Abstract
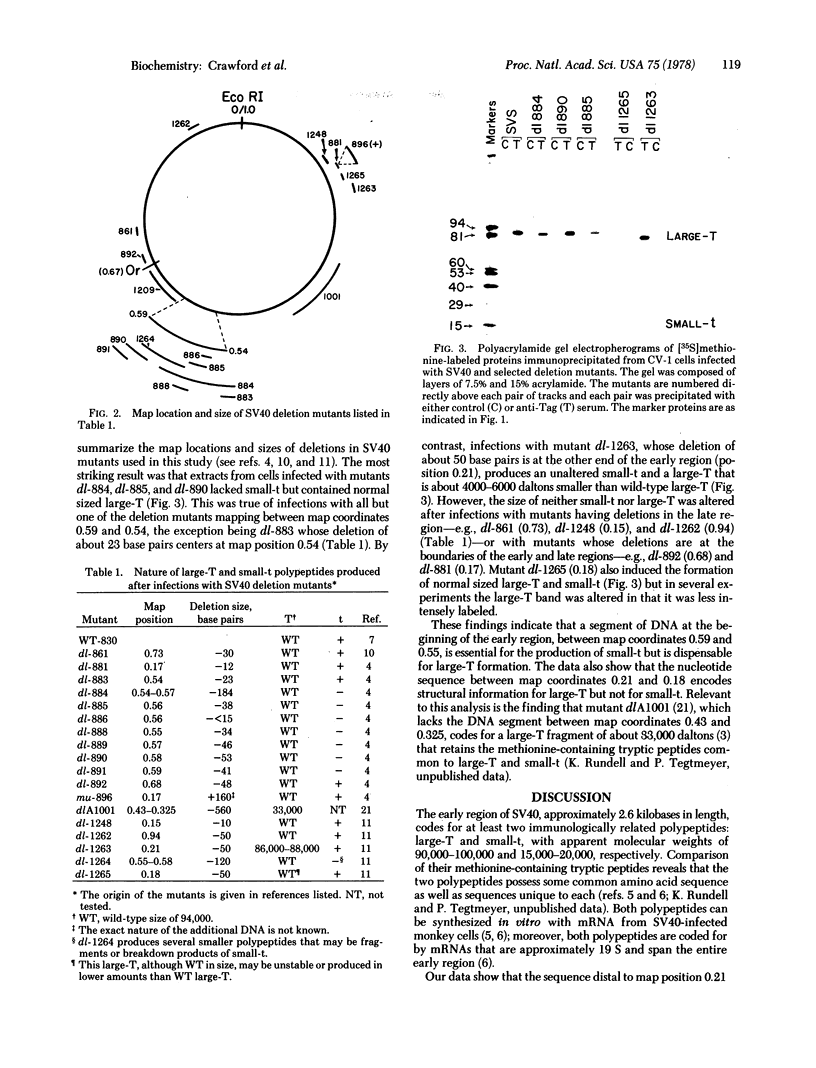
The early region of simian virus 40 codes for at least two immunologically related polypeptides: large-T and small-t, with apparent molecular weights of 90,000-100,000 and 15,000-20,000, respectively. Because small-t shares methionine-containing tryptic peptides with large-T, the two polypeptides are probably coded, in part, by a common nucleotide sequence. To locate the coding sequences for large-T and small-t in the DNA, the production of these proteins was examined after infection of CV-1 cells with wild-type and deletion mutants of simian virus 40. We found that a deletion at the distal portion of the early region alters the structure of large-T but not of small-t; but deletions within the region between map coordinates 0.59 and 0.55 result in an alteration or absence of small-t and a normal large-T. These findings have been rationalized by a model that proposes the existence of two early mRNAs, one coding for large-T and the other for small-t. Both mRNAs span virtually the entire early region; but the mRNA coding for large-T lacks the nucleotide sequence between map coordinates 0.59 and 0.54. We suggest that small-t is translated from the larger of the two mRNAs, beginning at or near its 5′ end and terminating at a termination codon at about map coordinate 0.54. Larger-T, on the other hand, is translated from the shorter mRNA, beginning at the same initiator codon, and, because of the deletion of the terminator codon at 0.54, translation proceeds to the terminator codon at or near map position 0.18.
Keywords: early deletion mutants, large-T and small-t polypeptides, spliced large-T mRNA
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Dhar R., Laub O., Horowitz M., Khoury G. Novel mechanism for RNA maturation: the leader sequences of simian virus 40 mRNA are not transcribed adjacent to the coding sequences. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3686–3690. doi: 10.1073/pnas.74.9.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget S. M., Moore C., Sharp P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Shenk T. E., Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Martin R. G. Complementation analysis of simian virus 40 mutants. J Virol. 1974 May;13(5):1101–1109. doi: 10.1128/jvi.13.5.1101-1109.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Landers T., Goff S. P., Manteuil-Brutlag S., Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J Virol. 1977 Oct;24(1):277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. J., Black P. H. Analysis of surface antigens on simian virus 40-transformed cells. II. Exposure of simian virus 40-induced antigens on transformed rabbit kidney and inbred hamster kidney cells by phospholipase C. J Natl Cancer Inst. 1973 Jul;51(1):115–134. doi: 10.1093/jnci/51.1.115. [DOI] [PubMed] [Google Scholar]
- Crawford L. V., Lane D. P. An immune complex assay for SV40 T antigen. Biochem Biophys Res Commun. 1977 Jan 10;74(1):323–329. doi: 10.1016/0006-291x(77)91411-5. [DOI] [PubMed] [Google Scholar]
- Del Villano B. C., Defendi V. Characterization of the SV40 T antigen. Virology. 1973 Jan;51(1):34–46. doi: 10.1016/0042-6822(73)90363-2. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Griffin B. E. Organization of the genomes of polyoma virus and SV40. Adv Cancer Res. 1977;24:67–113. doi: 10.1016/s0065-230x(08)61013-1. [DOI] [PubMed] [Google Scholar]
- Girardi A. J. Prevention of SV40 virus oncogenesis in hamsters. I. Tumor resistance induced by human cells transformed by SV40. Proc Natl Acad Sci U S A. 1965 Aug;54(2):445–451. doi: 10.1073/pnas.54.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Nathans D. The genome of simian virus 40. Adv Virus Res. 1977;21:85–173. doi: 10.1016/s0065-3527(08)60762-9. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. Deletion mutants of simian virus 40 generated by enzymatic excision of DNA segments from the viral genome. J Mol Biol. 1974 Oct 15;89(1):179–193. doi: 10.1016/0022-2836(74)90169-7. [DOI] [PubMed] [Google Scholar]
- Prives C., Gilboa E., Revel M., Winocour E. Cell-free translation of simian virus 40 early messenger RNA coding for viral T-antigen. Proc Natl Acad Sci U S A. 1977 Feb;74(2):457–461. doi: 10.1073/pnas.74.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild H., Lockwood M. P. Isolation and characterization of a heat-inducible simian virus 40 mutant. J Virol. 1976 Aug;19(2):374–381. doi: 10.1128/jvi.19.2.374-381.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K., Collins J. K., Tegtmeyer P., Ozer H. L., Lai C. J., Nathans D. Identification of simian virus 40 protein A. J Virol. 1977 Feb;21(2):636–646. doi: 10.1128/jvi.21.2.636-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWEET B. H., HILLEMAN M. R. The vacuolating virus, S.V. 40. Proc Soc Exp Biol Med. 1960 Nov;105:420–427. doi: 10.3181/00379727-105-26128. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Carbon J., Berg P. Construction and analysis of viable deletion mutants of simian virus 40. J Virol. 1976 May;18(2):664–671. doi: 10.1128/jvi.18.2.664-671.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staneloni R. J., Fluck M. M., Benjamin T. L. Host range selection of transformation-defective hr-t mutants of polyoma virus. Virology. 1977 Apr;77(2):598–609. doi: 10.1016/0042-6822(77)90485-8. [DOI] [PubMed] [Google Scholar]
- Takemoto K. K., Kirschstein R. L., Habel K. Mutants of simian virus 40 differing in plaque size, oncogenicity, and heat sensitivity. J Bacteriol. 1966 Oct;92(4):990–994. doi: 10.1128/jb.92.4.990-994.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegtmeyer P., Ozer H. L. Temperature-sensitive mutants of simian virus 40: infection of permissive cells. J Virol. 1971 Oct;8(4):516–524. doi: 10.1128/jvi.8.4.516-524.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya B., Weissman S. M. The early region of SV40 DNA may have more than one gene. Cell. 1977 Aug;11(4):837–843. doi: 10.1016/0092-8674(77)90295-1. [DOI] [PubMed] [Google Scholar]




