Abstract
The importance of the Kiss1 gene in the control of reproductive development is well documented. However, much less is known about the transcriptional regulation of Kiss1 expression in the hypothalamus. Critical for these studies is an accurate identification of the site(s) where Kiss1 transcription is initiated. Employing 5’-RACE PCR we detected a transcription start site (TSS1) used by the hypothalamus of rats, mice, nonhuman primates and humans to initiate Kiss1 transcription. In rodents, an exon 1 encoding 5’-untranslated sequences is followed by an alternatively spliced second exon, which encodes 5’-untranslated regions of two different lengths and contains the translation initiation codon (ATG). In nonhuman primates and humans exon 2 is not alternatively spliced. Surprisingly, in rat mediobasal hypothalamus (MBH), but not preoptic region (POA), an additional TSS (TSS2) located upstream from TSS1 generates an exon 1 longer (377 bp) than the TSS1-derived exon 1 (98 bp). The content of TSS1-derived transcripts increased at puberty in the POA and MBH of female rats. It also increased in the MBH after ovariectomy, and this change was prevented by estrogen. In contrast, no such changes in TSS2-derived transcript abundance were detected. Promoter assays showed that the proximal TSS1 promoter is much more active than the putative TSS2 promoter, and that only the TSS1 promoter is regulated by estrogen. These differences appear to be related to the presence of a TATA box and binding sites for transcription factors activating transcription and interacting with estrogen receptor alpha (ERα) in the TSS1, but not TSS2, promoter.
Keywords: Kiss1 gene, transcriptional start site, rats, hypothalamus, puberty
INTRODUCTION
Following the discovery ten years ago that mutations in the gene encoding the receptor GPR54 (now termed KISS1R) resulted in hypothalamic hypogonadism and pubertal failure in humans [1;2], much has been learned about the role of KISS1R and its ligands, the kisspeptins (encoded by the KISS1 gene), in the physiological control of reproduction (reviewed in [3;4]). Kisspeptins are now considered to have important roles in key aspects of reproductive function, including the control of puberty and the implementation of estrogen feedback mechanisms on gonadotropin-releasing hormone (GnRH) release (reviewed in [5;6]).
The first reports describing the potential role of hypothalamic kisspeptin neurons in the onset of puberty demonstrated that KISS1 mRNA expression increases with pubertal maturation in the POA-MBH of rats and the MBH of primates [7;8]. Subsequent studies in rodents demonstrated the existence of two populations of neurons expressing Kiss1 in the ventral forebrain. One of these populations is located in the anteroventral periventricular (AVPV) nucleus of the preoptic region; the other is located in the arcuate nucleus (ARC) of the medial basal hypothalamus [9–11]. The distribution of kisspeptin neurons in the hypothalamus of other species, including nonhuman primates (NHP), humans and sheep, is essentially the same [8;12–14]. These two populations are functionally different; while kisspeptin neurons of the AVPV respond to estrogen with increased synthesis of kisspeptin, production of kisspeptin is inhibited by estrogen in the ARC [15;16]. These and other observations have led to the concept that AVPV kisspeptin neurons are involved in the genesis of the preovulatory LH surge, whereas ARC kisspeptin neurons mediate the inhibitory effect of estrogen on gonadotropin release [16–18].
In contrast to the plethora of reports describing the physiological role of kisspeptin in reproductive function much less is known about the transcriptional control of the Kiss1 gene in the hypothalamus in general, and contribution of this control to the timing of puberty and the mechanisms mediating estrogen effects on the GnRH neuronal system in particular. Important for a precise analysis of the cis/ trans-acting factors and epigenetic molecules controlling Kiss1 transcription within the neuroendocrine brain is the identification of transcription start sites (TSSs) that may be used in this brain region to set in motion the Kiss1 transcriptional machinery. Thus far, the mouse is the only species in which the Kiss1 TSS has been identified using hypothalamic tissue [19]. The TSS of human KISS1 was described by four different groups [20–23], but using either placental tissue [21;23] or cell lines [20;22]. To our knowledge no studies have been published establishing the position of Kiss1 TSS in either the rat or NHP. Admittedly, the position of the Kiss1 TSS in different species can be estimated from public databases depicting the beginning of exon 1 (see for instance the track for human KISS1 in http://genome.ucsc.edu or follow the UCSC Genome browser custom track in http://fantom.gsc.riken.jp/5/suppl/Kanamori-Katayama_et_al_2011/). However, because the TSS presumed location is based on the alignment of transcripts sequences, instead of specific detection methods, such as Rapid Amplification of 5’cDNA Ends (5’-RACE) PCR, S1 nuclease protection assays or Cap Analysis of Gene Expression (CAGE) [24] the type and precise location of TSS(s) used by different tissues to initiate Kiss1 transcription remains uncertain. Even more important for the understanding of Kiss1 involvement in neuroendocrine reproductive function, the information currently available in databases does not address the potentially important issue of tissue-specific promoter usage and, in particular, whether promoter usage in the hypothalamus is similar to that known from non-neural tissues/cells.
In the present study we describe experiments aimed at determining the location of the TSSs employed by the hypothalamus of different species to initiate Kiss1 transcription, and use prepubertal female rats to determine the changes in alternative transcripts expression that occur during puberty and in response to both removal of ovarian steroids and estrogen replacement therapy.
MATERIAL AND METHODS
Animals
Upon arrival from the vendor (Charles River Laboratories international, Wilmington, MA) immature female Sprague Dawley rats were housed at the Oregon National Primate Research Center (ONPRC) vivarium under controlled conditions of temperature (23–25 C) and light (14-h light, 10-h dark; lights on from 0500–1900 h), with ad libitum access to standard chow (Purina laboratory chow; Ralston-Purina, St. Louis, MO) and tap water.
Female mice from the C57 strain (Jackson Labs, Bar Harbor, ME) were housed under similar conditions of temperature (23–25 C), but with a different photoperiod (12:12 light-dark cycle with lights on between 0600–1800 h), with standard pelleted food (LabDiet 5001, PMI Nutrition International Brentwood, St. Louis, MO) and water provided ad libitum.
The NHP brain tissues used in this study were obtained from mid-pubertal (3–4 years of age) female monkeys (Macaca mulatta) obtained through the ONPRC Necropsy Program.
All procedures performed using animals were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the ONPRC.
Tissue dissection
Dissection of the mediobasal hypothalamus (MBH) and preoptic area (POA) was as previously described [25]. In brief, the POA was dissected by a transverse cut behind the optic chiasm and two oblique cuts initiated on each lateral edge of the optic chiasm and intersecting at a point anterior to the decussation of the optic nerves. The MBH was dissected by two lateral cuts along the hypothalamic sulci, one posterior cut along the rostral border of the mammillary bodies and one anterior cut immediately behind the optic chiasm. The thickness of each fragment was about 1–2 mm. After dissection, the tissues were frozen on dry ice and stored at −85C until RNA extraction.
RNA extraction
Total RNA was extracted using the RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer’s protocol. To remove DNA contamination, the RNA samples were treated using a DNA-free DNase I kit (Ambion, Austin, TX). PCR analysis of each sample in the absence of reverse transcription showed no amplification, confirming the absence of contaminating genomic DNA in the RNA samples. RNA concentrations were determined by spectrophotometric analysis (Epoch, Winooski, VT), and RNA integrity was verified using a Bioanalyzer RNA nano kit (Agilent, Santa Clara, CA). Human hypothalamic total RNA was purchased from Ambion (Cat # 6864). The donor was a caucasian 23 year old male who died of cardiac arrest. There is no information about the donor’s reproductive function.
Experimental identification of Kiss1 TSS by SMART-5’-RACE
The position of the Kiss1 TSS was determined using the Switching Mechanism At 5′ end of RNA Transcript Rapid Amplification of cDNA Ends (SMART™ RACE cDNA Amplification kit, Clontech Laboratories, Mountain View, CA) according to the manufacturer’s recommendations. In brief, one microgram of total RNA from rat MBH, POA and placenta (PLA, BioChain Institute, Newark, CA), mouse and monkey MBH, and human hypothalamus (Ambion, Austin, TX) were reverse transcribed (RT) using a modified oligo(dT) primer, SMARTScribe Reverse Transcriptase, and the SMARTer II A Oligonucleotide, which anneals to the extended cDNA tail added by the SMARTScribe Reverse Transcriptase to the 3’ end of the first strand cDNA. The resulting cDNAs were amplified by two rounds of PCR using HotStart Taq polymerase (QIAGEN, Valencia, CA). For the first round, each 25 µl PCR mixture consisted of 1.25 µl RT product, 2.5 µl 10X buffer, 1 µl 10 mM dNTPs, 0.1 µl HotStar Taq Polymerase, 2.5 µl 10 X Universal Primer A Mix 0.4 µM from the SMARTer RACE kit, 0.5 µl gene specific primers (GSP, Table 1) and 1.25 µl DMSO. The Universal Primer (UP) recognizes the SMARTer II A Oligonucleotide sequence inserted at the 5’ end of the transcript during the RT reaction. For the second round, we used as input 1.25 µl of purified PCR product from the first round diluted 1:10, a nested universal primer (NUP), and nested gene-specific primers (NGSP, Table 1). Both rounds of PCR were performed on a Bio-Rad (Hercules, CA) C1000 Touch™ Thermal Cycler using the following touchdown PCR protocol: 5 cycles of 30 sec at 94 C and 3 min at 72 C, followed by 5 cycles of 30 sec at 94 C, 30 sec at 70 C and 3 min at 72 C, and 32 cycles of 30 sec at 94 C, 30 sec at 68 C, followed by a final extension of 3 min at 72 C. The second-round PCR products were then cloned into the plasmid pGEM-T (Promega, Madison, WI), and positive clones were sequenced from both ends, using M13 forward and M13 reverse primers to determine the initiation of Kiss1 mRNA transcription.
Table 1.
Primers used to identify by RACE-PCR the Kiss1 TSSs in rat, mouse, monkey and human
| Species | Accession # | Primers | Sequence 5’-3’ | Tissues | Kiss1 variants |
Amplicon (bp) |
|---|---|---|---|---|---|---|
| RAT | NM_181692.1 | rGSP | tagcgcaggccaaaggagttccagtt | MBH | E1a-E2b-E3 | 575 |
| E1b-E2a-E3 | 373 | |||||
| E1b-E2b-E3 | 296 | |||||
| rNGSP | taacgagttcctggggtccggactgt | POA | E1b-E2a-E3 | 370 | ||
| E1b-E2b-E3 | 293 | |||||
| PLA | E1-E2 | 210 | ||||
| MOUSE | NM_178260.3 | mGSP | tagcgcaggccgaaggagttccagtt | MBH | E1b-E2a-E3 | 351 |
| mNGSP | taacgagttcctggggtccggactgc | E1b-E2b-E3 | 276 | |||
| MONKEY | XM_001098284.2 | mkyGSP | ccgcttgccgaagcgcaggccgaaggagttccagtt | MBH | E1-E2-E3 | 317 |
| mkyNGSP | cggggccgagaggtccagggattccagctgc | |||||
| HUMAN | NM_002256.3 | hGSP | ccgcttgccgaagcgcaggccgaaggagttccagtt | MBH | E1-E2-E3 | 322 |
| hNGSP | cgggggccaggaggcccagggattctagctgc |
Semiquantitative RT-PCR
Five hundred nanograms of total RNA were reverse transcribed using the Omni RT kit (QIAGEN, Valencia, CA) following the manufacturer’s recommendations. Quantification of the different Kiss1 mRNA transcripts (E1a, E1b-E2a and E1b-E2b) was assessed by semiquantitative PCR, using the primer pairs and conditions shown in Table 2. As an internal control, PCR amplification of peptidylprolyl isomerase A (Ppia) mRNA was carried out in parallel. The PCR reactions consisted of a first denaturing step at 95 C for 5 min, followed by a variable number of cycles of amplification, each consisting of a denaturation step at 95 C for 30 sec, annealing for 30 sec, and extension at 72 C for 30 sec. A final extension step at 72 C for 5 min was also included. Annealing temperature was adjusted for each target and primer pair: 60 C for each Kiss1 mRNA variant and 55 C for Ppia mRNA. PCR-generated DNA fragments were resolved by electrophoresis in Tris-borate-buffered 1.5% agarose gels and visualized by ethidium bromide staining. Intensity of the RT-PCR signals was quantified by densitometric scanning using AlphaEaseFC software (Alpha Innotech, San Leandro, CA), and values for each Kiss1 mRNA transcript were normalized using the PCR signals derived from Ppia mRNA. The final values are expressed as Kiss1/Ppia mRNA ratios.
Table 2.
Primers used for semi-quantitative PCR analysis.
| Kiss1 gene variants |
Accession # | Primers | Sequence 5’-3’ | Amplicon (bp) |
PCR cycles |
|---|---|---|---|---|---|
| rKiss1E2f | atagaggaagcccaggagccagag | 289 | 35 | ||
| rKiss1E2r | acaggtgccatttttgccagtg | ||||
| E1b-E2b | JX139032.1 | rKiss1E2f | atagaggaagcccaggagccagag | 213 | 35 |
| rKiss1E2r | acaggtgccatttttgccagtg | ||||
| E1a | JX139030.1 | rKiss1E1af | atggtttctgggggcgagcaagtc | 249 | 35 |
| rKiss1E1ar | ttgagggccggagggagaagag | ||||
|
Housekeeping gene |
Accession # | Primers | Sequence 5’-3’ |
Amplicon (bp) |
PCR cycles |
| Ppia | M19533.1 | rPpiaf | ggcaaatgctggaccaaacacaa | 222 | 27 |
| rPpiar | ggtaaaatgcccgcaagtcaaaga |
In silico analysis of transcription factor binding sites in the 5’-flanking region of the Kiss1 gene
We used the TRANSFAC database of transcription factor binding site matrices and the accompanying MATCH tool [26;27] to predict the location of estrogen-responsive elements and binding sites for activators of Kiss1 transcription known to interact with estrogen receptor alpha (ERα) in a DNA segment comprising 1000 bp of the Kiss1 5’-flanking region. We used the high-quality matrices included in TRANSFAC’s non-redundant vertebrate profile and selected cutoffs to optimize the sum of false positive and false negative rates (minSUM).
Plasmids
To investigate the basal and estrogen-dependent changes in transcriptional activity of both E1a and E1b promoters, we cloned two genomic fragments containing either TSS2 (rKiss1 TSS2, −557 to −207) or TSS1 (rKiss1 TSS1, −259 to +71) into the luciferase reporter plasmid pGL2-Basic (Promega). The position of the TSS1 detected by 5’-RACE PCR (see figure 1b) was used as +1. To PCR amplify the DNA fragments containing TSS2 and TSS1 we used 100ng of rat genomic DNA and the primer pairs shown in Table 3. PCR amplification consisted of a first denaturing step at 95 C for 5 min, followed by 34 cycles of amplification, each consisting of a denaturation step at 95 C for 30 sec, annealing for 30 sec, and extension at 72 C for 30 sec. A final extension step at 72 C for 5 min was also included. PCR-generated DNA fragments were resolved by electrophoresis in Tris-borate-buffered 1.5% agarose gels and visualized by ethidium bromide staining. PCR products were column purified (QIAGEN, Valencia, CA) and ligated into the SacI-SmaI sites of pGL2-Basic using an In-Fusion HD Cloning Kit (Clontech). The sequences of the cloned DNA fragments were verified by sequencing.
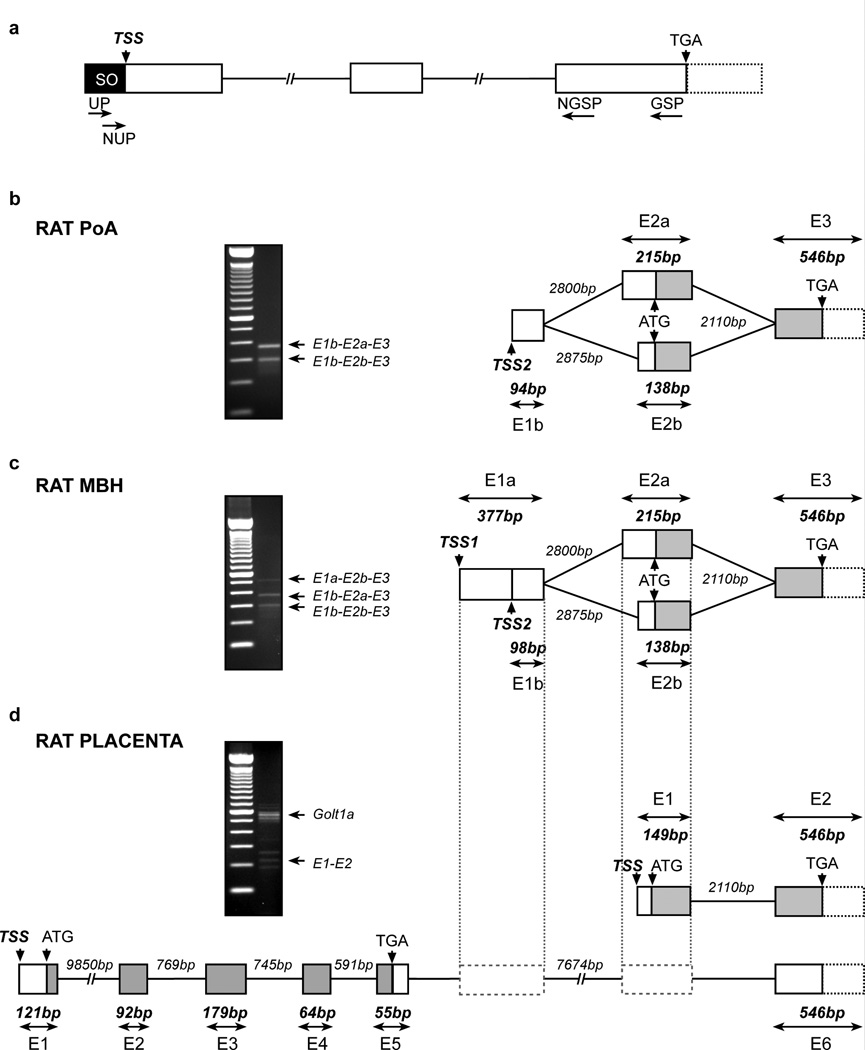
Figure 1.
Identification of Kiss1 TSS in the rat POA, MBH and placenta. (a) Schematic diagram showing locations of RACE-PCR primers used for the identification of Kiss1 TSSs. (b–d) Structure of the Kiss1 gene 5’-end and transcriptional variants detected by RACE-PCR in rat POA (b), MBH (c) and placenta (d). Representation of exon 3 (E3) in rat POA and MBH and exon 2 (E2) in rat placenta is based on the reported rat and mouse Kiss1 mRNA sequences (rat: Ensembl accession no. ENSRNOT00000035583; mouse: Ensembl accession no. ENSMUST00000007433). The gel images shown on each diagram depict the PCR products derived from RACE PCR using the nested universal primer (NUP) and the nested gene specific primer (NGSP) shown in panel a. Boxes and lines represent exons and introns, respectively. 5’-untranslated regions are represented by white boxes, coding regions are shown in gray and potential 3’-untranslated regions (88% homology with mouse sequence) are represented by white boxes with dotted outline. Numbers indicate the size of the exons and introns (in base pairs, bp). SO: Smarter Oligonucleotide; TSS: Transcription start site; UP: Universal primer; NUP: Nested universal primer; GSP: Gene specific primer; NGSP: Nested gene specific primer; ATG: Translation start codon; TGA: Translation stop codon; E1a: Long version of exon 1 in rat MBH; E1b: Short version of exon 1 in rat POA and MBH; E1: Exon1 in rat placenta; E2a: Long version of exon 2 in rat POA and MBH; E2b: Short version of exon 2 in rat POA and MBH; E2: Exon 2 in rat placenta; E1a-E2b-E3E1b-E2a-E3E1b-E2b-E3 and E1-E2: Transcriptional variants of Kiss1 mRNA detected by RACE-PCR in rat POA and MBH. In rat placenta Golt1a was detected as containing 6 exons: exon 1 to 5 (from ENSRNOT00000003918) and a sixth untranslated exon equivalent to Kiss1 exon 3.
Table 3.
Primers used to clone rat Kiss1 promoter regions containing either TSS2 or TSS1
| Kiss1 promoter regions |
Accession # | Primers | Sequence 5’-3’ | Amplicon (bp) |
PCR cycles |
|---|---|---|---|---|---|
| rKiss1 TSS2 | JX139030.1 | rKiss TSS2f | attctggtggaggccaatctg | 350 | 34 |
| rKiss TSS2r | ctctagcctcttgacacccc | ||||
| rKiss1 TSS1 | JX139030.1 | rKiss TSS1f | ttggggtacaatgagtctccagc | 330 | 34 |
| rKiss TSS1r | gagggagaagagccacttggg |
Cell culture, E2 stimulation and promoter assays
HEK 293T cells were grown in Dulbecco’s modified Eagle medium (DMEM, Sigma Chemicals, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT), 100µg/ml penicillin, and 100µg/ml streptomycin. For promoter assays, cells were seeded in 6 well plates (at a 2,000,000 cells/well), and grown in DMEM without phenol red or antibiotics, containing 10% charcoal-stripped FBS (Bioworld, Dublin, OH) and Na Pyruvate (1mM). Twenty-four hours after seeding, Lipofectamine 2000 (Invitrogen) was premixed for 30 min with the various reporter gene constructs at a ratio of 1 µg DNA to 2.5 µl transfection reagent before the mixture was added to the cells. Transfection efficiency was normalized by cotransfecting the plasmid CMV-Sport-β-gal (Invitrogen) at 10 ng/ml and determining β-galactosidase activity using the Tropix Galacto Reaction (ABI).
To examine the effect of estradiol (E2) on E1a and E1b promoter activity, HEK 293T cells were transiently transfected with the control plasmid (pGL2, 1µg/ml), 1µg of human pcDNA-ERa (a kind gift from Dr. Robert A. Shapiro, Department of Physiology and Pharmacology, Oregon Health and Science University, Portland, Oregon), and 1µg/ml of either rKiss1 TSS2-pGL2 or rKiss1 TSS1-pGL2. Twenty-four hours after transfection, the medium was replaced with fresh medium containing the E2 vehicle (ethanol, 0.1 µl/ml medium) or 100nM 17 β-Estradiol (Sigma). The cells were harvested 24 h later for luciferase and β-galactosidase assays. Luciferase activity was expressed as percent of the activity detected in control cells transfected with pGL2 and pcDNA-ERa.
Evaluation of sexual maturation
To determine the changes in hypothalamic expression of Kiss1 mRNA transcripts during female pubertal development, rats were euthanized at four different stages: early juvenile (EJ), late juvenile (LJ), early proestrus (EP) and late proestrus (LP), according to previously reported criteria [28;29]. According to these criteria, 21-d-old animals are considered to be in the early juvenile (EJ) phase of prepubertal development. At this time, the vagina is not yet patent, the uterine weight is 60 mg or less, and there is no intrauterine fluid. At 28d days of age, the rats are considered to be late juveniles (LJ); their vagina is closed and there is no intrauterine fluid accumulation. Pulsatile LH release in these animals is more pronounced in the afternoons [30]. As in humans and monkeys, this is the first hormonal manifestation of the increase in central drive that initiates puberty [31]. Older rats with a closed vagina, intrauterine fluid, and a uterine weight less that 200 mg are considered to be in the early proestrous (EP) phase of puberty. During this transition period the rising estrogen levels result in increased uterine weight, accumulation of intrauterine fluid [28], and generation of minisurges of LH secretion [30]. Finally, rats with a closed or open vagina, but showing a uterus filled with fluid and a uterine weight of at least 200 mg are considered to be in late proestrus, i.e. the phase of puberty when the first preovulatory surge of gonadotropins takes place. These animals are considered to be in mid-puberty because ovulation has not occurred yet. The first ovulation occurs the following day. All animals were euthanized between 1600 and 1700 h, and the MBH and POA were immediately dissected and collected as described above.
Ovariectomy and steroid treatment
Rats were ovariectomized (OVX) on postnatal day 23, under isoflurane anesthesia, via a dorsal approach. All animals were given carprofen (4 mg/kg) as an analgesic prior to surgery. Five days after OVX, the rats were injected with estradiol benzoate (E2) (100 µg/kg body weight, sc) (SIGMA, St Louis, MI) or vehicle alone (100 µl corn oil) and housed with intact animals (two OVX and two intact rats per cage). Starting on the same day (d 28), the intact animals were inspected daily for vaginal opening. Upon detection of swelling and hyperemia of the genital area (usually between postnatal day 30 and 32), both the intact rats entering puberty and the co-housed OVX animals were euthanized and the MBH was collected as described above.
Presentation of data and statistics
Semiquantitative PCR results are presented as the mean ± SEM from at least four animals per group. The data were first subjected to a normality test. Data passing this test, were analyzed by ANOVA followed by the Student-Newman-Keuls multiple comparison test (GraphPad PRISM version 5 software, USA). P ≤ 0.05 was considered significant.
RESULTS
Identification of the Kiss1 TSS in rat POA, MBH and placenta
To determine if the 5’-end of Kiss1 mRNA in rat hypothalamus is similar to that described in humans (GenBank accession no. AK291695.1) and mice (GenBank accession no. AB666166.1) we subjected total RNA isolated from rat POA and MBH to SMART 5’ RACE PCR. After reverse transcription using primers provided with the RACE PCR kit, the initial PCR reaction was performed using a reverse GSP complementary to a sequence located on exon 3 (Table 1) and the forward UP provided by the kit (Fig. 1a). This primer is complementary to an oligonucleotide sequence added to the 3’ end of the first-strand cDNA during the reverse transcription reaction. The products of this reaction were then subjected to nested PCR using a forward NUP provided by the kit and NGSPs (Fig. 1a; primers sequences listed in Table 1). The rat POA showed two different PCR products with approximate sizes of 373 (E1b-E2a-E3) and 296 bp (E1b-E2b-E3) (Fig. 1b). Rat MBH yielded a third PCR product with an estimated size of 575 bp (E1a-E2b-E3) (Fig. 1c). In POA, combination of the short exon E1b with either a long exon E2a or a short exon E2b results in two alternative transcripts. In MBH, in addition to these two variants a long exon E1a was found only in combination with short exon E2b. Based on the relative intensity of the PCR products it appears that the most abundant transcript is that containing the long E2a exon. The E1a-E2b-E3 transcript appeared to be the least expressed. Sequence and genomic analysis of these transcripts revealed that, as in humans and mice, the Kiss1 gene in rats is composed of three exons. However, in rat MBH transcription is initiated from two alternative TSSs (Fig.1c), which generate either a long (E1a: 377bp) or a short (E1b: 98bp) exon 1. In addition, the second exon (which contains the ATG translational initiation site) shows two 5´UTRs of different lengths, resulting in an exon of either 215 bp (E2a) or 138 bp (E2b) (Fig. 1c). Alternative use of these variants results in three different combinations of exons 1 and 2 (Fig. 1).
Because the only Kiss1 gene sequence reported in rat is from placenta (NM_181692.1), and this sequence lacks an experimentally verified TSS, we sought to determine if Kiss1 TSS(s) in placenta is similar to either the TSSs detected in rat MBH or the single TSS seen in rat POA. Although several PCR products were identified by SMART 5’ RACE PCR (Fig. 1d, gel image), only one of these products (E1-E2: 210 bp) corresponded to a Kiss1 transcript. All other PCR products were identified as segments of Golt1a (NM_001109070), which is located in chromosome 13q13 directly upstream from Kiss1 (Fig. 1d and Supplementary Table 1). This finding confirms the existence of the two Kiss1 exons previously described in rat placenta, and identifies the existence of a single TSS used by placental cells to initiate Kiss1 transcription (Fig. 1d). In addition, our results show that exon 3 of Kiss1 also serves as exon 6 in Golt1a (Fig. 1d).
Identification of the Kiss1 TSS in mouse and rhesus monkey MBH and human hypothalamus
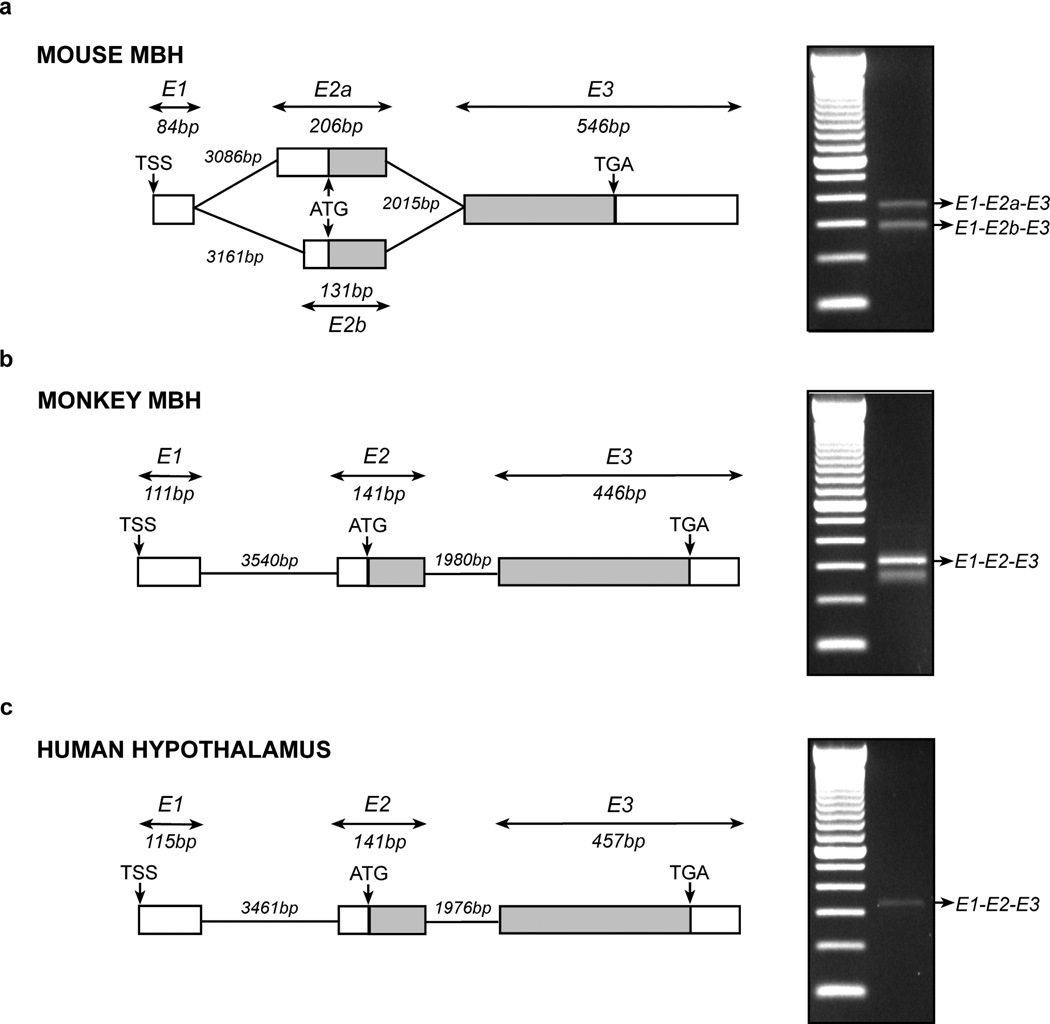
A single TSS has been shown to initiate transcription of the KISS1 gene in human placenta [21;23] and human cell lines [20;22]. With the exception of a very recent study [19] describing the Kiss1 genomic structure and TSS used in mouse hypothalamus (AB666166), it is not known if this or other potential TSSs initiate KISS1 transcription in the hypothalamus of human and nonhuman primates. To address this issue, we subjected total RNA extracted from the mouse and rhesus monkey MBH, and human hypothalamus to SMART 5’ RACE.
The results confirmed the existence of a major TSS in mouse MBH and the presence of two transcripts, both using the same TSS and containing a short (84 bp) exon 1 (Fig. 2a). Transcript 1 (E1-E2a-E3) is composed of exon 1, a long exon 2 (E2a) and the well-known exon 3. As in the rat MBH, transcript 2 (E1-E2b-E3) contains a short exon 2 (E2b) instead of the longer E2a exon. Both variants are reported in the Ensembl and NCBI genome databases (Ensembl ENSMUST00000007433 and GenBank AB666166), and were recently described in mouse hypothalamus [19].
Figure 2.
Identification of the Kiss1 TSS in the mouse and monkey MBH and human hypothalamus. (a–c) Structure of the Kiss1 gene 5’-end and transcriptional variants detected by RACE-PCR in mouse (a), monkey MBH (b) and human hypothalamus (c). Representation of exon 3 (E3) in mouse and monkey MBH and human hypothalamus is based on the reported Kiss1 mRNA sequences for each of these species (mouse: Ensembl accession no. ENSMUST00000007433; monkey: Ensembl accession no. ENSMMUT00000045110; human: Ensembl accession no. ENST00000367194). The gel images shown on the right of each diagram depict the PCR products derived from RACE PCR using the nested universal primer (NUP) and the nested gene specific primer (NGSP) shown in Figure 1, panel a. Boxes and lines represent exons and introns, respectively. Untranslated regions are represented by white boxes and coding regions are shown in gray. Numbers indicate the size of the exons and introns (in base pairs, bp). TSS: Transcription start site; ATG: Translation start codon; TGA: Translation stop codon; E1: Exon 1 in mouse and monkey MBH and human hypothalamus; E2a: Long version of exon 2 in mouse MBH; E2b: Short version of exon 2 in mouse MBH; E2: Exon 2 in mouse and monkey MBH and human hypothalamus; E1-E2a-E3E1-E2b-E3E1-E2-E3: Transcriptional variants of Kiss1 mRNA detected by RACE-PCR in mouse and/or monkey MBH and human hypothalamus.
In monkey MBH, nested PCR yielded two different bands (Fig. 2b, gel images). However, only the upper band contained a KISS1 sequence (E1-E2-E3) with the other band representing a non-specific PCR product. Only a single KISS1 transcript similar in size to the monkey transcript was detected in human hypothalamus (Fig. 2c, gel image). In both cases the sequence obtained corresponded to the KISS1 sequences reported in the Ensembl and NCBI genome databases (Human: Ensembl ENST00000367194 and GenBank AK291695.1; monkey: Ensembl ENSMMUT00000045110). Thus, as it occurs in human placenta and human cell lines, KISS1 mRNA is transcribed from a single TSS in both the monkey MBH and human hypothalamus.
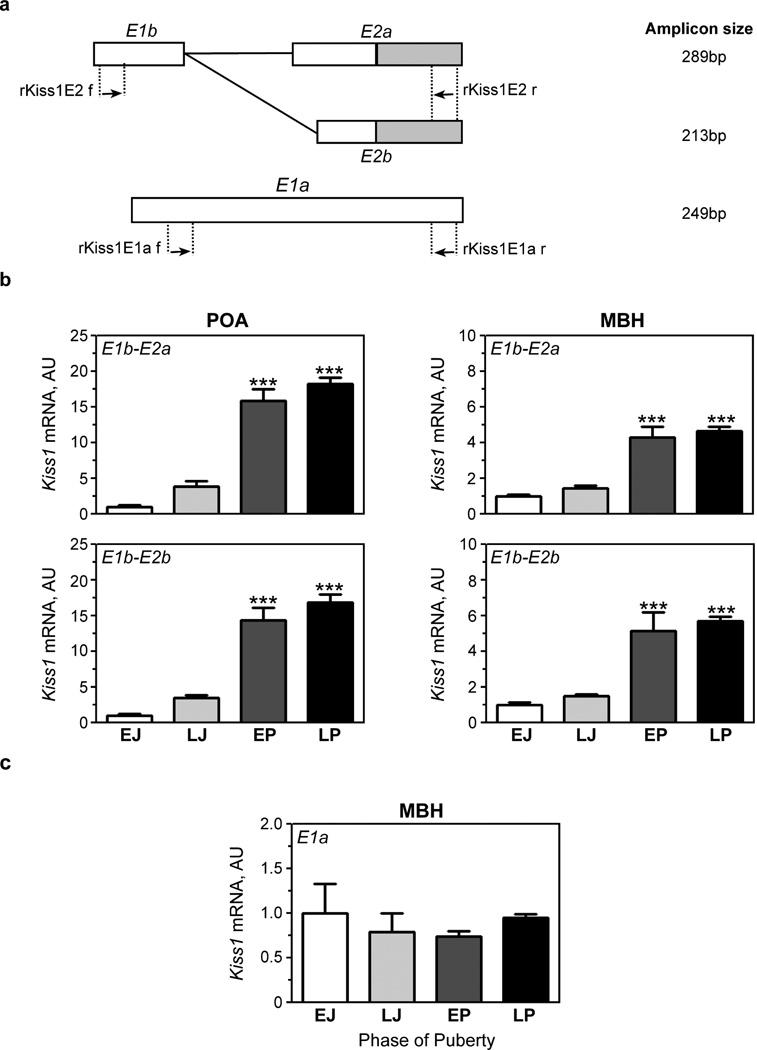
Expression profile of Kiss1 transcriptional variants in rat MBH and POA during female peripubertal development
We next used exon-specific primers (Fig. 3a) to define the expression profile of the three Kiss1 transcripts identified in the rat MBH during pre and peripubertal development of the female rat. The results showed that the abundance of the transcripts containing the combination E1b-E2a or E1b-E2b increased ~2-fold in POA and slightly less in MBH during juvenile development (EJ-LJ phases), and then more than 15-fold in POA and ~5-fold in MBH at the time of puberty (EP-LP phases) (Fig. 3b).
Figure 3.
Expression profile of Kiss1 mRNA transcriptional variants in rat POA and MBH during female peripubertal development. (a) Schematic diagram showing locations of RT-PCR primers used for the analysis of Kiss1 mRNA expression. Boxes and lines represent exons and introns, respectively. Untranslated regions are represented by white boxes and coding regions are shown in gray. (b) The abundance of E1b-E2a and E1b-E2b mRNA transcripts increases in both rat POA and MBH during pre-and peripubertal development (c) Expression of the alternatively transcribed E1a-containing transcript does not change during female peripubertal development. The results are expressed in arbitrary units (ratios between Kiss1 mRNA and Ppia mRNA). Vertical bars represent the mean ± SEM; n = 4–9 animals per group. ***, P < 0.001 vs. EJ and LJ groups (ANOVA followed by the Student-Newman-Keuls multiple comparison test). f = forward; r = reverse.
Because E1a, the long exon 1, was only detected by 5´RACE PCR in rat MBH, the expression analysis of Kiss1 transcripts containing this exon were carried out only in the MBH. Contrary to the puberty-related changes in E1b-E2a and E1b-E2b mRNA abundance observed earlier, no changes in E1a expression were detected throughout pre and peripubertal maturation (Fig. 3c), indicating that expression of this transcript is not developmental regulated.
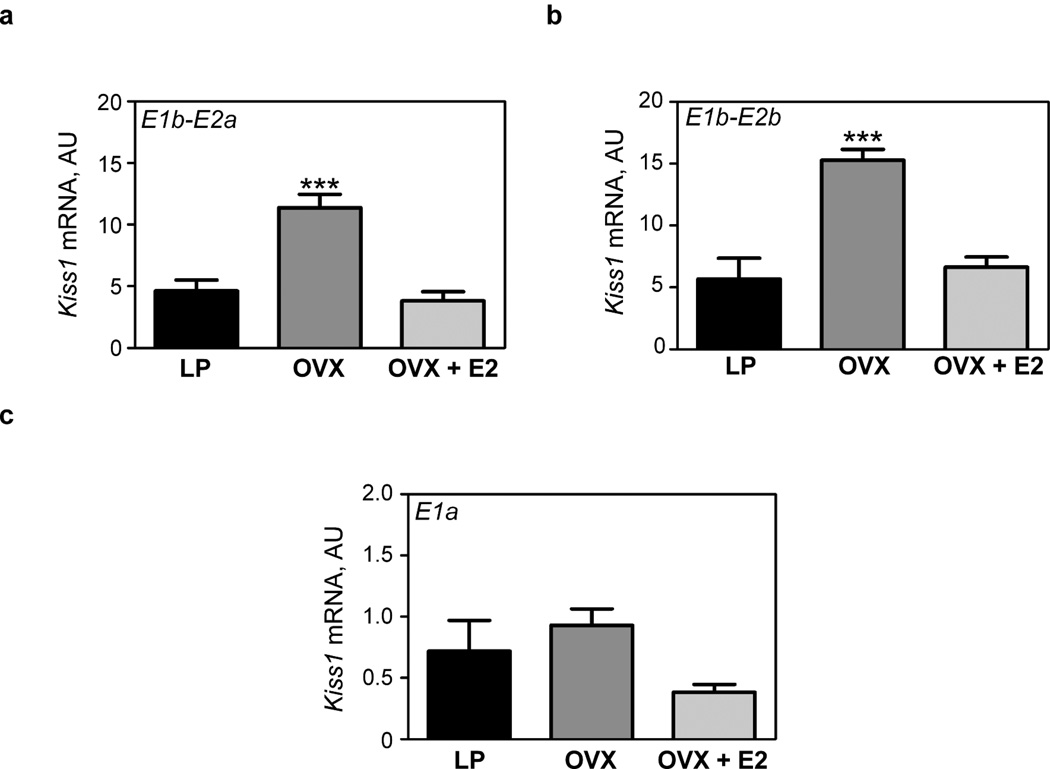
Changes in Kiss1 transcript abundance in the rat MBH in response to OVX and estradiol treatment at the expected time of puberty
Because estrogen has been repeatedly shown to repress Kiss1 expression in the MBH [16;32–34] we carried out a study to determine if OVX and treating OVX juvenile rats with estradiol would alter the expression of the three Kiss1 transcripts detected in rat MBH. As previously observed using primers that detect the Kiss1 coding region [33] or cRNA probes complementary to this region [15;16;32;34], OVX (on postnatal day 23) resulted 7–9 days later in a significant increase in both E1b-E2a and E1b-E2b-containing transcripts as compared with LP controls (Fig. 4a, b). A single EB injection (100µg/kg BW) administered to 28-d-old rats OVX five days earlier led to a significant decrease in the content of both transcripts 48 h later (Fig. 4a, b). In contrast, neither OVX nor EB treatment altered the abundance of E1a-containing transcripts (Fig. 4c), suggesting that neither the ovary nor estradiol regulates transcription of this variant. In keeping with this conclusion, basal levels of E1a-containing transcripts in intact LP rats were 10 times lower in the MBH than that of transcripts containing the short E1b exon (Fig. 4a–c).
Figure 4.
Expression profile of Kiss1 mRNA transcriptional variants in rat MBH of pubertal rats (30–32 days old), after ovariectomy (OVX) alone or OVX followed by E2 treatment. (a) The abundance of both E1b-E2a and (b) E1b-E2b mRNA transcripts increases after OVX and is restored to pre-OVX levels by E2 treatment. (c) The abundance of E1a-containing transcripts does not change after OVX or estrogen treatment. The results are expressed in arbitrary units (ratios between Kiss1 mRNA and Ppia mRNA). Vertical bars represent the mean ± SEM; n = 4–9 animals per group. ***, P < 0.001 vs. intact LP controls and OVX + E2 group (ANOVA followed by the Student-Newman-Keuls multiple comparison test).
The proximal promoter of E1b-containing transcripts, but not that of E1a, has the architecture of promoters displaying a single dominant TSS
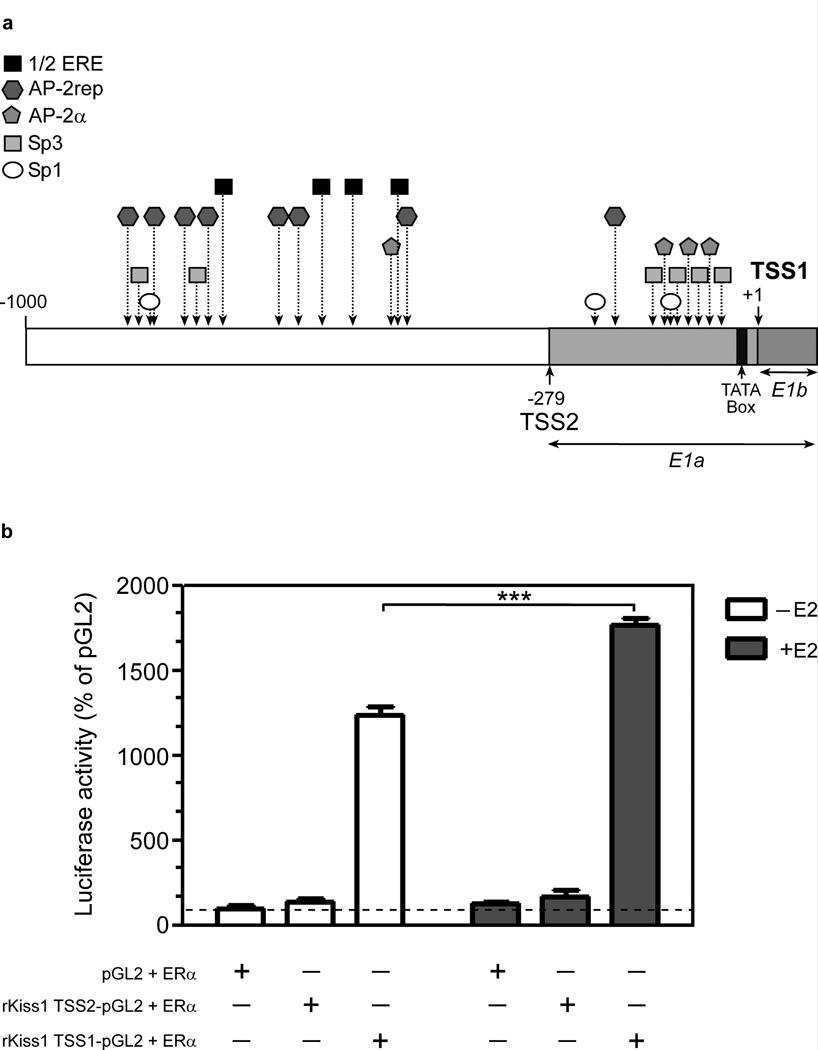
In silico analysis of the E1a and E1b proximal promoters demonstrated that the E1b promoter behaves like promoters exhibiting a single dominant TSS [24], with a preferential CA dinucleotide initiator, a TATA box signature near the TSS (−27 to −23 bp), and multiple binding sites for transcription factors able to tether ERα to the promoter, including Sp1, Sp3 and AP-2α [35–37], all of them located within 84 bp upstream of the TSS (Fig. 5a). The presence of these sites within −93 to 58 bp from the TSS of human KISS1 has been shown to be required for Sp1/AP-2α-mediated regulation of Kiss1 promoter activity [35;36]. Although the proximal promoter of E1a-containing transcripts also has a single dominant TSS, and like TSS1 uses the dinucleotide CA as initiator, it does not contain a TATA box. It also lacks Sp1, Sp3 and AP-2α binding sites near the TSS, with only one AP-2α binding site present at position −193 bp. The promoter region upstream, from TSS2 (−279bp) to 1000bp, contains a cluster of 4 binding sites for half-estradiol response elements (1/2ERE) and seven binding sites for AP-2 rep, a Kruppel-related zinc finger protein involved in gene repression [38;39]. The latter suggest that this region plays a role in repressing Kiss1 expression.
Figure 5.
The 5’-flanking region of rat Kiss1. (a) In silico prediction of recognition sites for transcription factors involved in the regulation of the Kiss1 gene in rats. The analysis of 1Kb upstream from the TSS1 detects several motifs implicated in estrogen-dependent and independent mediated transcriptional activation, including four incomplete EREs (1/2ERE), three SP1, six SP3, and four AP-2α sites. Binding sites for SP1 (n =2), SP3 (n = 4) and AP-2α (n =3) are overrepresented in the proximal promoter (−279 bp to +1) of TSS1, which also contains a canonical TATA box. In contrast, these binding sites are essentially absent from the proximal promoter of TSS2 (−558 to −279 bp from TSS1). An upstream genomic region located between −1000 to −600 bp (from TSS1) contains a cluster of binding sites (n = 7) for the transcriptional repressor AP-2rep, in addition to four half ERE sites. (b) HEK 293T cells were co-transfected with a luciferase vector containing either the proximal promoter region of E1a (rKiss1 TSS2-pGL2) or E1b (rKiss1 TSS1-pGL2) and a vector expressing estrogen receptor α (ERα). Twenty-four hs later they were exposed to 100 nM E2 and were collected after an additional 24h period for luciferase assays.
Transcriptional activity of the E1a and E1b promoters
To assess the basal activity and the potential effects of E2 on E1a and E1b promoter activity, HEK 293T cells were cotransfected with pcDNA-ERα and either rKiss1 TSS2-pGL2 or rKiss1 TSS1-pGL2. Twenty-four hours after transfection, cells were treated with 100 nM 17β-Estradiol or the vehicle (ethanol) as a control. In good agreement with our expression data (see Fig. 3 and Fig. 4), the results showed that basal activity of the E1b promoter was much greater (~10-fold) than that of E1a, and that only the E1b promoter was responsive to estradiol (Fig. 5b). However, contrary to the changes in E1b-containing transcripts in response to OVX and estradiol treatment observed in vivo, E2 increased - instead of decreasing - the transcriptional activity of the E1b promoter.
DISCUSSION
The present study identifies the start site used by the hypothalamus of rats, mice, nonhuman primates and humans to initiate transcription of the KISS1 gene. Our results indicate that in all these four species there is a single dominant KISS1 TSS on the 5’ end of a short exon1 (ranging from 84 bp in mouse hypothalamus to 115 bp in human hypothalamus). These findings are entirely consistent with earlier reports describing the structure of the KISS1 gene in human placenta [21;23], human cell lines [20;22], and mouse hypothalamus [19]. As already known, exon 1 encodes only untranslated sequences. This feature is similar to that of KISS1 mRNA in human placental tissue, but surprisingly, it differs from Kiss1 mRNA in rat placenta. Because exon 1 is not transcribed in this tissue, the TSS is located at the beginning of exon 2, which then becomes exon 1 and contains both the ATG translation initiation site and a short untranslated sequence.
In addition to this tissue-specific difference in promoter usage, the splicing pattern of the Kiss1 mRNA primary transcript is different in the hypothalamus of rodents as compared with primates. As recently reported for the mouse hypothalamus [19], the rat hypothalamus expresses two variants derived from the alternative splicing of exon 2, which contains the ATG translational start site. As a result, the 5’-untranslated region encoded by exon 2 displays two different lengths. In humans and nonhuman primates such splicing variants do not exist. The biological consequences of this difference are not known.
Perhaps the most surprising observation of our study is the presence in rat MBH, but not in rat POA or the MBH of mice, NHP and human hypothalamus, of an alternative TSS, which results in an exon 1 (E1a) that is 279 bp longer than the canonical exon 1 (E1b). Expression of transcripts containing the E1a exon is low and subjected to neither developmental control around the time of puberty nor steroid regulation. Specifically, the abundance of E1a-containing transcripts remains at low levels in the MBH during the juvenile-pubertal transition and does not change in response to either ovariectomy or estrogen treatment. Analysis of the underlying genomic structure provides a reasonable explanation for this behavior. While the proximal promoter of TSS1, the preferred TSS used by the hypothalamus of all four species analyzed, contains a canonical TATA box and a cluster of SP1, SP3, and AP-2α binding sites able to activate (SP1/AP-2α) or repress (SP3) Kiss1 transcription [35–37], the alternative TSS2 lacks these features. In addition to enhancing KISS1 promoter activity, Sp1 and AP-2α binding sites have been shown to physically interact with ERα to tether the receptor to DNA regions containing Sp1/AP-2α binding sites [35–37] in the absence of estrogen response elements (EREs) able to bind ERα.
Consistent with our expression data showing that E1b-containing transcripts are much more abundant than E1a-containing transcripts in rat MBH, we observed that the basal activity of the E1b promoter measured in reporter assays is strikingly greater than that of the E1a promoter. These results are in keeping with previous reports showing that human and mouse genomic regions equivalent to that comprising the E1b promoter fragment used in our transfection assays effectively drive Kiss1 transcription [19;35;36;40]. In fact, the strongest transcriptional activity of the mouse Kiss1 5’-flanking region resides in a segment immediately upstream from the TSS (−180 bp to +1) [19], similar to the rat E1b promoter (−259 bp to +71).
We also observed that only the activity of the E1b promoter changes in response to E2, but that in contrast to the in vivo expression data, promoter activity increases in response to estrogen. These findings are similar to earlier reports showing that human Kiss1 mRNA expression is up-regulated by E2 in ERα-positive GT1-7 [41;42] and 293T cells transfected with an ERα expression vector [41]. However, they differ from two other studies reporting that E2 either does not affect Kiss1 transcription in neural N7cells [19] or represses it in a MDA-MB-231 breast cancer cell line stably expressing ERα [22]. The reasons for these discrepancies are unknown.
A potential repressive effect of E2 on Kiss1 transcription may involve both interactions with Sp3 protein complexes [41] and loss of RNA polymerase II binding to the Kiss1 promoter [22]. It may not involve a direct interaction of ERα with SP1-containing complexes because estrogen can decrease KISS1 transcriptional activity in the absence of Sp1 sites [22], and the association of ERα to Sp1 appears to mediate estrogen-dependent activation of Kiss1 transcription instead of inhibition [35;36;41]. Future experiments are obviously required to address this important issue.
Our results show that the abundance of E1b-containing transcripts, measured by semi-quantitative PCR, increases not only in the POA but also the MBH during the juvenile-peripubertal period of the female rat, with maximal levels observed on the day of the first preovulatory surge of gonadotropins. The increase in Kiss1 mRNA content detected in the MBH is in keeping with earlier reports that also used qPCR to quantify changes in Kiss1 mRNA abundance in the female rat MBH at the time of puberty [33;43–45], but not with reports in which such changes were estimated in female mice using immunohistochemistry [11], in situ hybridization [46], or qPCR [47]. The reasons for this discrepancy are not readily apparent, especially because both PCR and in situ hybridization-based methods detect an increase in Kiss1 mRNA abundance in the MBH after ovariectomy and reversal of this change by estradiol treatment [16;33;34]. These considerations bring up another issue that has not been adequately addressed in the literature: why Kiss1 mRNA expression increases in the MBH [33;43–45] or remains unchanged [11;46;47] at puberty (when estradiol levels are high) but also increases after OVX (when estradiol levels are low)? Estrogen treatment of OVX animals is very effective in reducing Kiss1 mRNA levels in the MBH. Why it does not do that around the time of puberty? There must be another component or mechanism influencing the response of the Kiss1 gene to estrogen action.
The functional significance of an alternative TSS in rat MBH is unclear, as the resulting transcript does not appear to be developmental regulated or subject to ovarian feedback regulation. It does appear to be much less frequently used than the “canonical” TSS1, and although we could not detect it by RACE PCR in the rat POA, we have occasionally observed low levels of E1a-containing transcripts in the POA of some animals using the same primers that detect these transcripts in the rat MBH (data not shown). It is possible that because of its proximity to a cluster of AP-2rep binding sites located >500 bp upstream from TSS1 (but only 200–300 bp upstream from TSS2), transcription initiation at TSS2 is more susceptible to AP-2rep-mediated repression than that at TSS1. AP-2rep is a transcriptional inhibitor encoding a Krϋppel-related zinc finger protein with homology to the wt1/egr1 family of transcription factors [38].
Rather than being an exception, the use of alternative TSSs is a common feature of the mammalian genome as it is found in most protein-encoding genes [24;48]. Increasing evidence exists supporting the role of 5’-UTR’s in events such as RNA stability and translational efficiency [49;50], subcellular localization [51;52] as well as differential coupling to cellular signals [53;54]. Although we have not analyzed any of these features, they stand out as a possibility for future studies. The Kiss1 E1a and E1b promoters behave like most alternative promoters in that they generate alternative 5’ exons that splice into a common second exon [48]. Even if it not preferentially used, transcription initiated at TSS2 would necessarily be accompanied by recruitment of a host of accessory proteins required for transcription initiation and elongation [48;55]. The presence of these multiprotein complexes would limit the access of TATA binding proteins to this genomic region, thereby reducing the ability of these proteins to initiate transcription from TSS1. Based on these considerations it might be speculated that TSS2-initiated transcription, though weak, may serve to modulate the most active transcriptional activity of the TSS1 promoter, and/or play a role in terminating physiological episodes of TSS1-mediated transcriptional activation.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by a grant from the US National Science Foundation (NSF: IOS1121691) to S.R.O and a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (FP7-PEOPLE-2010-IOF) to J.M.C. HW was supported by NIH Training grant T32 HD007133.
REFERENCES
- 1.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 2.de Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinilla L, Aguilar E, Dieguez C, Millar RP, Tena-Sempere M. Kisspeptins and reproduction: physiological roles and regulatory mechanisms. Physiol Rev. 2012;92:1235–1316. doi: 10.1152/physrev.00037.2010. [DOI] [PubMed] [Google Scholar]
- 5.Tena-Sempere M. Deciphering puberty: novel partners, novel mechanisms. Eur J Endocrinol. 2012;167:733–747. doi: 10.1530/EJE-12-0669. [DOI] [PubMed] [Google Scholar]
- 6.Clarkson J. Effects of estradiol on kisspeptin neurons during puberty. Front Neuroendocrinol. 2013;34:120–131. doi: 10.1016/j.yfrne.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Navarro VM, Castellano JM, Fernández-Fernández R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology. 2004;145:4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- 8.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Increased hypothalamic GPR54 signaling: A potential mechanism for initiation of puberty in primates. Proc Natl Acad Sci U S A. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita M, Tsukamura H, Adachi S, Matsui H, Uenoyama Y, Iwata K, Yamada S, Inoue K, Ohtaki T, Matsumoto H, Maeda K. Involvement of central metastin in the regulation of preovulatory luteinizing hormone surge and estrous cyclicity in female rats. Endocrinology. 2005;146:4431–4436. doi: 10.1210/en.2005-0195. [DOI] [PubMed] [Google Scholar]
- 11.Clarkson J, Herbison AE. Postnatal development of kisspeptin neurons in mouse hypothalamus; sexual dimorphism and projections to gonadotropin-releasing hormone neurons. Endocrinology. 2006;147:5817–5825. doi: 10.1210/en.2006-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W, Jessen HM, Auger AP, Terasawa E. Postmenopausal increase in KiSS-1, GPR54, and luteinizing hormone releasing hormone (LHRH-1) mRNA in the basal hypothalamus of female rhesus monkeys. Peptides. 2009;30:103–110. doi: 10.1016/j.peptides.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith JT, Clay CM, Caraty A, Clarke IJ. KiSS-1 messenger ribonucleic acid expression in the hypothalamus of the ewe is regulated by sex steroids and season. Endocrinology. 2007;148:1150–1157. doi: 10.1210/en.2006-1435. [DOI] [PubMed] [Google Scholar]
- 14.Hrabovszky E, Ciofi P, Vida B, Horvath MC, Keller E, Caraty A, Bloom SR, Ghatei MA, Dhillo WS, Liposits Z, Kallo I. The kisspeptin system of the human hypothalamus: sexual dimorphism and relationship with gonadotropin-releasing hormone and neurokinin B neurons. Eur J Neurosci. 2010;31:1984–1998. doi: 10.1111/j.1460-9568.2010.07239.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, Clifton DK, Steiner RA. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology. 2005;146:2976–2984. doi: 10.1210/en.2005-0323. [DOI] [PubMed] [Google Scholar]
- 16.Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- 17.Clarkson J, d'Anglemont dT X, Moreno AS, Colledge WH, Herbison AE. Kisspeptin-GPR54 signaling is essential for preovulatory gonadotropin-releasing hormone neuron activation and the luteinizing hormone surge. J Neurosci. 2008;28:8691–8697. doi: 10.1523/JNEUROSCI.1775-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomikawa J, Uenoyama Y, Ozawa M, Fukanuma T, Takase K, Goto T, Abe H, Ieda N, Minabe S, Deura C, Inoue N, Sanbo M, Tomita K, Hirabayashi M, Tanaka S, Imamura T, Okamura H, Maeda K, Tsukamura H. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc Natl Acad Sci U S A. 2012;109:E1294–E1301. doi: 10.1073/pnas.1114245109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.West A, Vojta PJ, Welch DR, Weissman BE. Chromosome localization and genomic structure of the Kiss-1 metastasis suprressor gene (KISS1) Genomics. 1998;54:145–148. doi: 10.1006/geno.1998.5566. [DOI] [PubMed] [Google Scholar]
- 21.Mueller JK, Dietzel A, Lomniczi A, Loche A, Tefs K, Kiess W, Danne T, Ojeda SR, Heger S. Transcriptional regulation of the human KiSS1 gene. Mol Cell Endocrinol. 2011;342:8–19. doi: 10.1016/j.mce.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huijbregts L, de RN. KISS1 is down-regulated by 17beta-estradiol in MDA-MB-231 cells through a nonclassical mechanism and loss of ribonucleic acid polymerase II binding at the proximal promoter. Endocrinology. 2010;151:3764–3772. doi: 10.1210/en.2010-0260. [DOI] [PubMed] [Google Scholar]
- 23.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 24.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 25.Lomniczi A, Cornea A, Costa ME, Ojeda SR. Hypothalamic tumor necrosis factor-a converting enzyme (TACE) mediates excitatory amino acid-dependent neuron-to-glia signaling in the neuroendocrine brain. J Neurosci. 2006;26:51–62. doi: 10.1523/JNEUROSCI.2939-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ojeda SR, Urbanski HF. Puberty in the rat. In: Knobil E, Neill JD, editors. The Physiology of Reproduction. 2nd Edition. Vol 2. New York: Raven Press; 1994. pp. 363–409. [Google Scholar]
- 29.Ojeda SR, Skinner MK. Puberty in the rat. In: Neill JD, editor. The Physiology of Reproduction. 3rd Edition. San Diego: Academic Press/Elsevier; 2006. pp. 2061–2126. [Google Scholar]
- 30.Urbanski HF, Ojeda SR. The juvenile-peripubertal transition period in the female rat: Establishment of a diurnal pattern of pulsatile luteinizing hormone secretion. Endocrinology. 1985;117:644–649. doi: 10.1210/endo-117-2-644. [DOI] [PubMed] [Google Scholar]
- 31.Ojeda SR, Terasawa E. Neuroendocrine regulation of puberty. In: Pfaff D, Arnold A, Etgen A, Fahrbach S, Moss R, Rubin R, editors. Hormones, Brain and behavior. Vol 4. New York: Elsevier; 2002. pp. 589–659. [Google Scholar]
- 32.Kauffman AS, Navarro VM, Kim J, Clifton DK, Steiner RA. Sex differences in the regulation of Kiss1/NKB neurons in juvenile mice: implications for the timing of puberty. Am J Physiol Endocrinol Metab. 2009;297:E1212–E1221. doi: 10.1152/ajpendo.00461.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takase K, Uenoyama Y, Inoue N, Matsui H, Yamada S, Shimizu M, Homma T, Tomikawa J, Kanda S, Matsumoto H, Oka Y, Tsukamura H, Maeda KI. Possible role of oestrogen in pubertal increase of Kiss1/kisspeptin expression in discrete hypothalamic areas of female rats. J Neuroendocrinol. 2009;21:527–537. doi: 10.1111/j.1365-2826.2009.01868.x. [DOI] [PubMed] [Google Scholar]
- 34.Gottsch ML, Navarro VM, Zhao Z, Glidewell-Kenney C, Weiss J, Jameson JL, Clifton DK, Levine JE, Steiner RA. Regulation of Kiss1 and dynorphin gene expression in the murine brain by classical and nonclassical estrogen receptor pathways. J Neurosci. 2009;29:9390–9395. doi: 10.1523/JNEUROSCI.0763-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitchell DC, Abdelrahim M, Weng J, Stafford LJ, Safe S, Bar-Eli M, Liu M. Regulation of KiSS-1 metastasis suppressor gene expression in breast cancer cells by direct interaction of transcription factors activator protein-2alpha and specificity protein-1. J Biol Chem. 2006;281:51–58. doi: 10.1074/jbc.M506245200. [DOI] [PubMed] [Google Scholar]
- 36.Li D, Yu W, Liu M. Regulation of KiSS1 gene expression. Peptides. 2009;30:130–138. doi: 10.1016/j.peptides.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 37.Safe S. Transcriptional activation of genes by 17 beta-estradiol through estrogen receptor-Sp1 interactions. Vitam Horm. 2001;62:231–252. doi: 10.1016/s0083-6729(01)62006-5. [DOI] [PubMed] [Google Scholar]
- 38.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, Buettner R. Transcriptional regulation of the AP-2alpha promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen X, Hu Y, Jiang Y, Liu H, Zhu L, Jin X, Shan H, Zhen X, Sun L, Yan G, Sun H. Kruppel-like factor 12 negatively regulates human endometrial stromal cell decidualization. Biochem Biophys Res Commun. 2013;433:11–17. doi: 10.1016/j.bbrc.2013.02.078. [DOI] [PubMed] [Google Scholar]
- 40.Mitchell DC, Stafford LJ, Li D, Bar-Eli M, Liu M. Transcriptional regulation of KiSS-1 gene expression in metastatic melanoma by specificity protein-1 and its coactivator DRIP-130. Oncogene. 2007;26:1739–1747. doi: 10.1038/sj.onc.1209963. [DOI] [PubMed] [Google Scholar]
- 41.Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, Li X, Ning G, Wu X, Liu M. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148:4821–4828. doi: 10.1210/en.2007-0154. [DOI] [PubMed] [Google Scholar]
- 42.Jacobi JS, Martin C, Nava G, Jeziorski MC, Clapp C, Martinez dlE. 17-Beta-estradiol directly regulates the expression of adrenergic receptors and kisspeptin/GPR54 system in GT1-7 GnRH neurons. Neuroendocrinology. 2007;86:260–269. doi: 10.1159/000107770. [DOI] [PubMed] [Google Scholar]
- 43.Lomniczi A, Loche A, Castellano JM, Ronnekleiv OK, Bosh M, Kaidar G, Knoll JG, Wright H, Pfeifer GP, Ojeda SR. Epigenetic control of female puberty. Nature Neuroscience. 2013;16:281–289. doi: 10.1038/nn.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker DM, Kirson D, Perez LF, Gore AC. Molecular profiling of postnatal development of the hypothalamus in female and male rats. Biol Reprod. 2012;87:129. doi: 10.1095/biolreprod.112.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takumi K, Iijima N, Ozawa H. Developmental changes in the expression of kisspeptin mRNA in rat hypothalamus. J Mol Neurosci. 2011;43:138–145. doi: 10.1007/s12031-010-9430-1. [DOI] [PubMed] [Google Scholar]
- 46.Navarro VM, Ruiz-Pino F, Sanchez-Garrido MA, Garcia-Galiano D, Hobbs SJ, Manfredi-Lozano M, Leon S, Sangiao-Alvarellos S, Castellano JM, Clifton DK, Pinilla L, Steiner RA, Tena-Sempere M. Role of neurokinin B in the control of female puberty and its modulation by metabolic status. J Neurosci. 2012;32:2388–2397. doi: 10.1523/JNEUROSCI.4288-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gill JC, Wang O, Kakar S, Martinelli E, Carroll RS, Kaiser UB. Reproductive hormone-dependent and -independent contributions to developmental changes in kisspeptin in GnRH-deficient hypogonadal mice. PLoS ONE. 2010;5:e11911. doi: 10.1371/journal.pone.0011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 49.Pavithra L, Sreenath K, Singh S, Chattopadhyay S. Heat-shock protein 70 binds to a novel sequence in 5' UTR of tumor suppressor SMAR1 and regulates its mRNA stability upon Prostaglandin A2 treatment. FEBS Lett. 2010;584:1187–1192. doi: 10.1016/j.febslet.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 50.Zhao X, Chen J, Lei L, Hu G, Xiong Y, Xu J, Li Q, Yang X, Chang CC, Song B, Chang T, Li B. The optional long 5'-untranslated region of human ACAT1 mRNAs impairs the production of ACAT1 protein by promoting its mRNA decay. Acta Biochim Biophys Sin (Shanghai) 2009;41:30–41. doi: 10.1093/abbs/gmn004. [DOI] [PubMed] [Google Scholar]
- 51.Wang G, Guo X, Floros J. Differences in the translation efficiency and mRNA stability mediated by 5'-UTR splice variants of human SP-A1 and SP-A2 genes. Am J Physiol Lung Cell Mol Physiol. 2005;289:L497–L508. doi: 10.1152/ajplung.00100.2005. [DOI] [PubMed] [Google Scholar]
- 52.Gauss KA, Bunger PL, Crawford MA, McDermott BE, Swearingen R, Nelson-Overton LK, Siemsen DW, Kobayashi SD, Deleo FR, Quinn MT. Variants of the 5'-untranslated region of human NCF2: expression and translational efficiency. Gene. 2006;366:169–179. doi: 10.1016/j.gene.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 53.Capri M, Santoni MJ, Thomas-Delaage M, Ait-Ahmed O. Implication of a 5' coding sequence in targeting maternal mRNA to the Drosophila oocyte. Mech Dev. 1997;68:91–100. doi: 10.1016/s0925-4773(97)00130-5. [DOI] [PubMed] [Google Scholar]
- 54.Saunders C, Cohen RS. The role of oocyte transcription, the 5'UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- 55.Hartzog GA. Transcription elongation by RNA polymerase II. Curr Opin Genet Dev. 2003;13:119–126. doi: 10.1016/s0959-437x(03)00024-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.