and T3-leg in Figs. 3; S2). Because Ubx is expressed at higher levels in T3- relative to T2-legs, we tested whether these differences in expression levels contribute to the differential effect of Ubx in shaping the length of the two legs. To do this, we generated a series of mild to strong phenotypes by injecting low (0.1 μg/μl), moderate (1 μg/μl), or high (10 μg/μl) concentrations of Ubx double stranded RNA (ds-Ubx) to the mothers. First, we verified the specificity of Ubx RNAi, and whether Ubx depletion became stronger with increased amounts of injected ds-Ubx. Staining with the FP6.87 antibody allows us to assess the specificity of RNAi as ds-Ubx injection is expected to deplete Ubx but not Abd-A. In embryos obtained from mothers injected with a concentration of 0.1 μg/μl, FP6.87 staining still detects some Ubx protein in T3-legs in 66% of embryos examined, whereas no Ubx could be detected in T2-legs (Fig. 3B). In the 1 μg/μl and 10 μg/μl ds-Ubx injections, the percentage of embryos where Ubx was still detectable in T3-legs decreased to 39% and 26% respectively (Table S4) and the remaining embryo showed no Ubx expression in T3-legs (Fig. 3C). In embryos with strong Ubx depletion, the FP6.87 staining in the abdomen, which corresponds to Abd-A protein, became weaker because of the loss of Ubx expression there (Fig. 3C). Regardless of the injected concentration of ds-Ubx, Abd-A remains normally expressed in the abdomen (Fig. 3A–C). This indicates that there is a general correlation between the concentration of injected ds-Ubx and the severity of Ubx depletion. Therefore, increased ds-Ubx concentration generally results in stronger depletion of Ubx in the legs. Second, we wanted to know whether or not Ubx RNAi depletes Ubx transcripts in T2- and T3-legs with equal efficiency. We therefore analyzed Ubx transcript levels using the comparative transcriptomics data between legs dissected from wild type (WT) and Ubx RNAi dataset generated from a pool of samples injected with ~2 μg/μl concentration (Fig. 3D). In this dataset, the number of Ubx Reads Per Kilobase per Million (RPKM) is reduced from 32 in WT T2-legs to 17 in Ubx RNAi T2-legs, and from 162 in WT T3-legs to 92 in Ubx RNAi T3-legs (Fig. 3D). Therefore, RNAi depletes Ubx with equal efficiency in T2- and T3-legs, by 46.8% and 43.2% respectively. Combined, these experiments demonstrate that: (i) Ubx RNAi is specific and does not affect Abd-A; (ii) the efficiency of Ubx depletion generally increases with increased dose of injected ds-Ubx; and (iii) Ubx is depleted with equal efficiency in T2- and T3-legs.
To assess the effect of Ubx RNAi on the length of T2- and T3-legs, we dissected and measured each leg across a sample of embryos. In embryos laid by females injected with the lower 0.1 μg/μl concentration of ds-Ubx, T2-legs became shorter while T3-legs became longer compared to their respective control counterparts (Fig. 3E). In T2-legs of these embryos, we obtained a significant 38.6% and 16.3% shortening in the tarsus and tibia respectively, and a slight non-significant shortening effect in the femur (Fig. 3E; Table S5). In T3-legs of the same embryos, we obtained a significant 35.8% and 38.3% elongation in the tarsus and tibia respectively, and a slight non-significant shortening in the femur (Fig. 3E; Table S5). This demonstrates that when a relatively low concentration of ds-Ubx is injected, Ubx RNAi shortens T2- and elongates T3-legs in a significant manner. In embryos laid by females injected with 1 μg/μl ds-Ubx concentration, the shortening effect on T2-legs became stronger, whereas the lengthening effect on T3-legs became milder (Fig. 3E; Table S5). In T2-legs of these embryos, all segments are now significantly shortened by 49.9%, 20.8%, and 8.3% in the tarsus, tibia, and femur respectively (Fig. 3E; Table S5). This suggests that the more Ubx is depleted the shorter T2-legs develop. In contrast, the elongating effect on T3-legs was attenuated, especially in the tarsus, by the injection of the 1 μg/μl dose of ds-Ubx such that the tarsus is now elongated by only 17.5% (instead of 35.8%). In the tibia and the femur of T3-legs, the 1 μg/μl dose of ds-Ubx affected the length in a similar manner as the lower 0.1 μg/μl dose (Table S5). When an even higher 10 μg/μl dose of ds-Ubx was injected, all three segments of T2-legs were further shortened, whereas in T3-legs the lengthening of the tarsus was further attenuated (10.1% instead of 17.5% obtained with 1 μg/μl and 35.8% obtained with 0.1 μg/μl ds-Ubx concentrations; Table S5). These results suggest that mild depletion of Ubx causes T3-legs to grow longer, but a stronger depletion reverses, at least partially, this lengthening effect.
Ubx dose-dependent effect on leg length is linear in walking but non-linear in surface rowing semi-aquatic insects
To determine the correlation between the responses of T2- and T3-legs to Ubx RNAi, we analyzed the association between T2- and T3-leg length in each individual using a larger dataset, composed of all injected concentrations and additional samples (N=92). This analysis completes the previous, average-based, method because it considers the values of leg length of each embryo rather than a mean, it controls for the variation in phenotype penetrance observed (Table S4) and that is characteristic to the RNAi technique (Jaubert-Possamai et al., 2007; Kitzmann et al., 2013), and finally it reveals how the lengths of the two legs change with respect to each other within each embryo. If Ubx operates to increase or decrease leg length in a dose-dependent manner, we should expect a non-linear association between the responses of T2- and T3-legs to Ubx RNAi. Testing this prediction is based on three consistent observations: (i) control legs express higher levels of Ubx protein relative to their Ubx RNAi-treated counterparts (Fig. 3A–D); (ii) RNAi depletes Ubx transcripts equally in T2- and T3-legs (Fig. 3D); and (iii) the length of T2-legs consistently decreases with increased depletion of Ubx protein (Fig. 3E). Therefore, we can use the length of T2-legs as a proxy to infer Ubx levels, such that the shorter T2-legs are the lower Ubx protein levels are. Based on this, we plotted T2-leg length by T3-leg length using a dataset including a larger sample size than the sample used in Fig. 3E, to determine the nature of this association (Fig. 4). This analysis revealed that in Limnoporus, when T2-leg length was mildly reduced in response to Ubx RNAi, the increase in T3-leg length was substantial (Fig. 4A). Therefore, a slight depletion may have shifted the levels of Ubx in T3-legs from those sufficiently high to decrease, to those that are low enough to increase leg length. The shift in the morphology of Ubx RNAi-treated T3-legs to the likeness of control T2-legs provides further support to this conclusion. In contrast, when T2-leg length was severely reduced, the increase in T3-leg length became attenuated (Fig. 4A). This further confirms that a strong depletion brings the levels of Ubx in T3-legs even lower such that T3-leg length is now reduced. To determine whether or not all the segments within T3-legs are responsible for this non-linear response to Ubx RNAi, we analyzed the association between the responses of the tarsi, tibias, and femora individually (Fig. 4B–D). We found that the response of T3-leg tarsus (Fig. 4B), in particular, and the tibia (Fig. 4C), to a lesser degree, to be non-linear dose-dependent, whereas that of the femur is largely linear (Fig. 4D). Therefore in Limnoporus, the tarsus is primarily responsible for the overall non-linear response of T3-legs to Ubx RNAi. Altogether, these data suggest that in Limnoporus embryos, the lengthening/shortening effect of Ubx is dose-dependent and that the lengthening of T2-legs is due to the lower levels of Ubx and the shortening of T3-legs is due to the higher levels of Ubx.
Next, we wanted to test whether the dose-dependent requirement of Ubx to regulate leg length is a general feature of the semi-aquatic insects, or whether it is characteristic of species where relative leg length has been reversed. We therefore conducted a series of injection experiments with increasing doses of ds-Ubx in the two walking species, Microvelia and Mesovelia to determine if the strength of Ubx depletion would result in opposing effects on leg length. In both Microvelia (Fig. 4E–H) and Mesovelia (Fig. 4I–L), Ubx RNAi resulted in a consistent decrease of both T2- and T3-leg length in all injected concentrations. A plot of T2-leg length by T3-leg length in these two species revealed that, in contrast to Limnoporus, the lengths of the two legs decrease together in a linear manner in response to Ubx RNAi, regardless of the strength of this reduction. This linear relationship is present in all leg segments in the two species (Fig. 4E–L). Therefore, Ubx is required to increase the length of both T2- and T3-legs in these two species and no repressive role can be detected despite the apparent differences in Ubx levels between the two legs (Fig. 2E and F). Collectively, these findings suggest that the evolution of the dose-dependent activating-repressing effect of Ubx on leg growth is characteristic to the surface rowing water striders with derived relative leg length, but not other semi-aquatic insects that retain the ancestral state of leg length.
Ubx RNAi partially transforms T3-legs to the likeness of T2-legs
Because our experiment is based on parental RNAi, which depletes Ubx transcript starting early embryogenesis, we wanted to know the extent to which this experiment transforms one segment into the likeness of another. First, we looked for morphological and cellular attributes that could help differentiate thoracic segments from each other in normal animals. In Limnoporus, T1-legs are equipped with two distinctive combs; one sex comb and one grooming comb both located on the distal tibia (Fig. S3). Both T2- and T3-legs are similarly equipped with one grooming comb on the distal tibia (Fig. S3) (Khila et al., 2014). Therefore, while T2 and T3 can be clearly distinguished from T1 based on comb number, they are not distinguishable from each other based on this trait. We then stained Limnoporus embryos to reveal muscles, using the anti-Mef2 antibody (Sandmann et al., 2006), and nervous system, using the anti-HRP antibody (Jan and Jan, 1982; Paschinger et al., 2009), in an attempt to identify cellular characteristics that would help distinguish T2 from T3. Both antibodies revealed that muscular and nervous system structures are highly similar across all legs (Fig. S4). Therefore, the major observable difference between T2- and T3-legs in Limnoporus is their relative length and the length of the tarsi, tibias, and femora within each of them.
To examine the effect of Ubx RNAi on the development of these distinctive characteristics between segments across the three species, we quantified the degree of homeotic transformation in both mild and strong Ubx phenotypes. To do this, we selected a sample of five individuals representing the mildest and five representing the strongest Ubx phenotype presented in Fig. 4. In Limnoporus, because differences in length of the segments within legs are the only distinctive character that we could identify between T2-legs and T3-legs, we quantified the changes in the ratios of the tarsi, tibias, and femora in Ubx RNAi individuals in comparison with controls. We hypothesized that if the transformation of T3 towards T2 is complete, the ratio of each of the segments of Ubx RNAi T3-legs to control T2-legs should give an approximate value of 1. In normal Limnoporus embryos, the average ratio of T3-tarsus to T2-tarsus is 0.52, T3-tibia to T2-tibia is 0.58, and T3-femur to T2-femur is 0.9 (see Table S6A). We found in the sample representing the mildest Ubx RNAi phenotype that the average ratio of Ubx-T3-tarsus to control-T2-tarsus increases to the value of 0.7, the average ratio of Ubx-T3-tibia to control-T2-tibia increases to the value of 0.85 and the average ratio of Ubx-T3-femur to control-T2-femur decreases to the value of 0.86 (Table S6B). In the sample representing the strongest Ubx RNAi phenotype however, we found that the average ratio of Ubx-T3-tarsus to control-T2-tarsus decreases back to 0.57, the average ratio of Ubx-T3-tibia to control-T2-tibia decreases back to 0.78 and the average ratio of Ubx-T3-femur to control-T2-femur decreases to 0.71 (Table S6C). These changes in the ratios of leg segments with the differences in the strength of Ubx RNAi are consistent with the expression of Ubx in both legs. Mild Ubx RNAi probably brings the levels of Ubx protein in T3-legs down to the levels normally expressed in normal T2-legs, explaining the increase of the ratios towards the value of 1. This is the closest transformation of T3-legs towards T2-legs we observed. Conversely, strong Ubx RNAi depletes Ubx protein such that the remaining levels are even lower than those in normal T2-legs, explaining the decrease in the ratios of leg segments back away from 1. These data suggest that both mild and strong Ubx RNAi result in a partial transformation of T3-legs towards T2 identity. Aside from leg length, we were not able to detect any clear changes in the patterns of axons or muscles between T2- and T3-legs upon Ubx RNAi treatments in Limnoporus (Fig. S4).
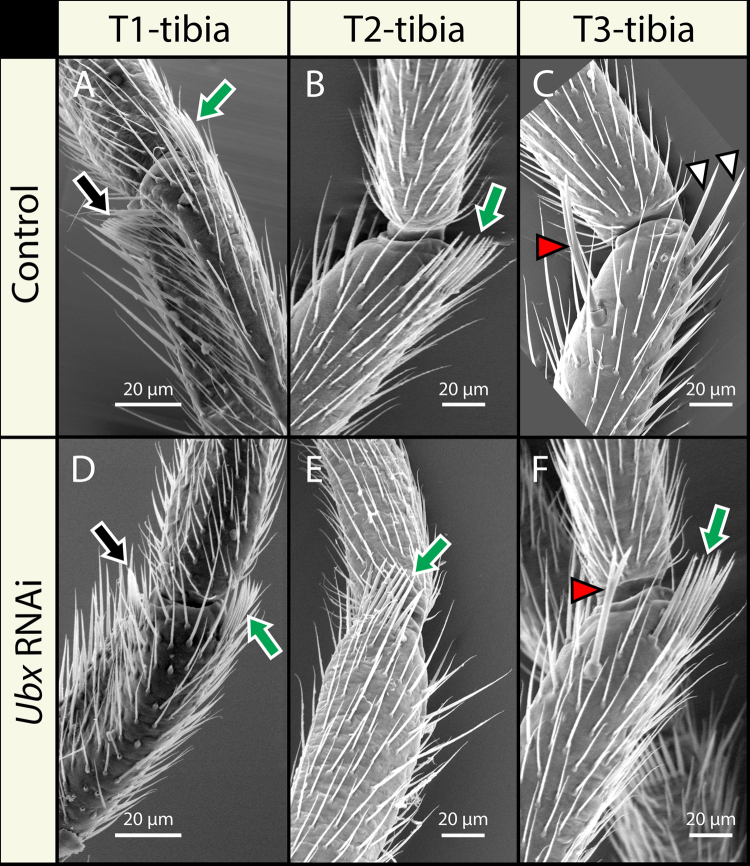
In contrast to Limnoporus, we were able to unambiguously identify a set of morphological attributes that distinguish T2-and T3-legs in both Microvelia and Mesovelia. Microvelia (Fig. 5) and Mesovelia (Fig. S5) (Khila et al., 2014) have two combs in T1 (arrows in Figs. 5A; S5A) and one comb in T2 (arrow in Figs. 5B; S5B), just like Limnoporus. However, unlike Limnoporus, both species have no combs at all in T3-legs (Figs. 5C; S5C). Instead, T3-legs bear large bristles on the tibia that distinguish them from T2-legs (white arrowheads in Figs. 5C and S5C). In addition in Microvelia, T3-legs bear a particular bristle that is forked in its distal part and that is not found on T2-legs (red arrowhead in Fig. 5C). In both Mesovelia and Microvelia, we assessed the degree of transformation between T2-legs and T3-legs based on the presence or absence of the grooming comb and the morphology of T3 distinctive bristles. In no case have we observed the development of a sex comb, in addition to the grooming comb, on T2-legs in any of the species including Limnoporus (Figs. 5E; S3E; S5E; Table S7). This is consistent with the well-known expression of the Hox gene sex combs reduced in T1 but not in the other thoracic segments across insects (Rogers et al., 1997). Like in Limnoporus, we found that Ubx RNAi causes a partial transformation of T3-legs towards T2-legs in these two species. In Microvelia, Ubx RNAi occasionally causes T3-legs to develop a grooming comb similar to the one found on T2-legs (green arrow in Fig. 5F). The formation of this comb is observed in all individuals representing the strongest, but not all those representing the mildest Ubx RNAi phenotype (Table S7). However, Ubx RNAi, whether mild or strong, never resulted in the loss of the large forked bristle (red arrowhead in Fig. 5F; Table S7). We observed a similar dynamics in Mesovelia with regards to the presence/absence of the comb and the size of the bristles on T3-legs (green arrow and white arrowheads in Fig. S5F; Table S7). Altogether, these data suggest that in Limnoporus the weaker the Ubx RNAi the closer the morphology adopted by T3-legs to T2-legs, and conversely the stronger the Ubx RNAi the farther the morphology adopted by T3-legs from T2-legs. However in both Mesovelia and Microvelia, this relationship is reversed such that the stronger the Ubx RNAi the more the T3-legs resembles T2-legs.
Fig. 5.
Ubx RNAi causes a partial homeotic transformation of T3-legs to T2-legs. Microvelia forelegs bear one sex comb (black arrow in A) and one grooming comb (green arrow in A), the T2-legs bear a single grooming comb (green arrow in B), and the T3-legs do not have any combs (C). T3-legs however bear additional thick bristles (white arrowheads in C) and a characteristic forked bristle (red arrowhead in C). Ubx RNAi does not affect the combs either in the T1-legs (D) or in the T2-legs (E). T3-legs in Ubx RNAi treatment now develop an ectopic comb (green arrow in F), lose the thick bristles, but retain their characteristic forked bristle (red arrowhead in F).
The spatial expression of leg patterning genes is unaffected in Ubx RNAi treatments
Ubx is known to regulate a set of important leg patterning genes, including decapentaplegic (dpp), hedgehog (hh), epidermal growth factor receptor (egfr), and wingless (wg) in insects (Estella and Mann, 2008; Estella et al., 2008; Grossmann and Prpic, 2012). Because of the changes in the spatial pattern and the levels of Ubx expression as well as its opposite effect on leg length throughout the evolution of the semi-aquatic insects, we wanted to know whether or not these changes in Ubx affected the expression of a set of important leg patterning genes. First, we interrogated our comparative leg transcriptome datasets generated from normal and Ubx RNAi treatments to determine if the levels of expression of these key patterning genes are affected by Ubx depletion. In untreated embryos, the levels of expression of all genes tested, as revealed by the obtained RPKM, are comparable between T2- and T3-legs (Table S8). In Ubx RNAi treatments, we detected an increase in the levels of expression of these genes ranging from subtle (15–20%) to double the expression levels, as revealed by their RPKM (Table S8). In this analysis, hh, egfr, exd, and wg showed over 50% increase in both T2- and T3-legs upon Ubx RNAi (Table S8). These data suggest that Ubx may regulate at various degrees the levels of expression of many developmental genes, consistent with Ubx role in flies (Pavlopoulos and Akam, 2011).
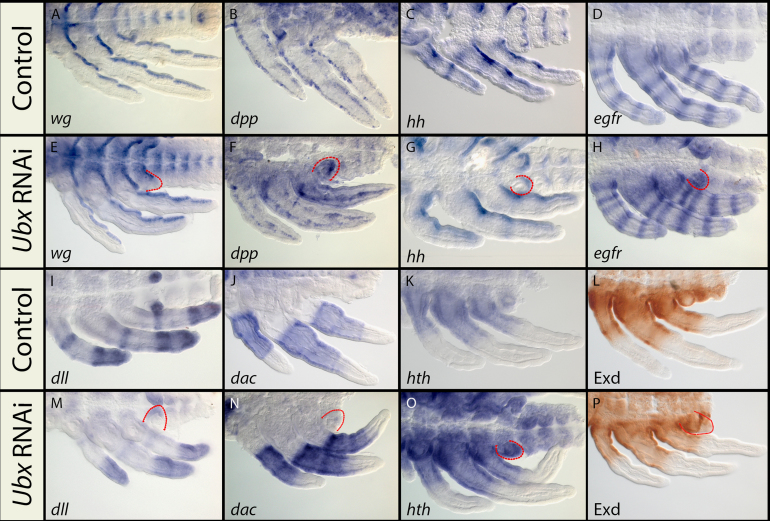
Next, we wanted to determine whether this change in the transcriptional levels affected the spatial patterns of expression of these genes in the various legs of Limnoporus embryos. First, we examined their expression patterns in normal embryos. in situ hybridization and immuno-staining on untreated embryos revealed that all these developmental genes show specific patterns of expression in the developing legs (Fig. 6). However, unlike Ubx (Fig. 2), all these genes are expressed in an invariable pattern in all legs. At mid-embryogenesis, wg mRNA is distributed in a ventral segmental pattern and extends along the proximal–distal axis of all thoracic appendages (Fig. 6A). dpp mRNA is distributed in a punctuated pattern along both anterior and posterior edges of the appendages (Fig. 6B). hh is strongly expressed in the posterior compartment, and fades towards the anterior (Fig. 6C). In addition to this posterior-to-anterior distribution, hh mRNA forms four relatively faint stripes along the proximal–distal axis. egfr is expressed in five distinct rings along the proximal–distal axis, which seem to prefigure the junctions between the five leg segments. In addition, a sixth egfr ring separates the claw-bearing tip of the leg from the proximal part of the tarsus (Fig. 6D). mRNA of the gene distal-less (dll), required for the specification of distal appendages across arthropods (Panganiban et al., 1994), is expressed in a conserved pattern consisting of a proximal ring, and uniformly throughout the distal tarsus (Fig. 6I). dachshund (dac), which specifies proximal appendages (Abzhanov and Kaufman, 2000; Angelini and Kaufman, 2004; Prpic et al., 2003, 2001), is also expressed in a conserved pattern consisting of a single band extending from the mid-femur until the boundary between the tibia and the tarsus (Fig. 6J). Both hth and Exd, two known co-factors that bind and direct Hox proteins to their specific target DNA (Joshi et al., 2007; Mann et al., 2009) are expressed in a highly similar pattern and their domains span the proximal segments from the coxa to the proximal half of the femur (Fig. 6K and L). Second, we revealed the spatial patterns of expression of all these leg patterning genes in the developing legs in both 1 μg/μl (Fig. 6E–H and M–P) and 10 μg/μl (not shown) Ubx RNAi treatments. Following this treatment, we were not able to detect any changes in the spatial patterns of expression of any of the eight patterning molecules in the legs (Fig. 6). This result indicates that, despite the effect of Ubx RNAi on their levels of expression, neither the mild nor the severe depletion of Ubx resulted in any obvious effect on the spatial expression of these leg patterning genes in Limnoporus. Therefore, the spatial patterns of these genes are likely regulated independently from Ubx during Limnoporus leg development.
Fig. 6.
Patterns of expression of Limnoporus developmental genes in the legs of control (A–D and I–L) and Ubx RNAi treatments (E–H and M–P). (A) wg is expressed along the posterior compartment of the three appendages. (B) dpp appears in a patched expression along the anterior posterior compartments of all legs. (C) hh is expressed as a stripe in the posterior compartment and fading towards the anterior side of all the legs, in addition to a weak ring expression. (D) egfr forms multiple proximo–distal stripes at the joints between the five leg segments. (E–H) patterns of the same genes in the 1 μg/μl Ubx RNAi treatments, showing no apparent change in the spatial patterns of these genes. (I) dll shows the classical ‘ring’ and ‘sock’ expression, in addition to the prominent expression in the first abdominal segment. (J) dac expression domain extends from the mid-femur until the end of the tibia. (K) hth and (L) Exd share the same pattern extending from the coxa until the mid-femur of all three legs. (M–P) patterns of the same genes in the 1 μg/μl Ubx RNAi treatments, showing no apparent change in the spatial patterns of these genes. All stainings represent mRNA (purple), except Exd, which represents the protein (brown). Highlighted in red is the ectopic limb bud that forms on the first abdominal segment, which is a phenotype characteristic to Ubx RNAi.
We also tested the possibility that Ubx might be downstream of some of the signaling molecules by examining the pattern of Ubx protein expression using the FP6.87 anti-UbdA antibody in wg and hh RNAi knockdown backgrounds. FP6.87 staining in wg and hh RNAi-treated embryos is unchanged in abdominal segments or in T2- and T3-legs despite the apparent developmental defects seen in these embryos (Fig. S6A–C). Altogether, these results suggest that Ubx may play a role in modulating the levels but not the spatial expression of leg patterning genes in Limnoporus.
Discussion
In this work, we present four main findings: (i) that generally in the semi-aquatic insects, Ubx is expressed at higher levels in T3- relative to T2-legs; (ii) that it is only in the derived Gerridae where the high levels of Ubx result in reduced T3-leg length; (iii) that in the surface rowing L. dissortis, both strong and mild depletion of Ubx result only in a partial transformation of T3 to the likeness of T2 segment; and (iv) that the changes in Ubx regulation and function have evolved in Limnoporus without any apparent effect on the spatial patterns of expression of a set of major leg patterning genes.
Changes in Ubx levels and their role in establishing leg allometry across the semi-aquatic insects
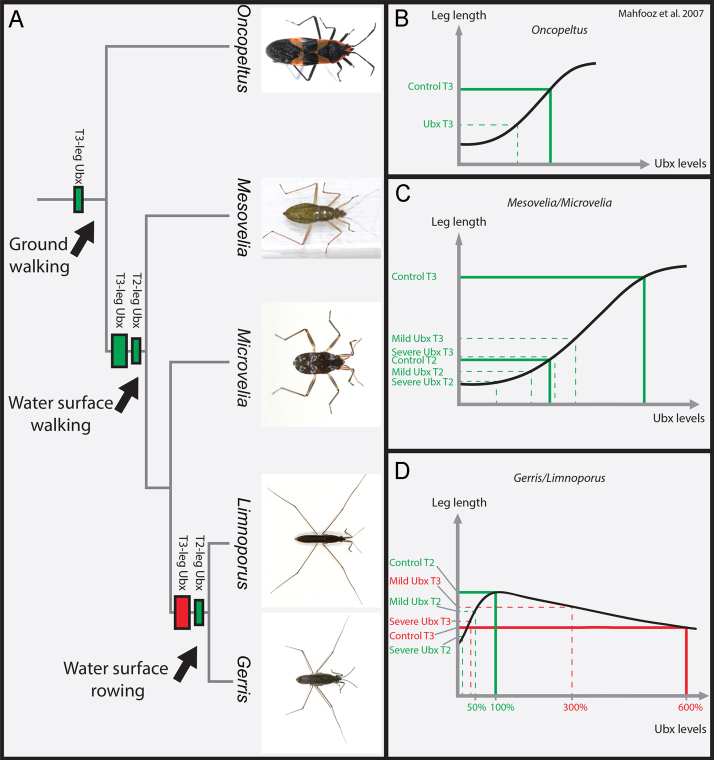
In the embryos of the water strider Limnoporus, Ubx dramatically increases the length of T2-legs whereas six to seven times higher levels of Ubx decrease the length of T3-legs, thereby reversing their relative length. Ubx RNAi embryos still exhibit basal levels of leg length indicating that a developmental ground state of leg growth is achieved without Ubx input. Therefore, this dose-dependent role of Ubx is required for fine-tuning relative length of T2- and T3-legs. Preliminary evidence indicates that most of the differences in leg length are due to differences in the rate of cell division (in preparation), and suggest that Ubx mediates its effect on relative leg length through differential control of cell proliferation in each leg. In the terrestrial heteropteran relative Oncopeltus (Fig. 7A), Ubx expression is restricted to T3-legs and is required for increasing their length (Fig. 7B; Mahfooz et al., 2007). Although we have not measured the levels in Oncopeltus, histochemistry revealing mRNA (Angelini et al., 2005) or protein (Mahfooz et al., 2007) seems to indicate that Ubx levels in T3-legs are low. It appears that during the split between semi-aquatic and terrestrial lineages, both the novel deployment of Ubx in T2-legs and the increase of its levels in T3-legs emerged at the same time, as this feature is common to all semi-aquatic species tested (Fig. 7A). At this early stage of the evolution of the group, Ubx was still required to increase leg length and the increase in Ubx protein levels did not reverse this lengthening effect (Fig. 7C). This is clearly illustrated in the basally branching Mesovelia and the derived Microvelia, both of which retain the ancestral state of relative leg length (Andersen, 1982; Damgaard, 2008; Damgaard et al., 2005), and where the response of the legs to Ubx RNAi is linear (Fig. 7C). Although this first change in Ubx regulation at the base of the Gerromorpha resulted only in a subtle global increase in the length of both T2- and T3-legs, it has also provided a genetic potential that set the stage for further diversification of the group. This genetic potential was exploited by the Gerridae, such as L. dissortis (Fig. 7D), through the emergence of a novel sensitivity to the pre-existing high Ubx levels in T3-legs (Fig. 4). We conclude that this sensitivity of leg tissue to Ubx levels is a derived feature that emerged in surface-rowing species, and distinguishes them from the ancestral state inherited from the common ancestor of the semi-aquatic insects (Fig. 7A). Therefore, adaptive morphological traits can evolve through fine-tuning the levels of expression of key developmental genes early during embryogenesis and the emergence of tissue sensitivity to changes in these levels. This may have facilitated the change in leg function from walking to rowing structures, and has been instrumental in the specialization and radiation of the Gerridae in various open water niches, from small ponds to the vast surfaces of the great oceans.
Fig. 7.
Evolution of Ubx levels of expression and its role in shaping adaptive leg allometry. (A) Phylogenetic relationships between terrestrial Heteroptera, (Oncopeltus), water surface walking (Mesovelia and Microvelia), and water surface rowing (Gerris and Limnoporus) semi-aquatic insects. Narrow green rectangle represents the ancestral state where Ubx expression was confined to T3-legs and functioned to lengthen them. Large green rectangle in T3- and narrow green rectangle in T2-legs represent the change at the base of the semi-aquatic insects consisting of increased Ubx levels in T3- and deployment of Ubx in T2-legs. At this stage, Ubx lengthens both T2- and T3-legs. Large red rectangle indicates the emergence of T3-leg sensitivity to high Ubx levels, which now shortens T3-legs. This characteristic distinguishes surface-rowing semi-aquatic insects from their surface-walking relatives. (B) Ubx role and RNAi response in Oncopeltus T3-legs. (C) Ubx role in lengthening both T2- and T3-legs is linear in the basally branching Mesovelia and the derived Microvelia, both of which are surface walkers. (D) Model for the dose-dependent role of Ubx in increasing/decreasing leg length. Percentages of Ubx levels were extracted from a comparative transciptome between T2- and T3-legs in normal and Ubx RNAi-treatments. Approximate leg length values were taken from Fig. 3D. Levels of Ubx in T2-legs are used as a reference at 100%, and levels in T3-legs are at least six times higher (600%) based on the comparative transcriptome. At highest levels Ubx shortens whereas at lower levels Ubx lengthens the legs.
Dose-dependent switch of Ubx role between increasing and decreasing leg length
The molecular interactions underlying the different responses of Limnoporus T3-legs and those of Microvelia and Mesovelia to high levels of Ubx remain to be investigated. The response of the legs in Limnoporus to Ubx RNAi suggests that there is a dose threshold, and when Ubx exceeds this threshold the growth-suppressing effect ensues. By knocking down Ubx, we have shifted the levels of Ubx protein in T3-legs close to or below this threshold, thereby increasing their length (Figs. 3E; 4A; 7D). In control T2-legs, Ubx levels are already below this threshold and RNAi brings these levels even further down, which explains the consistent decrease in T2-leg length with increased ds-Ubx injected concentration (Figs. 3E; 4A; 7D). A number of molecular mechanisms have been previously described and might explain these distinct responses of leg tissues to Ubx dose. In the fly Drosophila, early gradient and gap genes can act as activators or repressors depending on their concentration (Sauer and Jackle, 1991; Schulz and Tautz, 1994). At low concentration, the transcription factor Kruppel (Kr), for example, remains a monomer and activates its target DNA, whereas at high concentration Kr forms homodimers and represses the same target (Sauer and Jackle, 1993). Many Hox proteins, including sex combs reduced (Papadopoulos et al., 2012), Abd-A (Hudry et al., 2011), and Ubx itself (Samir Merabet, personal communication) are known to form homodimers. It is therefore possible that a mechanism, similar to that of Kr, involving the formation of homodimers at high Ubx concentration results in the opposite effect on leg length relative to low Ubx concentration. The formation of heterodimers between Ubx and its known co-factors might also explain these differences in tissue response to Ubx levels. Our results, however, provide evidence that exclude possible heterodimer formation between Ubx and the two known co-factors Exd and Hth in regulating leg length. Both Exd and Hth patterns of expression are confined to the proximal segments, reminiscent to their pattern of expression in fly appendages (Casares and Mann, 2000). The non-linear bimodal response to Ubx RNAi concerns distal leg segments where these two co-factors are not expressed, and therefore, inconsistent with a possible cooperation between these factors and Ubx in this tissue. This is also the case in fly leg development, where Ubx acts without any cooperation with Exd and Hth (Casares and Mann, 1998; Galant et al., 2002). In addition to co-factors, Ubx is known to mediate its role in transcriptional regulation through cooperation with a number of transcription factors called Hox collaborators (Mann et al., 2009). These collaborators provide direct input into a Hox-regulated element without necessarily binding the element in cooperation with the Hox gene (Mann et al., 2009). It is conceivable that such collaborators may be part of the dose-dependent differential response of each leg to Ubx. Other additional mechanisms may be in play to mediate this dose-dependent positive-to-negative shift in Ubx effect. The degree of occupancy of Ubx binding sites within its target sequences, dependent on Ubx protein levels, might dictate the nature of transcriptional regulation of these target genes, as is the case in flies (Galant and Carroll, 2002; Hersh and Carroll, 2005). Finally, it is possible that differences in Ubx affinity to its various targets may explain this bimodal response of the two legs to Ubx dose. Ubx may have high affinity to growth-promoting and low affinity to growth-suppressing targets. In this case, at low concentration, Ubx binding to the high-affinity growth-promoting partners prevails, and the outcome of this interaction increases cell proliferation, as is the case of T2-legs in Limnoporus. On the other hand, when Ubx levels exceed a certain dose the excess of Ubx protein is now free to interact with low affinity growth-suppressing partners. This type of interaction results in reduced cell proliferation and shortened legs, as is the case of Limnoporus T3. This interpretation is supported by the observation, in the fly Drosophila, that Ubx can act both as an activator and as a repressor, and that some target genes respond only at high Ubx concentration (Feinstein et al., 1995; Pavlopoulos and Akam, 2011).
The difference in length between T2 and T3-legs is primarily grounded in cell number, reflecting increased cell proliferation in T2- compared to T3-legs (in preparation). This is in contrast to the size difference in Drosophila halters compared to wing, which is mostly grounded on cell shape changes rather than cell proliferation or volume (Roch and Akam, 2000). Consistent with these cellular differences between T2- and T3-legs, we found in our comparative transcriptome that Ubx regulates a large number of genes involved in various cellular processes. These include cell cycle, cell death, cell differentiation, response to stress, and catabolic processes (not shown). Future work will examine the cellular nature of Ubx effect on leg length and dissect the role of its target genes in modulating the differential rate of cell proliferation between T2- and T3-legs.
Ubx, segment identity, and adaptive morphological evolution
It is striking that such substantive changes in Ubx regulation and requirement in shaping leg allometry have emerged throughout the evolution of the semi-aquatic insects without altering the spatial expression of major players of early pattern formation. The role of the transcription factors and signaling molecules we examined in growth and patterning is well documented (Baena-Lopez et al., 2012, 2003; Crickmore and Mann, 2006; Estella et al., 2008; Theisen et al., 1996; Zecca and Struhl, 2002). Leg patterning genes may contribute to the dramatic growth of Limnoporus legs and could have constituted a target of Ubx during the evolution of its novel role in reversing the length of T2- relative to T3-legs. It is possible that these genes generate the ground state of leg growth during development, and that Ubx comes to modulate this ground state and generate the adaptive relative leg length required for surface rowing. The interactions through which Ubx mediates this fine-tuning of relative leg length may involve a battery of genes that are required for growth but not pattern formation. It seems that there has been a change in the landscape of Ubx targets in repressing tissue growth, at least when this role is compared between Limnoporus T3-legs and Drosophila halteres where Ubx exerts a similar effect (Crickmore and Mann, 2006; Pavlopoulos and Akam, 2011; Roch and Akam, 2000). This observation raises an important question: why is Ubx׳s regulation of the spatial expression of this set of genes so subtle, if any? A plausible explanation is the early timing at which Ubx intervenes to set leg allometry in water striders. Embryogenesis is too early for the evolution of profound rearrangements in the network composed of major regulators of pattern formation (Davidson and Erwin, 2010; Wilkins, 2002). Alteration of these networks, by the novel changes in Ubx role and regulation, would have had disruptive effects that would not have been tolerated by natural selection (Davidson and Erwin, 2010; Wilkins, 2002). Throughout the semi-aquatic insects, either with the ancestral or the derived state of relative leg length (Fig. 7), the change in Ubx expression has not introduced a drastic change in morphology. Our results clearly outline yet another example, in addition to Ubx role in the crustacean Parhyale (Pavlopoulos et al., 2009), that the evolutionary transition to the derived state of relative leg length, adapted for water surface rowing, involved a gradual process that started with an initial change in Ubx expression already in the common ancestor of the semi-aquatic insects (Fig. 7). These observations come to further support the view that Ubx acts as a micromanager in establishing organ morphogenesis during development and evolution (Akam, 1998b, 1998c; Castelli-Gair and Akam, 1995; Stern, 1998, 2003). Hox genes may not only act as master binary switches of segment identity, but can rather accumulate changes throughout evolution, the sum of which leads to incremental changes in segment morphology, ‘without the need to invoke the selective advantage of hopeful monsters (Akam, 1998b).’ A similar observation involving the effect of Ubx levels on morphological diversification, at the micro-evolutionary scale, is known in flies. Across distinct fly species, the presence of trichomes on the femur is associated with low levels of Ubx, whereas the absence of trichomes is associated with high levels of Ubx (Stern, 1998). Our data show that, at a macro-evolutionary scale, this flexibility in the regulation, function, and most likely diversity of targets, can generate the remarkable phenotypic diversity that selection acts upon within specific ecological environments.
Acknowledgments
We thank Ehab Abouheif for hosting AK and PR in his lab during earlier steps of this project, Michalis Averof, Samir Merabet, Dali Ma, and Emilia Santos, and three anonymous reviewers for comments on the manuscript. We thank Kelly Langlais for technical support and E. Furlong, R. White, and Developmental Studies Hybridoma Bank for providing the anti-Mef2, anti-UbdA and Exd antibodies respectively, all of which cross-react in various semi-aquatic insect species. We thank John Colbourne for help with generating Limnoporus full transcriptome used as a reference, and Lois Taulelle and Hervé Gilquin for providing access to computing resources in the Pôle Scientifique de Modélisation Numérique (PSMN) at the ENS Lyon. Scanning electron microscopy was performed in the facility of the Centre Technologique des Microstructures at the Université Claude Bernard Lyon1. This work was funded through an ATIP-Avenir from CNRS and an ERC-CoG # 616346 to AK and a PhD Fellowship from Université Claude Bernard Lyon1 (0103/2010) to PR.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2014.05.021.
Appendix A. Supporting information
Supplementary data
References
- Abzhanov A., Kaufman T.C. Homologs of Drosophila appendage genes in the patterning of arthropod limbs. Dev. Biol. 2000;227:673–689. doi: 10.1006/dbio.2000.9904. [DOI] [PubMed] [Google Scholar]
- Abzhanov A., Protas M., Grant B.R., Grant P.R., Tabin C.J. Bmp4 and morphological variation of beaks in Darwin׳s finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Akam M. Hox genes in arthropod development and evolution. Biol. Bull. 1998;195:373–374. doi: 10.2307/1543151. [DOI] [PubMed] [Google Scholar]
- Akam M. Hox genes, homeosis and the evolution of segment identity: no need for hopeless monsters. Int. J. Dev. Biol. 1998;42:445–451. [PubMed] [Google Scholar]
- Akam M. Hox genes: from master genes to micromanagers. Curr. Biol. 1998;8:R676–R678. doi: 10.1016/s0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- Andersen N.M. A Comparative Study of Locomotion on the Water Surface in Semiaquatic Bugs (Insecta, Hemiptera, Gerromorpha) (Vidensk. Meddr dansk naturh. Foren) 1976. pp. 337–396. [Google Scholar]
- Andersen N.M. Scandinavian Science Press Ltd.; Klampenborg, Denmark: 1982. The Semiaquatic Bugs (Hemiptera: Gerromorpha) [Google Scholar]
- Angelini D.R., Kaufman T.C. Functional analyses in the hemipteran Oncopeltus fasciatus reveal conserved and derived aspects of appendage patterning in insects. Dev. Biol. 2004;271:306–321. doi: 10.1016/j.ydbio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Angelini D.R., Liu P.Z., Hughes C.L., Kaufman T.C. Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera) Dev. Biol. 2005;287:440–455. doi: 10.1016/j.ydbio.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez L.A., Nojima H., Vincent J.P. Integration of morphogen signalling within the growth regulatory network. Curr. Opin. Cell Biol. 2012;24:166–172. doi: 10.1016/j.ceb.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Baena-Lopez L.A., Pastor-Pareja J.C., Resino J. Wg and Egfr signalling antagonise the development of the peripodial epithelium in Drosophila wing discs. Development. 2003;130:6497–6506. doi: 10.1242/dev.00884. [DOI] [PubMed] [Google Scholar]
- Baker N.E. Transcription of the segment-polarity gene wingless in the imaginal discs of Drosophila, and the phenotype of a pupal-lethal wg mutation. Development. 1988;102:489–497. doi: 10.1242/dev.102.3.489. [DOI] [PubMed] [Google Scholar]
- Bronikowski A.M. Experimental evidence for the adaptive evolution of growth rate in the garter snake Thamnophis elegans. Evolution. 2000;54:1760–1767. doi: 10.1111/j.0014-3820.2000.tb00719.x. [DOI] [PubMed] [Google Scholar]
- Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Casares F., Mann R.S. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Casares F., Mann R.S. A dual role for homothorax in inhibiting wing blade development and specifying proximal wing identities in Drosophila. Development. 2000;127:1499–1508. doi: 10.1242/dev.127.7.1499. [DOI] [PubMed] [Google Scholar]
- Castelli-Gair J., Akam M. How the Hox gene Ultrabithorax specifies two different segments: the significance of spatial and temporal regulation within metameres. Development. 1995;121:2973–2982. doi: 10.1242/dev.121.9.2973. [DOI] [PubMed] [Google Scholar]
- Crickmore M.A., Mann R.S. Hox control of organ size by regulation of morphogen production and mobility. Science. 2006;313:63–68. doi: 10.1126/science.1128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore M.A., Mann R.S. The control of size in animals: insights from selector genes. Bioessays. 2008;30:843–853. doi: 10.1002/bies.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard J. Phylogeny of the semiaquatic bugs (Hemiptera—Heteroptera, Gerromorpha) Insect Syst. Evol. 2008;39:30. [Google Scholar]
- Damgaard J., Andersen N.M., Meier R. Combining molecular and morphological analyses of water strider phylogeny (Hemiptera—Heteroptera, Gerromorpha): effects of alignment and taxon sampling. Syst. Entomol. 2005;30:289–309. [Google Scholar]
- Davidson E.H., Erwin D.H. Evolutionary innovation and stability in animal gene networks. J. Exp. Zool. B: Mol. Dev. Evol. 2010;314:182–186. doi: 10.1002/jez.b.21329. [DOI] [PubMed] [Google Scholar]
- Duboule D., Tarchini B., Zakany J., Kmita M. Tinkering with constraints in the evolution of the vertebrate limb anterior-posterior polarity. Novartis. Found. Symp. 2007;284:130–137. doi: 10.1002/9780470319390.ch9. (discussion 138–141, 158–163) [DOI] [PubMed] [Google Scholar]
- Emlen D.J., Warren I.A., Johns A., Dworkin I., Lavine L.C. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337:860–864. doi: 10.1126/science.1224286. [DOI] [PubMed] [Google Scholar]
- Estella C., Mann R.S. Logic of Wg and Dpp induction of distal and medial fates in the Drosophila leg. Development. 2008;135:627–636. doi: 10.1242/dev.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C., McKay D.J., Mann R.S. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into distalless during Drosophila leg development. Dev. Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P.G., Kornfeld K., Hogness D.S., Mann R.S. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galant R., Carroll S.B. Evolution of a transcriptional repression domain in an insect Hox protein. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- Galant R., Walsh C.M., Carroll S.B. Hox repression of a target gene: extradenticle-independent, additive action through multiple monomer binding sites. Development. 2002;129:3115–3126. doi: 10.1242/dev.129.13.3115. [DOI] [PubMed] [Google Scholar]
- Gibson G. Epistasis and pleiotropy as natural properties of transcriptional regulation. Theor. Popul. Biol. 1996;49:58–89. doi: 10.1006/tpbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- Grossmann D., Prpic N.M. Egfr signaling regulates distal as well as medial fate in the embryonic leg of Tribolium castaneum. Dev. Biol. 2012;370:264–272. doi: 10.1016/j.ydbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Hersh B.M., Carroll S.B. Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development. 2005;132:1567–1577. doi: 10.1242/dev.01737. [DOI] [PubMed] [Google Scholar]
- Hu D., Bush J.W. The hydrodynamics of water-walking arthropods. J. Fluid Mech. 2010;644:5–33. [Google Scholar]
- Hu D.L., Chan B., Bush J.W. The hydrodynamics of water strider locomotion. Nature. 2003;424:663–666. doi: 10.1038/nature01793. [DOI] [PubMed] [Google Scholar]
- Hudry B., Viala S., Graba Y., Merabet S. Visualization of protein interactions in living Drosophila embryos by the bimolecular fluorescence complementation assay. BMC Biol. 2011;9:5. doi: 10.1186/1741-7007-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan L.Y., Jan Y.N. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc. Natl. Acad. Sci. USA. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert-Possamai S., Le Trionnaire G., Bonhomme J., Christophides G.K., Rispe C., Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Passner J.M., Rohs R., Jain R., Sosinsky A., Crickmore M.A., Jacob V., Aggarwal A.K., Honig B., Mann R.S. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh R., Weinzierl R.O., White R.A., Akam M. Homeotic gene expression in the locust Schistocerca: an antibody that detects conserved epitopes in Ultrabithorax and abdominal-A proteins. Dev. Genet. 1994;15:19–31. doi: 10.1002/dvg.1020150104. [DOI] [PubMed] [Google Scholar]
- Khila A., Abouheif E., Rowe L. Evolution of a novel appendage ground plan in water striders is driven by changes in the Hox gene Ultrabithorax. PLoS Genet. 2009;5:e1000583. doi: 10.1371/journal.pgen.1000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khila A., Abouheif E., Rowe L. Function, developmental genetics, and fitness consequences of a sexually antagonistic trait. Science. 2012;336:585–589. doi: 10.1126/science.1217258. [DOI] [PubMed] [Google Scholar]
- Khila A., Abouheif E., Rowe L. Comparative functional analyses of ultrabithorax reveal multiple steps and paths to diversification of legs in the adaptive radiation of semi-aquatic insects. Evolution. 2014 doi: 10.1111/evo.12444. 〈 10.1111/evo.12444〉 (published online) [DOI] [PubMed] [Google Scholar]
- Kitzmann P., Schwirz J., Schmitt-Engel C., Bucher G. RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genomics. 2013;14:5. doi: 10.1186/1471-2164-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin D.W., Werren J.H. Evolution of shape by multiple regulatory changes to a growth gene. Science. 2012;335:943–947. doi: 10.1126/science.1215193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfooz N., Turchyn N., Mihajlovic M., Hrycaj S., Popadic A. Ubx regulates differential enlargement and diversification of insect hind legs. PloS One. 2007;2:e866. doi: 10.1371/journal.pone.0000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R.S., Lelli K.M., Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek A.P., Rose D.J. Differential recruitment of limb patterning genes during development and diversification of beetle horns. Proc. Natl. Acad. Sci. USA. 2009;106:8992–8997. doi: 10.1073/pnas.0809668106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nijhout H.F., Grunert L.W. The cellular and physiological mechanism of wing-body scaling in Manduca sexta. Science. 2010;330:1693–1695. doi: 10.1126/science.1197292. [DOI] [PubMed] [Google Scholar]
- O’Farrell P.H. How metazoans reach their full size: the natural history of bigness. In: Michael Hall M.R., Thomas George, editors. Cell Growth: Control of Cell Size. Cold Spring Harbor Press; NY: 2003. [Google Scholar]
- Panganiban G., Nagy L., Carroll S.B. The role of the distal-less gene in the development and evolution of insect limbs. Curr. Biol. 1994;4:671–675. doi: 10.1016/s0960-9822(00)00151-2. [DOI] [PubMed] [Google Scholar]
- Papadopoulos D.K., Skouloudaki K., Adachi Y., Samakovlis C., Gehring W.J. Dimer formation via the homeodomain is required for function and specificity of sex combs reduced in Drosophila. Dev. Biol. 2012;367:78–89. doi: 10.1016/j.ydbio.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Paschinger K., Rendic D., Wilson I.B. Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj. J. 2009;26:385–395. doi: 10.1007/s10719-008-9155-3. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A., Akam M. Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:2855–2860. doi: 10.1073/pnas.1015077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos A., Kontarakis Z., Liubicich D.M., Serano J.M., Akam M., Patel N.H., Averof M. Probing the evolution of appendage specialization by Hox gene misexpression in an emerging model crustacean. Proc. Natl. Acad. Sci. USA. 2009;106:13897–13902. doi: 10.1073/pnas.0902804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpic N.M., Janssen R., Wigand B., Klingler M., Damen W.G. Gene expression in spider appendages reveals reversal of exd/hth spatial specificity, altered leg gap gene dynamics, and suggests divergent distal morphogen signaling. Dev. Biol. 2003;264:119–140. doi: 10.1016/j.ydbio.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Prpic N.M., Wigand B., Damen W.G., Klingler M. Expression of dachshund in wild-type and distal-less mutant Tribolium corroborates serial homologies in insect appendages. Dev. Genes Evol. 2001;211:467–477. doi: 10.1007/s004270100178. [DOI] [PubMed] [Google Scholar]
- Roch F., Akam M. Ultrabithorax and the control of cell morphology in Drosophila halteres. Development. 2000;127:97–107. doi: 10.1242/dev.127.1.97. [DOI] [PubMed] [Google Scholar]
- Rogers B.T., Peterson M.D., Kaufman T.C. Evolution of the insect body plan as revealed by the sex combs reduced expression pattern. Development. 1997;124:149–157. doi: 10.1242/dev.124.1.149. [DOI] [PubMed] [Google Scholar]
- Sandmann T., Jensen L.J., Jakobsen J.S., Karzynski M.M., Eichenlaub M.P., Bork P., Furlong E.E. A temporal map of transcription factor activity: mef2 directly regulates target genes at all stages of muscle development. Dev. Cell. 2006;10:797–807. doi: 10.1016/j.devcel.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jackle H. Concentration-dependent transcriptional activation or repression by Kruppel from a single binding site. Nature. 1991;353:563–566. doi: 10.1038/353563a0. [DOI] [PubMed] [Google Scholar]
- Sauer F., Jackle H. Dimerization and the control of transcription by Kruppel. Nature. 1993;364:454–457. doi: 10.1038/364454a0. [DOI] [PubMed] [Google Scholar]
- Schulz C., Tautz D. Autonomous concentration-dependent activation and repression of Kruppel by hunchback in the Drosophila embryo. Development. 1994;120:3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- Stern D.L. A role of Ultrabithorax in morphological differences between Drosophila species. Nature. 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.L. The Hox gene Ultrabithorax modulates the shape and size of the third leg of Drosophila by influencing diverse mechanisms. Dev. Biol. 2003;256:355–366. doi: 10.1016/s0012-1606(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Stern D.L., Emlen D.J. The developmental basis for allometry in insects. Development. 1999;126:1091–1101. doi: 10.1242/dev.126.6.1091. [DOI] [PubMed] [Google Scholar]
- Stillwell R.C., Dworkin I., Shingleton A.W., Frankino W.A. Experimental manipulation of body size to estimate morphological scaling relationships in Drosophila. J. Vis. Exp. 2011;56:1–4. doi: 10.3791/3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. Genes controlling segmental specification in the Drosophila thorax. Proc. Natl. Acad. Sci. USA. 1982;79:7380–7384. doi: 10.1073/pnas.79.23.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen H., Haerry T.E., O’Connor M.B., Marsh J.L. Developmental territories created by mutual antagonism between wingless and decapentaplegic. Development. 1996;122:3939–3948. doi: 10.1242/dev.122.12.3939. [DOI] [PubMed] [Google Scholar]
- Tseng M., Rowe L. Sexual dimorphism and allometry in the giant water strider, Gigantometra gigas. Can. J. Zool. 1999;77:923–929. [Google Scholar]
- White R.A.H., Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984;39:163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]
- Wilkins A.S. Sinauer Associates, Inc.; Sunderland: 2002. The Evolution of Developmental Pathways. [Google Scholar]
- Zecca M., Struhl G. Control of growth and patterning of the Drosophila wing imaginal disc by EGFR-mediated signaling. Development. 2002;129:1369–1376. doi: 10.1242/dev.129.6.1369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data