Abstract
Both biological and social sciences have identified contributing factors to human health. However, health outcomes are unlikely to equal a simple sum of these identified factors. This article makes an attempt to put together the information, methods, and technologies that relate to health outcomes from biological, behavioral, and social disciplines. Much of this information was obtained by controlling for the variations of the factors in “other” disciplines. For example, genetic factors were controlled for in identifying the behavioral determinants of health. Looking forward, better understandings of health outcomes may require exploiting the interactions of health determinants that were identified from different disciplines. We propose the concept of “systems health” studies, which take health outcomes as the outputs of a system, where the inputs and their interactions from multiple disciplines are considered.
INTRODUCTION
Human health has been studied by inadvertently isolated disciplines, including biology, behavioral and social sciences. This reductionist approach effectively narrowed down the parameters to consider, facilitated technology developments, and led to the identification of important contributing factors in each discipline to human health. These achievements prompted this abridged overview of what is known and unknown about health. From prevention and intervention perspectives, it would be good to know what parameters in biological, behavioral, and social domains can be adjusted for promoting health outcomes. Towards this goal, a systems approach that considers multiple domains of information and their interactions is likely to be instrumental.
BIOLOGICAL DETERMINANTS OF HEALTH
Genomic factors associated with physiological traits and health outcomes
Early evidence of genetic contribution of health outcomes came from familial diseases, including cystic fibrosis [1–3] and others. These diseases are associated with and often caused by polymorphisms in the genome, sometimes a single nucleotide polymorphism (SNP). Not only the missense SNPs that changed protein coding sequences, but also the SNPs that changed cis-regulatory sequences such as enhancers can alter health outcomes [4]. These Mendelian genomic loci represent the best known genomic determinants of health outcomes [5,6]. Nevertheless, a larger variety of physiological traits are associated with combinations of alleles at multiple genomic loci, and thus considered complex traits [7]. Genome-wide association studies (GWAS) were carried out to identify risk loci of the genome to a variety of common diseases, including cardiovascular, mental, and autoimmune diseases [8–11]. Common physiological traits have also been mapped, including height, body mass index (BMI), HDL cholesterol, and others [11]. A recently added dimension of research is mapping the variability in drug responses to genomic loci and alleles [12]. SNP microarray has been the primary technical platform used in GWAS studies [13,14], partially due to its cost advantage to genome sequencing.
Personal genome sequencing allowed for identification of rare and common genomic variants associated with a disease. Sequencing based analyses were carried out around two major experimental designs. The first is the comparison of genomes within an individual. Two early studies on a skin cancer patient [15] and a lung cancer patient [16] sequenced and compared the “normal” genomes of their lymphoblast cells and the “abnormal” genomes of metastasized tumors. Comparing the normal and abnormal genomes of the same patient allowed for identification of cancer associated somatic mutations. Single-cell sequencing technology enabled sequencing the genomes or partial genomes of dozens and even hundreds of single cells within a tumor or a normal tissue [17,18]. Using the frequencies of a mutation or a copy number variation (CNV), multiple teams traced the earlier somatic genomic changes in the cancer cells [19–21]. These early changes were reasoned to be “driver” changes that have causal relationships with tumorigenesis [22]. Surprisingly, normal neurons in the brain possess mosaic CNVs within the same person, posing a challenge to redefine the concept of a “normal” genome [18].
The second major experimental design is to sequence individual genomes from a patient cohort, and then summarize the shared genomic variations among these patients. Examples include but not restricted to sequencing Autism cohorts [23–25] and cancer cohorts [26][27][28][29]. A major lesson learned from these studies is that there are much fewer shared mutations [23] or CNVs [24] among patients of similar diagnoses than previously expected. Considering the hallmarks of cancer are perturbations of molecular pathways, a bioinformatics approach was developed to aggregate mutations on genes of the same pathway and then cluster patients based on these aggregated impacts of mutations and molecular pathways [30]. This approach stratified prostate cancer patients into subgroups using genomic sequences alone, nevertheless these subgroups exhibited different survival time.
Beyond genomic sequences
Omics, the study of a total collection of a molecular species, revealed substantial amounts of health-related information beyond the genomes. The transcriptome, epigenome, proteome, and metabolome were four major data strata that exhibited strong associations with diseases. Before the accomplishment of the human genome project, genome-wide gene expression data were used to classify two types of leukemia (AML, ALL) [31]. Most strikingly the physicians changed their diagnosis on one child in this study, after seeing the transcriptome based results. With this proof of principle, gene expression differences have been extensively utilized to narrow down disease candidate genes.
The epigenome, the chemical modifications on histones and DNA, appears to strongly correlate with cellular behaviors and thus with health outcomes. The genome-wide distributions of as few as 1–3 histone modifications were effective in predicting prognosis of multiple cancer types [32]. Almost every cancer being analyzed exhibited hyper- and hypo- modified regions in the chromosome of the tumor cells [32–34]. Perhaps most strikingly, DNA methylation levels at a defined set of genomic locations are strongly predictive of human aging [35,36]. Mechanistically, the epigenome interacts with the genome to modulate the personal specific transcription factor binding, and thus directly contributes to personal variation of gene expression [37]. Moreover, the temporal changes of the epigenomic modifications (e.g. 5-hmC) during a biological process are predictive of gene expression changes [38]. Because the epigenome is jointly determined by the genomic sequence and the environmental signals, and the epigenome is likely to be less sensitive than the transcriptome to transient environmental changes, the epigenome may serve as a preferred molecular layer for quantifying personal responses to interventions.
Protein levels often correlate well with physiological outcomes, and sometimes with the subtypes of a disease. Breast cancer is a case in point. The protein levels of estrogen receptor, progesterone receptor, and Her2 are currently used to stratify patients for prognosis and treatment purposes. Thus, high-throughput protein identification technologies, especially mass spectrometry (mass-spec) hold the potentials for becoming diagnostic tools [39]. Serum proteomic profiling is a heavily pursued approach for identifying disease biomarkers, catalyzed by increasing sensitivity and specificity of mass spec.
Microbiome, our very close neighbors
“Within the body of a healthy adult, microbial cells are estimated to outnumber human cells ten to one” [40]. The diversity and the social structure of the microbial community in humans could hold the key to unexplained parts of health outcomes. The metagenomics methods that utilize deep genomic sequencing to identify the microbial species as well as their relative population sizes have started to reveal the personal variations of oral, nasal, skin, gastrointestinal, and urogenital microbiomes. Emerging data have suggested associations between the compositions of gut microbiome with obesity [41], inflammatory bowel diseases (IBDs) [42], colon cancer [43], sepsis [44], and other diseases [45]. Metagenomic analyses may offer a new approach to test outstanding hypotheses of disease etiology, including but not restricted to the hygiene hypothesis of asthma [46] and the intestine-toxin hypothesis of fatty liver diseases [47].
Environmental and behavioral impacts to biological determinants
Behaviors can impact health outcomes through modulating every layer of hitherto mentioned biological factors, including the genome, epigenome, transriptome, protein interactions and signal transduction, and gut microbiome (Figure 1). Sun tanning and smoking can stimulate somatic genomic mutations which are found in skin and lung cancers [15,16]. Smoking also induces DNA methylation at specific genomic loci [48]. Physical activities correlate with breast cancer survival at least partially by modulating the epigenome [49]. Early experience of children correlate with gene expression in the brain [50]. Dietary restriction may affect insulin receptor signaling and is reproducibly correlated with longevity [51]. Alcohol overconsumption results in the secretion of pro-inflammatory cytokines and alters gut microbiome, which may be causally linked to liver diseases [47]. We proceed to summarize the framework and methods for studying the behavioral and social determinants of health.
Figure 1.
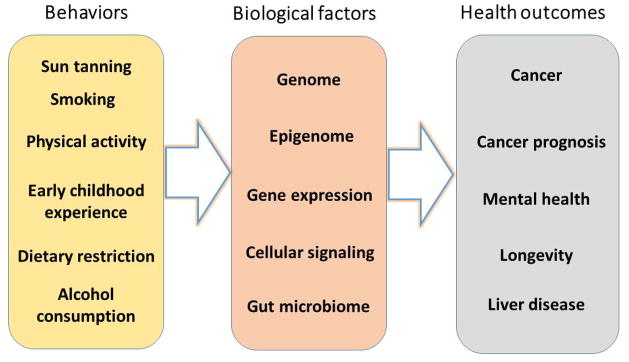
Examples of behavior-related health outcomes that were mediated by biological factors.
BEHAVIORAL AND SOCIAL DETERMINANTS OF HEALTH
Conceptual frameworks
Humans are social beings. A typical social science approach to identifying health determinants is to analyze the correlation between the variations of social environment and that of the health outcomes. It is suggested that the “conditions in which people are born, grow, live, work and age” correlated to health outcomes, and “these circumstances are shaped by the distribution of money, power, and resources at global, national and local levels” [52]. Dahlgren and Whitehead in 1991 proposed a conceptual framework for summarizing social determinants of health [53], including four highly interconnected categories: 1) age, gender, and constitutional factors, 2) individual lifestyles, 3) social interactions and communities, and 4) socioeconomic, cultural and environmental conditions.
Social and economic development has produced physical products that can modulate behaviors, and in turn affect health outcomes. The man-made context for human activities has been formalized into the notion of built environment [54], which may include local facilities, infrastructures, and food environment. By incorporating the built environment, Barton, Grant and Guise categorized the determinants of health into seven groups [55].
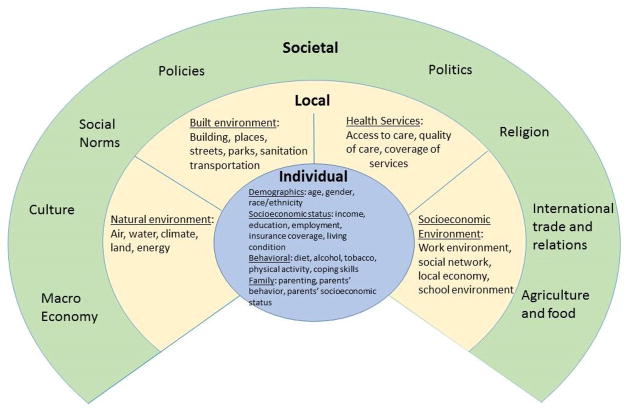
Adding together the proposed factors by Dahlgren and Whitehead [53] and Barton et al. [55] would provide a relatively comprehensive list of behavioral and social factors that should be considered in health studies. However, from a prevention or intervention perspective, we recognize that not all factors are equal in their potentials to be changed for promoting better health. Some factors can be changed by personal determinations, whereas some may require policy and economic reforms. For this reason we propose a three-tier system to summarize and categorize behavioral and social factors that may contribute to health outcomes (Figure 2). The first tier includes the factors directly associated with the individuals. The second tier includes the local environment where individuals live and work. The third tier includes macro environmental factors. The factors within each tier can interact with each other and may independently or collectively interact with the factors of other tiers.
Figure 2.
Three layers of behavioral and social determinants of health. Adapted from Dahlgren & Whitehead 1991 and Barton et al. 2005.
The ongoing research of health-related behavioral and social studies can fit nicely into the framework as described above. An example is the studies of obesity, the prevalence of which has increased considerably during the past several decades [56]. Some studies assessed individual and local characteristics associated with obesity [57–60], and suggested that individual socioeconomic and lifestyle profile, local food access, and physical activity environment have significant impacts. Some other studies looked at macro level determinants (third tier), and reported that, price and taxation policies on food and other relevant products accounted for the variation in obesity in addition to individual level factors, and the level of urbanization mattered as well [61–64].
Analysis methods
In contrast to the flexibility of generating experimental data from model organisms, ethical and practical constraints limit the capacity of experimentations that involve human subjects. The ethical and practical constraints often require a correlation of the outcome and the factors of interest as preliminary evidence for trials. Moreover, such correlations would be likely to increase the cost effectiveness of the designed trials. Observational data, in which human subject are observed in natural circumstances that cannot be controlled, is therefore often used to detect correlations of health and explanatory factors, understand interactions of factors, and provide support for hypothesis testing. Examples of observational data include surveys that sample individual units from a population and ask a number of questions to the respondents at one time point (cross-sectional survey) or repeated time points (longitudinal survey); behavioral data extracted by electronic technologies such as pedometers, smartphones, or internet; government administrative or surveillance data; and data collected by public or private entities such as medical claims. Data from different sources can be merged to enable a more thorough look of the determinants at multiple levels of a given health outcome. Statistical power and representativeness of the data are essential, and they can be achieved by large sample size and careful sampling of the study units.
A well-known problem of observational data is the presence of confounding factors, which simultaneously contribute to the health outcome with the factors of interest [65]. To isolate the effects of the factors of interest, statistical and epidemiological approaches are often employed on the observational data to make the confounding factors “under control”. For example, multivariate regression analysis is often used to determine the function that describes how a vector of factors responds to the changes in others and how they collectively affect the health outcome of interest. Factor analysis and principle components analysis allow researchers to create a new set of synthetic variables and investigates the contribution of each set to the outcome [66]. Cluster analysis groups population into clusters so that individuals within cluster share more similar characteristics than individuals in other clusters. Multilevel models are appropriate when observational data are organized at multiple levels, such as individuals nested in local communities, which are further nested in states and countries [67]. Case control studies are used to compare two groups that differ in health outcomes but are otherwise similar, and cohort studies track a cohort of population over time to observe the development of health outcomes [65].
The inferences derived from observational data are often “correlational”, because confounding factors are not able to be observed in the observational data but are correlated with the health outcomes being studied [68]. Biological factors are typical “unobserved heterogeneities” acknowledged in behavioral and social sciences. Other examples are individual preferences, intellectual capabilities, and coping skills. To correct for omitted variable bias and establish “causal” inferences, experimental studies such as randomized trials are still the “gold standard” when applicable. Randomized controlled trials balance the observed and unobserved factors by randomly allocating human subjects to receive one or other of the alternative interventions. Any change between intervention and control group, therefore, is attributable to the intervention alone. The 1971–1982 RAND’s Health Insurance Experiment assigned thousands of people to different health insurance plans. It concluded that the cost sharing reduced medical expenditures, and in general, the reduction in service had no adverse consequences on participants’ health [69–71]. The result of this experiment has encouraged the restructuring of private insurance and the promotion in managed care.
When randomized controlled trials are not considered, quasi-experimental design is often perceived as an alternative [72]. The study design is similar to randomized controlled trials, but lacks the component of random assignment. As a result, the characteristics of groups are not equivalent at baseline and any differences observed after the intervention may not be solely due to the intervention received [72]. A few statistical and econometrical strategies have been developed to address this concern of internal validity. Developed by Paul Rosenbaum and Donald Rubin [73], propensity score matching attempts to statistically mimic the random assignment of the intervention by creating a control sample that has comparable observed characteristics to the intervention sample. Instrumental variable is an observed variable, usually policy changes, that does not affect health outcome itself, but is correlated with the endogenous explanatory variables conditional on other covariates. It allows consistent estimates of the regression relationship when the explanatory variables are correlated with the error terms [74]. In regression discontinuity design [75], a threshold is selected above or below which an intervention is assigned. The causal correlation between the intervention and the health outcome is obtained by comparing people lying closely on two sides of the threshold. Panel analysis utilizes longitudinal observational data which collects repeated measures over time and over the same individuals [68]. The unobserved factors, varying non-stochastically or stochastically over time, can be modeled or differenced out. The health behaviors and health outcomes of siblings or twins are compared assuming that genetic factors are shared and any variation observed would be attributable to environmental contexts.
Key modifiable behavioral and social determinants
Special interests were given to the health determinants that are modifiable by policies or interventions. Six key modifiable determinants have been proposed. These include modifiable health risk behaviors, namely physical inactivity, poor nutrition, tobacco use, and excessive alcohol use, account for much of the burden in morbidity and mortality (CDC). Education is another modifiable health determinant. Quasi-experimental studies have demonstrated causal influences of educational policies on health outcomes and that the improvement of education attainment can lead to improvement in health. Economic stability is causally correlated with health in two ways [76]: through a direct impact on material conditions and through an impact on social participation. Social environment such as social capital, family structure, discrimination and civic participation affects health outcome by modifying the inter-personal relationship, beliefs and perceptions. Health services, including access to care, insurance coverage, and quality of care, are services directly dealing with the diagnosis and treatment of disease, and the promotion, maintenance and restoration of health (WHO). Built environment modifies the physical environment that we live and work. It is associated with health behaviors such as physical activity and alcohol use, and health outcomes such as obesity and depression [77].
FUTURE DIRECTIONS: A SYSTEMS HEALTH APPROACH
A holistic view of “systems health”
An alternative to the reductionist approach of studying biological and social factors of health in separation is a holistic approach where “all things considered”. We take the liberty to propose a name “systems health” for this emerging interdisciplinary research area. Systems health concerns about any biological, behavioral, social factors as well as their interactions that affect health outcomes. Its major goal is to derive mechanistic and predictive models of health outcomes, and major applications are in prevention, intervention, and prognosis.
The central question of systems health is how gene-environment interactions relate to health. This is a Nature vs. Nurture question [78] with special emphases (Figure 3). First, human health is the outcome of interest. Second, the modifiable social and behavior factors receive special attentions. Third, interventions and preventative procedures can be thought of as feedbacks. One success of this type was made by Caspi and colleagues, who discovered that a polymorphism in the gene encoding for a serotonin transporter protein modulates the impact of life stress to depression [79]. This finding and others led to the idea that the heterogeneities of health outcomes are primarily determined by behavioral and social factors; however personal genomic variation modifies the extent of this correlation [80–82].
Figure 3.
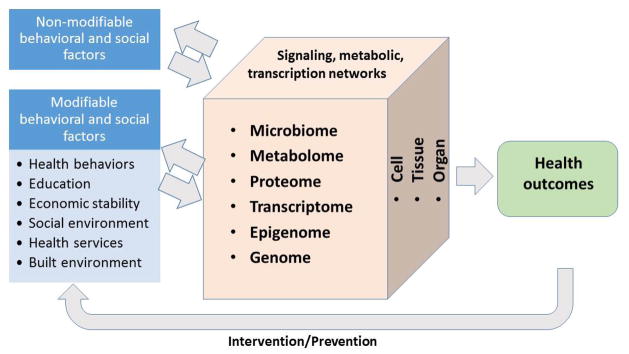
“Systems health” studies gene-environment interactions, with special emphases on the health outcomes and the modifiable input factors.
Challenges in data collection
The major challenges of systems health studies lie in collecting interdisciplinary data and devising new research methods. Ethical and monetary cost issues need to be addressed. On the data end, a few population surveys on health care topics, including the repeated cross-sectional National Health and Nutrition Examination Survey, have incorporated laboratory module to collect respondents’ physiological data. Add Health, a survey that followed approximately 20,000 US adolescents through their adulthood, is currently genotyping 12,000 subjects of its cohort [83].
Electronic equipment including pedometers, personal digital assistants (PDA), and portable personal computers have started to contribute data to behavioral science, opening the possibility of collecting personal data at real time. Electronic data of personal physical, dietary, and social networking activities are among the first to be reported [84–86]. These devices and systems may reduce monetary and time costs of traditional surveys [87], but will certainly bring new challenges in data handling and analyses.
Candidate approaches for “systems health” studies
It is likely too early to speculate the most effective methods, but two general approaches showed promises. The first is to leverage the knowledge of the other disciplines. An example of the success is the process of revealing the mechanistic link between smoking and lung cancer. The prevalence of lung cancer mortality increased dramatically from 5.3% of cancer deaths in men and 2.0% in women in the 1930s to 33.1% in men and 22.8% in women in the 1990s [88], posting a great puzzle to health researchers. Epidemiological studies reported that cigarette consumption, increased from 54 cigarettes per capita per year to 4166 in 1960s, has significantly contributed to the lung cancer epidemic. These evidences led to the first U.S. Surgeon General’s Report in 1964 that “cigarette smoking is causally related to lung cancer in men” [89]. Guided by this knowledge, the genomic mutations resulted from tobacco exposure were analyzed and documented [16], revealing the chain of events from smoking to genomic mutations and then to cancer development.
Biological knowledge may lend a hand to behavioral and social research of health outcomes as well. It has been four decades since the 1973 report [90] on the effects of shift work on worker health [90–94]. The next steps may take advantage of the biological findings on gene expression [95] and metabolic outcomes [96] of the perturbation of circadian rhythms and their mechanistic links to depression and obesity. Furthermore, the data on the role of Melatonin on the regulation of human circadian rhythms and sleep [97] are perhaps worth considering in future contemplation of intervention strategies.
Another approach is stratification, where the information of one discipline is used to stratify the data analysis of another discipline. For example, stratifying the recruits by the polymorphisms of their neural transporter genes may increase the chances of detecting associations in a case control study of stress and depression where the sample size is limited by monetary cost. A drawback of stratification is the potential difficulty of recruiting sufficient respondents in a stratum. This difficulty may have to be addressed by an integrated analysis.
Integrated data analysis may deploy machine learning methods that take both genomic and survey data as inputs to learn information about the variation of health outcomes. These methods are relatively mature as long as the desired data are collected for all subjects. Incomplete data will likely to be a major challenge, in which case some individuals would only have either social or genomic data. The computational methods that aim for unbiased estimates under the missing data scenario can be useful [98].
HIGHLIGHTS.
“Systems health” is an emerging interdisciplinary field.
Systems health integrates biological, behavioral, social factors and their interactions.
Special emphases are given to health outcomes and modifiable social and behavior factors.
Intervention and preventative procedures can be regarded as feedbacks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N, et al. Identification of the cystic fibrosis gene: chromosome walking and jumping. Science. 1989;245:1059–1065. doi: 10.1126/science.2772657. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 5.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 9.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev Genet. 2009;10:318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly AK. Genome-wide association studies in pharmacogenomics. Nat Rev Genet. 2010;11:241–246. doi: 10.1038/nrg2751. [DOI] [PubMed] [Google Scholar]
- 13.Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, Johnson T, Castillo BA, Barnard J, Baumert J, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O’Brien ET, Poulter NR, et al. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688–700. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasance ED, Stephens PJ, O’Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. Discovered DNA copy number variation in different cells of the same postmortem human brain with a single-cell genome sequencing technology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman S, Howarth KD, Greenman CD, Bignell GR, Tavare S, Edwards PA. The relative timing of mutations in a breast cancer genome. PLoS One. 2013;8:e64991. doi: 10.1371/journal.pone.0064991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Podlaha O, Riester M, De S, Micho F. Evolution of the cancer genome. Trends Genet. 2012;28:155–163. doi: 10.1016/j.tig.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24*.Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. Identified de novo DNA copy number variations in autism spectrum disorder patients from whole-genome sequencing of monozygotic twins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nat Methods. 2013;10:1108–1115. doi: 10.1038/nmeth.2651. A computational method to cluster personal genomes of cancer patients, producing patient groups with different survival rates. This method can potentially be used for prognosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 32.Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am J Cancer Res. 2012;2:589–597. [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35**.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. A proof of principle that DNA methylation is a molecular clock of human ageing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. A proof of principle that DNA methylation is a molecular clock of human ageing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CC, Xiao S, Xie D, Cao X, Song CX, Wang T, He C, Zhong S. Understanding variation in transcription factor binding by modeling transcription factor genome-epigenome interactions. PLoS Comput Biol. 2013;9:e1003367. doi: 10.1371/journal.pcbi.1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Yu P, Xiao S, Xin X, Song CX, Huang W, McDee D, Tanaka T, Wang T, He C, Zhong S. Spatiotemporal clustering of the epigenome reveals rules of dynamic gene regulation. Genome Res. 2013;23:352–364. doi: 10.1101/gr.144949.112. Derived rules on how temporal epigenomic changes may regulate gene expression changes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics. 2004;3:367–378. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 42.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. MBio. 2013:4. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stewart CJ, Marrs EC, Nelson A, Lanyon C, Perry JD, Embleton ND, Cummings SP, Berrington JE. Development of the preterm gut microbiome in twins at risk of necrotising enterocolitis and sepsis. PLoS One. 2013;8:e73465. doi: 10.1371/journal.pone.0073465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey CD, Celedon JC. The hygiene hypothesis and asthma. Curr Opin Pulm Med. 2005;11:14–20. doi: 10.1097/01.mcp.0000145791.13714.ae. [DOI] [PubMed] [Google Scholar]
- 47.Hartmann P, Chen WC, Schnabl B. The intestinal microbiome and the leaky gut as therapeutic targets in alcoholic liver disease. Front Physiol. 2012;3:402. doi: 10.3389/fphys.2012.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng H, Irwin ML, Lu L, Risch H, Mayne S, Mu L, Deng Q, Scarampi L, Mitidieri M, Katsaros D, et al. Physical activity and breast cancer survival: an epigenetic link through reduced methylation of a tumor suppressor gene L3MBTL1. Breast Cancer Res Treat. 2012;133:127–135. doi: 10.1007/s10549-011-1716-7. [DOI] [PubMed] [Google Scholar]
- 50.Child NSCotD. Early Experiences Can Alter Gene Expression and Affect Long-Term Development. 2010. [Google Scholar]
- 51.Wang ZQ, Floyd ZE, Qin J, Liu X, Yu Y, Zhang XH, Wagner JD, Cefalu WT. Modulation of skeletal muscle insulin signaling with chronic caloric restriction in cynomolgus monkeys. Diabetes. 2009;58:1488–1498. doi: 10.2337/db08-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.WHO. What are social determinants of health? World Health Organization; [Google Scholar]
- 53.Dahlgren G, Whitehead M. Policies and strategies to promote social equity in health. Institute for Futures Studies; 1991. [Google Scholar]
- 54.Roof K, Oleru N. Public health: Seattle and King County’s push for the built environment. J Environ Health. 2008;71:24–27. [PubMed] [Google Scholar]
- 55.Barton H. A Health Map for Urban Planners: Towards a Conceptual Model for Healthy, Sustainable Settlements. Built Environment (1978-) 2005;31:339–355. [Google Scholar]
- 56.Sturm R. Increases in clinically severe obesity in the United States, 1986–2000. Arch Intern Med. 2003;163:2146–2148. doi: 10.1001/archinte.163.18.2146. [DOI] [PubMed] [Google Scholar]
- 57.Reidpath DD, Burns C, Garrard J, Mahoney M, Townsend M. An ecological study of the relationship between social and environmental determinants of obesity. Health Place. 2002;8:141–145. doi: 10.1016/s1353-8292(01)00028-4. [DOI] [PubMed] [Google Scholar]
- 58.Black JL, Macinko J. The changing distribution and determinants of obesity in the neighborhoods of New York City, 2003–2007. Am J Epidemiol. 2010;171:765–775. doi: 10.1093/aje/kwp458. [DOI] [PubMed] [Google Scholar]
- 59*.Gordon-Larsen P, Nelson MC, Page P, Popkin BM. Inequality in the built environment underlies key health disparities in physical activity and obesity. Pediatrics. 2006;117:417–424. doi: 10.1542/peds.2005-0058. By using national representative population survey and geocoded data of recreational facilities, this paper found that lower socioeconomic status and minorities had reduced access to facilities, which then led to decreased physical activity and increased overweight. [DOI] [PubMed] [Google Scholar]
- 60.Sallis JF, Glanz K. Physical activity and food environments: solutions to the obesity epidemic. Milbank Q. 2009;87:123–154. doi: 10.1111/j.1468-0009.2009.00550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cummins S, Macintyre S. Food environments and obesity--neighbourhood or nation? Int J Epidemiol. 2006;35:100–104. doi: 10.1093/ije/dyi276. [DOI] [PubMed] [Google Scholar]
- 62.Chou SY, Grossman M, Saffer H. An economic analysis of adult obesity: results from the Behavioral Risk Factor Surveillance System. J Health Econ. 2004;23:565–587. doi: 10.1016/j.jhealeco.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 63.Powell LM, Chaloupka FJ. Food prices and obesity: evidence and policy implications for taxes and subsidies. Milbank Q. 2009;87:229–257. doi: 10.1111/j.1468-0009.2009.00554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caballero B. The global epidemic of obesity: an overview. Epidemiol Rev. 2007;29:1–5. doi: 10.1093/epirev/mxm012. [DOI] [PubMed] [Google Scholar]
- 65.Aschengrau A, Seage GR. Essentials of Epidemiology in Public Health. Jones & Bartlett Publishers; 2013. [Google Scholar]
- 66.Bartholomew DJ, Steele F, Galbraith J, Moustaki I. Analysis of Multivariate Social Science Data. Taylor & Francis; 2008. [Google Scholar]
- 67.Bryk S, Raudenbush WSA. Hierarchical linear models : applications and data analysis methods. Sage Publications; 2002. [Google Scholar]
- 68.Arellano M. Panel Data Econometrics. Oxford University Press; 2003. [Google Scholar]
- 69.Relman AS. The Rand health insurance study: is cost sharing dangerous to your health? N Engl J Med. 1983;309:1453. doi: 10.1056/NEJM198312083092310. [DOI] [PubMed] [Google Scholar]
- 70.Newhouse JP. A summary of the Rand Health Insurance Study. Ann N Y Acad Sci. 1982;387:111–114. doi: 10.1111/j.1749-6632.1982.tb17166.x. [DOI] [PubMed] [Google Scholar]
- 71.Hay JW, Ricardo-Campbell R. Rand Health Insurance study. Lancet. 1986;2:106. doi: 10.1016/s0140-6736(86)91639-9. [DOI] [PubMed] [Google Scholar]
- 72.DiNardo J. Natural experiments and quasi-natural experiments. In: Durlauf SN, Blume LE, editors. The New Palgrave Dictionary of Economics. 2. Palgrave Macmillan; 2008. [Google Scholar]
- 73.ROSENBAUM PR, RUBIN DB. The central role of the propensity score in observational studies for causal effects. Biometrika. 1983;70:41–55. [Google Scholar]
- 74.Imbens G, Angrist J. Identification and estimation of local average treatment effects. Econometrica. 1994;62:467–476. [Google Scholar]
- 75.Thistlewaite D, Campbell D. Regression-Discontinuity Analysis: An alternative to the ex post facto experiment. Journal of Educational Psychology. 1960;51:309–317. [Google Scholar]
- 76.Marmot M. The influence of income on health: views of an epidemiologist. Health Aff (Millwood) 2002;21:31–46. doi: 10.1377/hlthaff.21.2.31. [DOI] [PubMed] [Google Scholar]
- 77.Renalds A, Smith TH, Hale PJ. A systematic review of built environment and health. Fam Community Health. 2010;33:68–78. doi: 10.1097/FCH.0b013e3181c4e2e5. [DOI] [PubMed] [Google Scholar]
- 78.Galton F. English men of science: their nature and nurture. Cass; 1970. [Google Scholar]
- 79.Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, et al. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- 80**.Reiss D, Leve LD, Neiderhiser JM. How Genes and the Social Environment Moderate Each Other. Am J Public Health. 2013;103:S111–S121. doi: 10.2105/AJPH.2013.301408. This article proposes four major mechanisms to explain the interaction of social environment and gene and its impact on health, and discusses the barriers and promises of current studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81**.Jackson FLC, Niculescu MD, Jackson RT. Conceptual Shifts Needed to Understand the Dynamic Interactions of Genes, Environment, Epigenetics, Social Processes, and Behavioral Choices. Am J Public Health. 2013;103:S33–S42. doi: 10.2105/AJPH.2013.301221. Using case-study examples, this article presents that the simultaneous assessment of genetic, epigenetic and environmental data can strengthen the behavioral and social health research by providing causal pathways and expanding parameters of the predictive modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82**.Roetker NS, Page CD, Yonker JA, Chang V, Roan CL, Herd P, Hauser TS, Hauser RM, Atwood CS. Assessment of Genetic and Nongenetic Interactions for the Prediction of Depressive Symptomatology: An Analysis of the Wisconsin Longitudinal Study Using Machine Learning Algorithms. Am J Public Health. 2013;103:S136–S144. doi: 10.2105/AJPH.2012.301141. Integrating population survey data with genetic testing data, this article uses machine learning algorithms to explore the interactions of genetic, environmental, social and behavioral factors and their impact on depressive symptoms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83*.Harris KM, Halpern CT, Hussey J, Whitsel EA, Killeya-Jones L, Tabor J, Elder G, Hewitt J, Shanahan M, Williams R, et al. Social, Behavioral, and Genetic Linkages from Adolescence Into Adulthood. Am J Public Health. 2013;103:S25–S32. doi: 10.2105/AJPH.2012.301181. Using Add Health, a large sample following adolescents into their adulthood, this article illustrates how the integration of population survey and genotyping can enhance social and behavioral research in understanding the interactions of genetic and social determinants and their influences on health. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.King AC, Ahn DK, Oliveira BM, Atienza AA, Castro CM, Gardner CD. Promoting Physical Activity Through Hand-Held Computer Technology. Am J Prev Med. 2008;34:138–142. doi: 10.1016/j.amepre.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ngo J, Engelen A, Molag M, Roesle J, García-Segovia P, Serra-Majem L. A review of the use of information and communication technologies for dietary assessment. Br J Nutrition. 2009;101:S102–S112. doi: 10.1017/S0007114509990638. [DOI] [PubMed] [Google Scholar]
- 86.Carrasco JA, Hogan B, Wellman B, Miller EJ. Collecting social network data to study social activity-travel behavior: an egocentric approach. Env Plan B. 2008;35:961. [Google Scholar]
- 87.King JD, Buolamwini J, Cromwell EA, Panfel A, Teferi T, Zerihun M, Melak B, Watson J, Tadesse Z, Vienneau D, et al. A novel electronic data collection system for large-scale surveys of neglected tropical diseases. PLoS One. 2013;8:e74570. doi: 10.1371/journal.pone.0074570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wingo PA, Cardinez CJ, Landis SH, Greenlee RT, Ries LA, Anderson RN, Thun MJ. Long-term trends in cancer mortality in the United States, 1930–1998. Cancer. 2003;97:3133–3275. doi: 10.1002/cncr.11380. [DOI] [PubMed] [Google Scholar]
- 89.Smoking and Health. 1964 [Google Scholar]
- 90.Taylor PJ. The effects of shift work on worker health. IMS Ind Med Surg. 1973;42:13–19. [PubMed] [Google Scholar]
- 91.Vogel M, Braungardt T, Meyer W, Schneider W. The effects of shift work on physical and mental health. J Neural Transm. 2012;119:1121–1132. doi: 10.1007/s00702-012-0800-4. [DOI] [PubMed] [Google Scholar]
- 92.Bambra CL, Whitehead MM, Sowden AJ, Akers J, Petticrew MP. Shifting schedules: the health effects of reorganizing shift work. Am J Prev Med. 2008;34:427–434. doi: 10.1016/j.amepre.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 93.Waterhouse JM, Folkard S, Minors DS. Effects of a change in shift-work on health. Occup Med (Lond) 1993;43:167–168. doi: 10.1093/occmed/43.3.167. [DOI] [PubMed] [Google Scholar]
- 94.Poole CJ, Evans GR, Spurgeon A, Bridges KW. Effects of a change in shift work on health. Occup Med (Lond) 1992;42:193–199. doi: 10.1093/occmed/42.4.193. [DOI] [PubMed] [Google Scholar]
- 95.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Choudary PV, Cartagena P, Barchas JD, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Froy O. Metabolism and circadian rhythms--implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 97.Cajochen C, Krauchi K, Wirz-Justice A. Role of melatonin in the regulation of human circadian rhythms and sleep. J Neuroendocrinol. 2003;15:432–437. doi: 10.1046/j.1365-2826.2003.00989.x. [DOI] [PubMed] [Google Scholar]
- 98.Little RJA, Rubin DB. Statistical analysis with missing data. Wiley; 2002. [Google Scholar]