Abstract
Mitophagy is a process of mitochondrial turnover through lysosomal mediated autophagy activities. This review will highlight recent studies that have identified mediators of mitophagy in response to starvation, loss of mitochondrial membrane potential or perturbation of mitochondrial integrity. Furthermore, we will review evidence of mitophagy dysfunction in various human diseases and discuss the potential for therapeutic interventions that target mitophagy processes.
1. Introduction
Mitochondria are important for cell metabolism and ATP generation via oxidative phosphorylation through the electron transport chain, fatty acid oxidation, and TCA cycle. In addition, mitochondria are responsible for their own DNA replication, transcription and translation of mitochondrial proteins, import of nuclear-encoded proteins, calcium storage, production and scavenging of oxidants, signaling, and sequestration of pro-apoptotic proteins to sustain cell survival (Newmeyer and Ferguson-Miller, 2003; Mitchell et al., 2013a; Dodson et al., 2013). Maintenance of a healthy mitochondrial population is essential for cellular function and survival, and is controlled by balancing biogenesis and turnover of mitochondria via autophagy (mitophagy) (Lee et al., 2012;Dodson et al., 2013;Zhang, 2013;Hill et al., 2012) This review will focus on the mechanisms by which mitochondria are removed by mitophagy, and highlight evidence that mitophagy dysregulation contributes to human diseases.
2. Molecular mechanisms of mitochondrial quality control through mitophagy
2.1 Mitophagy mediated by adaptor proteins
Autophagy is a lysosomal mediated degradation program (Lee et al., 2012;Dodson et al., 2013;Zhang 2013;Hill et al., 2012). The term “mitophagy” was officially proposed by JJ Lemasters in 2005 to describe a selective process of mitochondrial degradation by autophagy (Lemasters, 2005). Since then the molecular mechanisms of selective mitophagy have been described in some detail, particularly in yeast. The mitochondrial outer membrane protein Uth1p was the first protein identified to be essential for autophagic removal of mitochondria in yeast in response to rapamycin or nitrogen starvation, while dispensable for non-specific autophagy activities (Kissova et al., 2004). Another example is Aup1p, which is located in the mitochondrial intermembrane space and is important for mitophagy during growth in stationary phase (Tal et al., 2007). It has been shown that Atg11 is dispensable for non-specific autophagy but essential for selective autophagy. Atg32 localizes to the outer mitochondrial membrane in response to starvation or when cells reach the post-log phase in non-fermentable glycerol medium (Kanki et al., 2009;Okamoto et al., 2009). An increase in Atg32 levels occurs in post-log phase, and is mitigated by N-acetylcysteine, suggesting regulation of Atg32 levels by intracellular glutathione (Okamoto et al., 2009). Atg32 also interacts with Atg8 and Atg11, thus recruiting mitochondria to the autophagosome (Kanki et al., 2009;Okamoto et al., 2009). The C-terminus on Atg11 interacts with the N-terminus on Atg32 when Atg32 is phosphorylated at Ser-114 and 119 by casein kinase 2, and it has been shown that the phosphorylation is also dependent on Pbs1-Hog1 activities (Aoki et al., 2011;Kanki et al., 2013). Proteolytic cleavage of the C-terminus of Atg32 by mitochondrial i-AAA protease Yme1 also enhances the interaction between Atg32 and Atg11 (Wang et al., 2013). These findings demonstrate critical protein-protein interactions in yeast that allows for specific, regulated removal of mitochondria using the autophagic machinery (Fig.1).
Figure 1. Adaptor proteins in mitophagy.
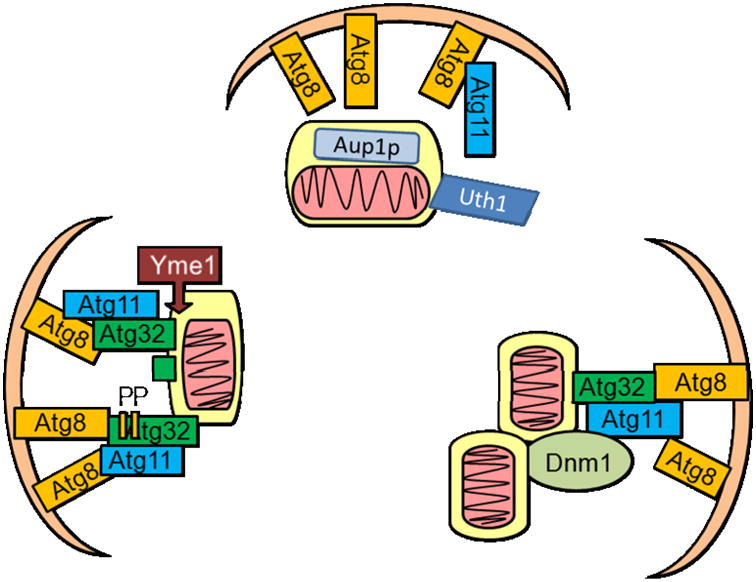
In yeast, selective autophagy of mitochondria is mediated by mitochondrial targeted proteins such as Uth1, Aup1p, or Atg32. While the mechanisms of interaction between Aup1p and Uth1 and the autophagy machinery are still unclear, more is known regarding Atg32 function in mitophagy. Under starvation conditions, Atg32 localizes to the mitochondria and interacts with Atg8 and Atg11 to bring mitochondria to the autophagosome. Phosphorylation of Atg32 at Ser-114 and Ser-119, or cleavage of Atg32 by Yme1, facilitates its interaction with Atg11. Atg11 interaction with mitochondrial fission protein Dnm1 and recruitment of fission complex to the mitochondria are also required for mitophagy in response to nitrogen starvation.
Less is known about selective adaptor proteins in mammalian cells, and there is currently no known mammalian homolog of Uth1 or Atg32. It has been observed that during mammalian erythrocyte maturation, NIX targets to the mitochondria, binds LC3 or LC3 homologs, and plays an important role in mitophagy (Sandoval et al., 2008;Novak et al., 2010). Additionally, BNIP3 and FUNDC1 also contain LC3 interacting regions and play a role in mitophagy under hypoxic conditions (Zhang et al., 2008;Liu et al., 2012). In cells undergoing high rates of oxidative phosphorylation, the GTPase RHEB can be recruited to the mitochondria, where it interacts with NIX and LC3 to promote mitophagy and maintain an active and healthy mitochondrial pool (Melser et al., 2013).
2.2 Mitophagy regulation by mitochondrial dynamics and membrane potential
Mitochondrial dynamics are emerging as an important aspect of cellular physiology, and play an important role in regulating mitophagy (Twig and Shirihai, 2011). In yeast, the Atg32/Atg11 complex recruits the fission machinery in response to nitrogen starvation through an interaction between Atg11 and Dnm1 (Fig.1). Mutations of the Atg11-interaction domain on Dnm1 protein decreased mitophagy (Mao et al., 2013). In mammalian cells, mitophagy has been shown to occur coordinately with fission events in hepatocytes (Kim and Lemasters, 2011). Inhibition of mitochondrial fission by either Fis1 siRNA or a dominant negative form of DRP-1 prevents mitophagy in INS-1 β-cells (Twig G et al., 2008). In mouse embryonic fibroblasts (MEFs), elimination of either glutamine or amino acids from the growth medium, but not elimination of glucose or serum, resulted in mitochondrial tubulation. MEFs with Opa1 or Mfn2 gene knockout exhibited increased mitophagy during starvation (Rambold et al., 2011). The mechanisms through which changes of fission or fusion due to Drp1, Opa1 or Mfn2 knockdown or knockout impact mitophagy may involve changes not only in mitochondrial morphology but also in mitochondrial membrane potential and bioenergetics (Frank et al., 2001;Olichon et al., 2003;Duvezin-Caubet et al., 2006;Narendra et al., 2008). Recent evidence also suggests that FIS1 may act downstream of DRP-1, in response to calcium ionophores in C.elegans, or to Antimycin A or CCCP in HCT116 cells, to initiate formation of nascent mitophagosomes by participation in the mitochondrion-associated membrane (MAM) complex (Shen et al., 2014). These studies demonstrate that mitochondrial fission/fusion machinery plays a pivotal role in the regulation of mitophagy.
In addition to fission and fusion, mitochondrial membrane potential plays an important role in mitophagy. In rat hepatocytes, serum deprivation has been shown to induce mitochondrial depolarization and engulfment by autophagosomes (Elmore et al., 2001). How mitochondrial depolarization leads to mitophagy has been extensively studied in the context of understanding potential pathogenic mechanisms of Parkinson's disease, and it has been found that recessive genes identified in familial Parkinson's disease, PARKIN, PINK1 and DJ-1, encode proteins involved in mitophagy. Studies using Drosophila models and established human cell lines have found that upon depolarization, PINK1 (PTEN–induced putative kinase 1) is imported into the mitochondria by the Translocase of the Outer Membrane (TOM) and Translocase of the Inner Membrane (TIM) complexes. PINK1 then anchors at the inner mitochondrial membrane (IMM) where it is processed by the rhomboid protease of the IMM, presenilin associated rhomboid-like protease (PARL) (Michiorri et al., 2010), and degraded by mitochondrial processing peptidases (Jin et al., 2010). However, when mitochondrial membrane potential is lost, PINK1 is not cleaved by PARL, and thus accumulates in the mitochondria (Jin et al., 2010).
Following accumulation of PINK1, the cytosolic E3 ubiquitin ligase PARKIN is recruited to the mitochondria, where it ubiquitinates various mitochondrial proteins (Geisler et al., 2010b; Narendra et al., 2010b; Vives-Bauza et al., 2010). Endogenous PARKIN has been shown to ubiquitinate mitochondrial localized MFN1 and MFN2 in both Drosophila and human cells (Gegg et al., 2010;Poole et al., 2010;Rakovic et al., 2011;Ziviani et al., 2010;Tanaka et al., 2010), while overexpressed PARKIN can also ubiquitinate VDAC (Voltage Dependent Anion Channel), components of the mitochondrial transport TOM complex, fission protein FIS1, pro-apoptotic protein BAK, mitochondrial movement Rho GTPases (MIRO) 1 and 2, and mitochondrial hexokinase (Yoshii et al., 2011;Chan et al., 2011;Okatsu et al., 2012;Sarraf et al., 2013). These ubiquitinated mitochondrial proteins can either be degraded by the proteasome, thus coordinating mitochondrial shape, cell metabolism, and mitochondrial movement with mitophagy, or be recognized by an LC3-and ubiquitin-binding autophagy adaptor protein p62/SQSTM1 thereby promoting mitophagy (Tanaka et al., 2010;Geisler et al., 2010a;Lee et al., 2010;Narendra et al., 2010a). Recent studies provided evidence that PARKIN interaction with AMBRA1, PINK1 interaction with BECN1, or sterile α and TIR motif containing 1 (SARM1) and tumor necrosis factor receptor-associated factor 6 (TRAF6), may also play a role in mitophagy (Van et al., 2011;Michiorri et al., 2010;Murata et al., 2013). These studies suggest an essential role of PINK1 and PARKIN in mediating mitophagy of depolarized mitochondria (Fig.2).
Figure 2. Mitophagy in response to loss of membrane potential.
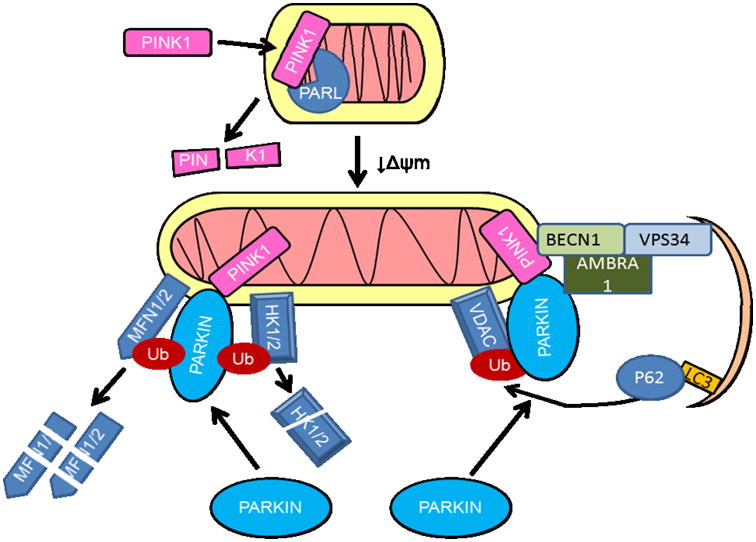
In healthy mitochondria PINK1 (PTEN – induced putative kinase 1) is targeted to the mitochondrial inner membrane where it is cleaved by the rhomboid protease of the IMM, presenilin associated rhomboid-like protease (PARL), and degraded. In stressed mitochondria with depleted membrane potential (↓Δψm), PARL is inactivated, and PINK1 accumulates in the mitochondria, recruiting the E3 ubiquitin ligase PARKIN. PARKIN can ubiquitinate Mitofusins 1 and 2 (MFN1 and 2), hexokinases, TOM complex components, FIS1, BAK, MIRO as well as VDAC. The ubiquitinated proteins are either degraded by the proteasome, or recognized by ubiquitin and LC3-binding autophagy adaptor protein p62/SQSTM1 which recruits them to autophagosomes. PARKIN-AMBRA1 interaction, or PINK1-BECN1 interaction can also facilitate mitophagy.
In parallel with the PINK1/PARKIN pathway, DJ-1 is also involved in removal of damaged mitochondria (Thomas et al., 2011). DJ-1 levels increase at mitochondria following oxidative damage in both fibroblasts and neurons, with mitochondrial removal dependent on PARKIN and possibly PINK1 (Joselin et al., 2012). DJ-1 knockout neuroblastoma cells exhibit reduced mitochondrial membrane potential and increased mitochondrial fragmentation that is reversible upon treatment with glutathione (McCoy and Cookson, 2011). These models demonstrate the importance of key mitophagy genes in maintaining proper mitochondrial health and function, as well as preventing damage associated with an increased production of free radical species.
It should be noted that PINK1 and PARKIN translocation to damaged mitochondria either by direct depolarization or exposure to other mitochondrial toxins is highly dependent on cell type, levels of PINK1 and PARKIN expression, and growth conditions (Rakovic et al., 2013;Rakovic et al., 2010;Van, V et al., 2011;Cai et al., 2012;Vives-Bauza et al., 2010). Additional factors, including AF-6 (Haskin et al., 2013), and FBXO7 (Burchell et al., 2013), have been identified as regulating mitophagy by direct interactions with PARKIN.
Several Genome-wide screens have further identified other mitophagy regulators (Orvedahl et al., 2011;Lefebvre et al., 2013;Hasson et al., 2013). In a screen to detect genes required for virophagy, 96 were found to play a role in PARKIN-mediated mitophagy (Orvedahl et al., 2011). From this screen, functions of one particular protein, SMURF1 (SMAD specific E3 ubiquitin protein ligase 1 which encodes a HECT-domain ubiquitin ligase), have been validated in mouse knockouts as they accumulate damaged mitochondria in the heart, brain and liver (Orvedahl et al., 2011). Another independent screen has identified ATPIF1/IF1 (ATPase inhibitory factor 1) as promoting collapse of ΔΨ and enabling PARKIN recruitment to the mitochondria and mitophagy by blocking ATPase activity (Lefebvre et al., 2013). A third genome-wide siRNA screen further identified factors, including TOMM7, SIAH7, HSPA1L and BAG4, that can either enhance or decrease PARKIN accumulation at the mitochondria (Hasson et al., 2013). Validation in additional cell lines confirmed that TOMM7 is not required for TOM-mediated general protein import, but plays an important role in anchoring PINK1 when ΔΨ is depleted, in both HCT116 cells and in human iPSC derived tyrosine hydroxylase positive neurons. SIAH3 knockdown in BE(2)-M17 neuroblastoma cells increased both PINK1 accumulation and PARKIN translocation to depolarized mitochondria. Independent of PINK1 protein accumulation, HSPA1L enhances, whereas BAG4 prevents, PARKIN translocation to the mitochondria by direct interaction with PARKIN (Hasson et al., 2013).
Downstream of PARKIN activation, it has been found that TBC1D15, a mitochondrial Rab GTPase-activating protein (Rab-GAP), inhibits excessive Rab7 activity, and associates with both the mitochondria through binding FIS1 and the isolation membrane through binding to LC3 family proteins (Yamano et al., 2014). In addition to TBC1D15, TBC1D17 interacts with FIS1 and TBC1D15, and facilitates TBC1D15 function (Yamano et al., 2014).
2.3 Mitophagy regulation by MTOR, mitochondrial membrane lipids and mitochondrial protein modifications
Recent studies have also identified additional mechanisms of mitophagy regulation in response to mtDNA damage, mitochondrial toxins, loss of iron, mitochondrial protein modification or viral infections. In bovine aortic endothelial cells, exposure to hemin led to mitochondrial dysfunction and activation of mitophagy (Higdon et al., 2012). In cybrid cells with a variety of pathogenic mtDNA mutations, loss of mitochondrial membrane potential alone could not trigger mitophagy. But when coupled with the inhibition of MTOR by rapamycin, mitophagy occurred (Gilkerson et al., 2012). In Akita+/Ins2-derived β-cells, a mutation in the insulin 2 gene led to ER stress and mitochondrial damage, which was associated with increased DRP1 and decreased MFN1, p62 and PARKIN (Mitchell et al., 2013b). In primary human fibroblasts, iron chelation induced mitophagy, which was independent of PINK1 or PARKIN, but interestingly was dependent on glycolysis (Allen et al., 2013).
Starvation-induced PI3K/AKT activities block the translocation of DRAM to mitochondria by direct physical interactions between p-AKT and DRAM, thereby attenuating mitophagy (Liu et al., 2014). Additional factors, including HMGB1 and HSPB1 enable mitophagy, while the exact mechanism of their actions are yet to be defined (Tang et al., 2011). Interestingly, PINK1 can also engage mitophagy of energetically healthy mitochondria in response to expression of unfolded proteins in the mitochondrial matrix, or when LONP1, a mitochondrial protease, is knocked down (Jin and Youle, 2013). In addition, autophagosome-independent, PARKIN and PINK1-dependent lysosome-mediated degradation of mitochondria has been reported (McLelland et al., 2014).
In primary cortical neurons and SH-SY5Y cells, rotenone, staurosporine, and 6-hydroxydopamine caused externalization of the mitochondrial lipid cardiolipin to the mitochondrial surface, which is then recognized by LC3 to signal removal by mitophagy. When cardiolipin synthesis or transport is blocked by siRNA-mediated knockdown of cardiolipin synthase or phospholipid scramblase-3, mitochondria are no longer engulfed by the autophagosome, indicating that cardiolipin externalization is necessary for mitophagy in response to these toxins (Chu et al., 2013).
In HepG2 cells, siRNA mediated knockdown of GCN5L1, an essential component of the mitochondrial acetyltransferase program, leads to an increase of mitochondrial associated LC3-II, p62 and protein ubiquitination, as well as an increase of LAMP-1 colocalization with the mitochondria, and decreased mitochondrial mass and protein levels (Webster et al., 2013). The GCN5L1 knockdown induced mitophagy is dependent on SIRT3, ATG5 and p62, but not PARKIN (Webster et al., 2013). The whole cell levels of p-S6K, p62 and LC3-II/LC3-I are unchanged in response to GCN5L1 knockdown, indicating a specific effect on mitophagy (Webster et al., 2013). Interestingly, the increased mitophagy in response to GCN5L1 depletion led to an increased resistance to mitochondrial stressors such as rotenone and paraquat (Webster et al., 2013).
3. Mitophagy in human diseases
Here we summarize evidence that mitophagy is important for mitochondrial quality control in vivo, and that mitophagic deficiencies exacerbate pathologies in disease situations, with a main focus on Parkinson's disease, cancer, heart and liver diseases (Table 1).
Table 1. Evidence that mitophagy is involved in Parkinson's disease, cancer, heart and liver diseases.
| Mitophagy dysregulation | Pathology | References |
|---|---|---|
| DJ-1 deficient PD patients | Mitochondrial dysfunction in lymphoblasts | (Irrcher et al., 2010) |
| PINK1 deficient PD patients | Mitochondrial dysfunction in fibroblasts | (Abramov et al., 2011) |
| PARKIN deficient PD patient | Mitochondrial dysfunction in iPSC-derived neurons | (Imaizumi et al., 2012) |
| PARKIN mutation or down-regulation | Glioblastoma, breast cancer, ovarian cancer, sporadic colorectal cancer, hepatocellular carcinoma, and pancreatic tumors | (Letessier et al., 2007;Fujiwara et al., 2008;Veeriah et al., 2010;Poulogiannis et al., 2010;Sun et al., 2013) |
| PINK1 mRNA expression | Survival prognostic marker in adrenocortical (ACT) tumors | (de Reynies A. et al., 2009;Fragoso et al., 2012) |
| GABARAPL1 down-regulation | breast tumors | (Berthier et al., 2010) |
| Impairment of fission | Tumorigenic lung epithelial A549 cells with decreased apoptosis, autophagy and mitophagy | (Thomas and Jacobson 2012) |
| Mice deficient in Atg5 in adult heart, Bnip3 or Nix | Accumulate damaged or dysfunctional mitochondria and develop cardiac dysfunction | (Nakai et al., 2007;Dorn 2010) |
| Mice deficient in Pink1 | Early onset cardiac hypertrophy | (Billia et al., 2011) |
| Mfn2 deficiency in mouse cardiomyocytes | Decreased depolarization-induced PARKIN translocation to the mitochondria, suppressed mitophagy, accumulation of abnormal mitochondria, dilated cardiomyopathy | (Chen and Dorn 2013) |
| PARKIN | Involved in survival in response to hemodynamic stress, infarction, doxorubicin toxicity, and ischemia; as well as cardioprotection of simvastatin | (Nakai et al., 2007;Kubli et al., 2013;Huang et al., 2011;Hoshino et al., 2013;Andres et al., 2013) |
| DNM1L | Involved in mitophagic depletion of mitochondria in response to environmental liver toxin cadmium | (Pi et al., 2013). |
| MFN1 and MFN2 | Involved in mitophagy in response to acetaminophen-induced liver injury and may play a role in compensatory proliferation of cells adjacent to the necrotic centrilobular area | (Ni et al., 2013) |
| BNIP3 deficiency | Increased mitochondrial mass, increased mitochondrial populations with a loss of membrane potential, abnormal morphology, decreased mitochondrial function, and increased inflammation and steatohepatitis | (Glick et al., 2012) |
3.1 Mitophagy in Parkinson's disease
Mutations of mitophagy genes PARKIN, PINK1, and DJ-1 have been associated with autosomal recessive Parkinson's disease (Trinh and Farrer, 2013;Lubbe and Morris, 2013). Mitochondrial dysfunction has also been reported in lymphoblasts carrying DJ-1mutations (Irrcher et al., 2010), fibroblasts carrying PINK1 mutations (Abramov et al., 2011), and in induced pluripotent stem cell (iPSC)-derived neurons carrying PARKIN mutations (Imaizumi et al., 2012).
Pesticides that target the mitochondria, such as rotenone and paraquat, have been associated with increased risk of developing Parkinson's disease (Tanner et al., 2011). As discussed above in 2.3, rotenone induces mitophagy in SH-SY5Y cells and primary neurons, with externalization of cardiolipin acting as the signal to remove damaged mitochondria (Chu et al., 2013). Externalized cardiolipin is recognized and bound by LC3 to recruit mitochondria to the autophagosome (Chu et al., 2013). This mechanism of mitophagy is independent of mitochondrial depolarization, as well as PINK1 or p62 recruitment to the mitochondria, and may represent a more controlled removal mechanism for mitochondrial turnover (Chu et al., 2013). MPP+, a metabolite of MPTP which induces parkinsonism in humans, has been shown to induce mitophagy in neuroblastoma SH-SY5Y cells in a MAPK, ATG5, ATG7, and ATG8 dependent, but BECN1 independent manner (Zhu et al., 2007).
3.2 Mitophagy in cancer
Mitochondrial quality control is important, as dysfunctional mitochondria may contribute to accumulation of reactive oxygen species (ROS) (Murphy, 2009), which can then damage nuclear and mtDNA, inducing genome instability and tumor initiation. In addition, mitophagy alteration in the host stromal microenvironment may also play a role in nutrient recycling and promotion of cancer growth. For example, cigarette smoke promotes DNA damage, autophagy and mitophagy in stromal fibroblasts, leading to the secretion of L-lactate and ketone bodies to support neighboring cancer cells to proliferate (Salem et al., 2013). Finally, significant alteration of levels of proteins that impact mitophagy has been found in various tumors.
PARKIN has been proposed as a tumor suppressor gene as it is mutated and down-regulated in different tumors, such as glioblastoma, breast cancer, ovarian cancer, sporadic colorectal cancer, hepatocellular carcinoma, and pancreatic tumors (Letessier et al., 2007;Fujiwara et al., 2008;Veeriah et al., 2010;Poulogiannis et al., 2010;Sun et al., 2013). Another mitophagy effector, PINK1, induced a decrease in cell growth in soft agar when overexpressed in MCF-7 cells (Berthier et al., 2011). PINK1 mRNA expression has also been proposed to serve as a survival prognostic marker in adrenocortical (ACT) tumors (de Reynies A. et al., 2009;Fragoso et al., 2012).
BNIP3 has been shown to be silenced in pancreatic cancer compared to normal tissue (Erkan et al., 2005;Okami et al., 2004). It has been proposed that BNIP3 and mitophagy protects cells against tumorigenesis by controlling intracellular levels of ROS, but when cells undergo a transformation event, the BNIP3 expression is lost. This leads to the accumulation of damaged mitochondria, increased ROS, genomic instability and progression to a more advanced stage of pancreatic cancer (Lu and Harrison-Findik, 2013). NIX/BNIP3L (BNIP3-like protein) expression is a factor of good prognosis for astrocytomas (AS, grade II). In hepatocellular carcinomas (HCC), BNIP3L and BNIP3 silencing has been linked to a poor prognosis (Calvisi et al., 2007). Interestingly, the silencing of these genes in these tumors has been correlated to hypermethylation.
GABARAPL1 (GABAA receptor associated protein like 1), one of the ATG8 homologues in mammals (Le Grand et al., 2011), has been shown to be associated with autophagosomes (Chakrama et al., 2010) and to interact with NIX to participate in the specific degradation of mitochondria in reticulocytes (Novak et al., 2010). It has also been demonstrated that GABARAPL1 expression is decreased in breast tumors and that a high expression of its mRNA is associated with a better outcome for lymph node-positive breast cancer patients (Berthier et al., 2010).
Mitochondrial fission is increased in human lung cancer cells (Rehman et al., 2012) and invasive breast carcinoma (Zhao et al., 2013). Impairment of fission in tumorigenic lung epithelial A549 cells decreased apoptosis, autophagy and mitophagy (Thomas and Jacobson, 2012). It should be noted that although mitophagy genes are frequently down-regulated in many tumors, and their activation has been shown to inhibit cancer cell proliferation or tumor growth in nude mice, few have directly tested whether tumorigenesis or tumor growth is directly linked to mitophagy. This will need to be investigated in future studies.
3.3 Mitophagy in diseases in the heart and liver
Heart failure, aging, cardiomyopathy, and ischemia-reperfusion injury are all associated with mitochondrial dysfunction, indicating that mitochondrial quality control through mitophagy may play an important role in limiting cellular damage (Dutta et al., 2012). Mice deficient in Atg5 in the adult heart, Bnip3 or Nix, accumulate damaged or dysfunctional mitochondria and develop cardiac dysfunction (Nakai et al., 2007;Dorn, 2010). Mice deficient in Pink1 develop early onset cardiac hypertrophy (Billia et al., 2011). Recent studies also found that Mfn2 deficiency in mouse cardiomyocytes prevented depolarization-induced PARKIN translocation to the mitochondria and suppressed mitophagy, leading to accumulation of morphologically and functionally abnormal mitochondria, and dilated cardiomyopathy (Chen and Dorn, 2013),
PARKIN is also important for survival in response to hemodynamic stress (Nakai et al., 2007). Parkin deficient mice exhibit decreased mitophagy, accumulation of dysfunctional mitochondria, and decreased survival after infarction (Kubli et al., 2013). Ischemia-induced mitophagy appears to involve BNIP3, and is inhibited by p53-mediated induction of TIGAR (TP53-induced glycolysis and apoptosis regulator) (Hoshino et al., 2012). Doxorubicin-induced mitophagy has also been shown to depend on PARKIN and inhibited by cytosolic p53 binding to PARKIN (Hoshino et al., 2013). In response to ischemic preconditioning, PARKIN and p62 are translocated to the mitochondria, and PARKIN is required for cardioprotection (Huang et al., 2011). A cholesterol lowering drug, simvastatin, increased PARKIN translocation to the mitochondria, as well as loss of mitochondrial connectivity and membrane TOM70 protein in HL-1 cells. In vivo, simvastatin protected against coronary artery ligation induced infarct, and the protection was diminished in PARKIN deficient mice, suggesting that cardioprotection of simvastatin is mediated by PARKIN (Andres et al., 2013). Mitophagy may also be important to control inflammation, as a recent study reported that mtDNA that escaped from autophagy led to Toll-like receptor (TLR) 9-mediated inflammatory responses in cardiomyocytes, myocarditis and dilated cardiomyopathy (Oka et al., 2012).
In the liver, mitophagy contributes to mitochondrial loss and bioenergetic deficit in response to the environmental toxin cadmium, which is mediated by upregulation and translocation of DNM1L to the mitochondria, followed by mitochondrial fragmentation and colocalization of mitochondria with the lysosomes (Pi et al., 2013). It has also been suggested that MFN1 or MFN2-dependent mitochondrial spheroid formation is important for mitophagy in response to acetaminophen-induced liver injury and may play a role in compensatory proliferation of cells adjacent to the necrotic centrilobular area (Ni et al., 2013). BNIP3 deficiency also led to increased mitochondrial mass, increased mitochondrial populations with a loss of membrane potential, abnormal morphology, decreased mitochondrial function, and increased inflammation and steatohepatitis (Glick et al., 2012).
4 Conclusions/Future Directions
Targeted removal of mitochondria via mitophagy is essential in maintaining mitochondrial quality, cell viability and homeostasis. Understanding of mitophagic mechanisms and their regulation in different tissues and cells under healthy and stressed conditions will help us better understand disease pathogenesis and develop more effective therapeutic approaches.
Organelle Facts.
Mitophagy is important for mitochondrial quality control
Mitophagy is regulated by interactive adaptor proteins in the autophagy pathway
Mitophagy is regulated by mitochondrial dynamics
Mitophagy is regulated by mitochondrial depolarization, PINK1 and PARKIN
Mitophagy dysregulation contributes to disease pathologies
Acknowledgments
This work was supported by NIHR01-NS064090 and a VA merit award (to JZ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Abramov AY, Gegg M, Grunewald A, Wood NW, Klein C, Schapira AH. Bioenergetic consequences of PINK1 mutations in Parkinson disease. PLoS ONE. 2011;6:e25622. doi: 10.1371/journal.pone.0025622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AM, Hernandez G, Lee P, et al. Mitophagy is Required for Acute Cardioprotection by Simvastatin. Antioxid Redox Signal. 2013 doi: 10.1089/ars.2013.5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Kanki T, Hirota Y, et al. Phosphorylation of Ser114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-02-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthier A, Navarro S, Jimenez-Sainz J, et al. PINK1 displays tissue-specific subcellular location and regulates apoptosis and cell growth in breast cancer cells. Hum Pathol. 2011;42:75–87. doi: 10.1016/j.humpath.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Berthier A, Seguin S, Sasco AJ, et al. High expression of gabarapl1 is associated with a better outcome for patients with lymph node-positive breast cancer. Br J Cancer. 2010;102:1024–1031. doi: 10.1038/sj.bjc.6605568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci U S A. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchell VS, Nelson DE, Sanchez-Martinez A, et al. The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat Neurosci. 2013;16:1257–1265. doi: 10.1038/nn.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A, Sheng ZH. Spatial Parkin Translocation and Degradation of Damaged Mitochondria via Mitophagy in Live Cortical Neurons. Curr Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvisi DF, Ladu S, Gorden A, et al. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrama FZ, Seguin-Py S, Le Grand JN, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013 doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Reynies A, Assie G, Rickman DS, et al. Gene expression profiling reveals a new classification of adrenocortical tumors and identifies molecular predictors of malignancy and survival. J Clin Oncol. 2009;27:1108–1115. doi: 10.1200/JCO.2008.18.5678. [DOI] [PubMed] [Google Scholar]
- Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn GW. Mitochondrial pruning by Nix and BNip3: an essential function for cardiac-expressed death factors. J Cardiovasc Transl Res. 2010;3:374–383. doi: 10.1007/s12265-010-9174-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Calvani R, Bernabei R, Leeuwenburgh C, Marzetti E. Contribution of impaired mitochondrial autophagy to cardiac aging: mechanisms and therapeutic opportunities. Circ Res. 2012;110:1125–1138. doi: 10.1161/CIRCRESAHA.111.246108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem. 2006;281:37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Erkan M, Kleeff J, Esposito I, et al. Loss of BNIP3 expression is a late event in pancreatic cancer contributing to chemoresistance and worsened prognosis. Oncogene. 2005;24:4421–4432. doi: 10.1038/sj.onc.1208642. [DOI] [PubMed] [Google Scholar]
- Fragoso MC, Almeida MQ, Mazzuco TL, et al. Combined expression of BUB1B, DLGAP5, and PINK1 as predictors of poor outcome in adrenocortical tumors: validation in a Brazilian cohort of adult and pediatric patients. Eur J Endocrinol. 2012;166:61–67. doi: 10.1530/EJE-11-0806. [DOI] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Marusawa H, Wang HQ, et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010a;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Treis A, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010b;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- Gilkerson RW, de Vries RL, Lebot P, et al. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet. 2012;21:978–990. doi: 10.1093/hmg/ddr529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Zhang W, Beaton M, et al. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012;32:2570–2584. doi: 10.1128/MCB.00167-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskin J, Szargel R, Shani V, et al. AF-6 is a positive modulator of the PINK1/parkin pathway and is deficient in Parkinson's disease. Hum Mol Genet. 2013;22:2083–2096. doi: 10.1093/hmg/ddt058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson SA, Kane LA, Yamano K, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013 doi: 10.1038/nature12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higdon AN, Benavides GA, Chacko BK, et al. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. Am J Physiol Heart Circ Physiol. 2012;302:H1394–H1409. doi: 10.1152/ajpheart.00584.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BG, Benavides GA, Lancaster JR, et al. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012 doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Matoba S, Iwai-Kanai E, et al. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- Hoshino A, Mita Y, Okawa Y, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning Involves Selective Mitophagy Mediated by Parkin and p62/SQSTM1. PLoS ONE. 2011;6:e20975. doi: 10.1371/journal.pone.0020975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y, Okada Y, Akamatsu W, et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrcher I, Aleyasin H, Seifert EL, et al. Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joselin AP, Hewitt SJ, Callaghan SM, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- Kanki T, Kurihara Y, Jin X, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–794. doi: 10.1038/embor.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol Cell Physiol. 2011;300:C308–C317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubli DA, Zhang X, Lee Y, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288:915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Grand JN, Chakrama FZ, Seguin-Py S, et al. GABARAPL1 (GEC1): Original or copycat? Autophagy. 2011;7 doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V, Du Q, Baird S, et al. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013;9 doi: 10.4161/auto.25413. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Letessier A, Garrido-Urbani S, Ginestier C, et al. Correlated break at PARK2/FRA6E and loss of AF-6/Afadin protein expression are associated with poor outcome in breast cancer. Oncogene. 2007;26:298–307. doi: 10.1038/sj.onc.1209772. [DOI] [PubMed] [Google Scholar]
- Liu K, Shi Y, Guo XH, et al. Phosphorylated AKT inhibits the apoptosis induced by DRAM-mediated mitophagy in hepatocellular carcinoma by preventing the translocation of DRAM to mitochondria. Cell Death Dis. 2014;5:e1078. doi: 10.1038/cddis.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- Lu SZ, Harrison-Findik DD. Autophagy and cancer. World J Biol Chem. 2013;4:64–70. doi: 10.4331/wjbc.v4.i3.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbe S, Morris HR. Recent advances in Parkinson's disease genetics. J Neurol. 2013 doi: 10.1007/s00415-013-7003-2. [DOI] [PubMed] [Google Scholar]
- Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy MK, Cookson MR. DJ-1 regulation of mitochondrial function and autophagy through oxidative stress. Autophagy. 2011;7 doi: 10.4161/auto.7.5.14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S, Chatelain EH, Lavie J, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013;17:719–730. doi: 10.1016/j.cmet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Michiorri S, Gelmetti V, Giarda E, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- Mitchell T, Chacko B, Ballinger SW, Bailey SM, Zhang J, Darley-Usmar V. Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochem Soc Trans. 2013a;41:127–133. doi: 10.1042/BST20120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T, Johnson MS, Ouyang X, et al. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita+/Ins2-derived beta-cells. Am J Physiol Endocrinol Metab. 2013b;305:E585–E599. doi: 10.1152/ajpendo.00093.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Sakaguchi M, Kataoka K, Huh NH. SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Mol Biol Cell. 2013;24:2772–2784. doi: 10.1091/mbc.E13-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, Youle RJ. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010a;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010b;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Ni HM, Williams JA, Jaeschke H, Ding WX. Zonated induction of autophagy and mitochondrial spheroids limits acetaminophen-induced necrosis in the liver. Redox Biol. 2013;1:427–432. doi: 10.1016/j.redox.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Hikoso S, Yamaguchi O, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Cancer Res. 2004;64:5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Okatsu K, Iemura S, Koyano F, et al. Mitochondrial hexokinase HKI is a novel substrate of the Parkin ubiquitin ligase. Biochem Biophys Res Commun. 2012;428:197–202. doi: 10.1016/j.bbrc.2012.10.041. [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Jr, Xiao G, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi H, Xu S, Zhang L, et al. Dynamin 1-like-dependent mitochondrial fission initiates overactive mitophagy in the hepatotoxicity of cadmium. Autophagy. 2013;9:1780–1800. doi: 10.4161/auto.25665. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulogiannis G, McIntyre RE, Dimitriadi M, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci U S A. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Kottwitz J, et al. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLoS ONE. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakovic A, Grunewald A, Seibler P, et al. Effect of endogenous mutant and wild-type PINK1 on Parkin in fibroblasts from Parkinson disease patients. Hum Mol Genet. 2010;19:3124–3137. doi: 10.1093/hmg/ddq215. [DOI] [PubMed] [Google Scholar]
- Rakovic A, Shurkewitsch K, Seibler P, et al. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem. 2013;288:2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, et al. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AF, Al-Zoubi MS, Whitaker-Menezes D, et al. Cigarette smoke metabolically promotes cancer, via autophagy and premature aging in the host stromal microenvironment. Cell Cycle. 2013;12:818–825. doi: 10.4161/cc.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Liu M, Hao J, et al. Parkin deficiency contributes to pancreatic tumorigenesis by inducing spindle multipolarity and misorientation. Cell Cycle. 2013;12:1133–1141. doi: 10.4161/cc.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner CM, Kamel F, Ross GW, et al. Rotenone, paraquat, and Parkinson's disease. Environ Health Perspect. 2011;119:866–872. doi: 10.1289/ehp.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, Jacobson MR. Defects in mitochondrial fission protein dynamin-related protein 1 are linked to apoptotic resistance and autophagy in a lung cancer model. PLoS ONE. 2012;7:e45319. doi: 10.1371/journal.pone.0045319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, McCoy MK, Blackinton J, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013 doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Shirihai OS. The interplay between mitochondrial dynamics and mitophagy. Antioxid Redox Signal. 2011;14:1939–1951. doi: 10.1089/ars.2010.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van HC, Cornelissen T, Hofkens H, et al. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31:10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van L V, Arnold B, Cassady SJ, Chu CT, Burton EA, Berman SB. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum Mol Genet. 2011;20:927–940. doi: 10.1093/hmg/ddq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Taylor BS, Meng S, et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Jin M, Liu X, Klionsky DJ. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy. 2013;9 doi: 10.4161/auto.26281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BR, Scott I, Han K, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013 doi: 10.1242/jcs.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011 doi: 10.1074/jbc.M110.209338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang J. Autophagy and Mitophagy in Cellular Damage Control. Redox Biol. 2013;1:19–23. doi: 10.1016/j.redox.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang J, Yu M, et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]


