SUMMARY
Silencing of the MLH1 gene is frequently seen in sporadic cancers. We report that hypoxia causes decreased H3K4 methylation at the MLH1 promoter via the H3K4 demethylases, LSD1 and PLU-1, and promotes long-term silencing of the promoter in a pathway that requires LSD1. Knockdown of LSD1 or its co-repressor, CoREST, also prevents the re-silencing (and cytosine DNA methylation) of the endogenous MLH1 promoter in RKO colon cancer cells following transient reactivation by the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-dC). The results demonstrate that hypoxia is a critical driving force for silencing of MLH1 through chromatin modification and indicate that the LSD1/CoREST complex is essential for MLH1 silencing.
INTRODUCTION
DNA mismatch repair (MMR) is necessary for genome stability, and inherited defects in MMR are linked to hereditary non-polyposis colorectal cancer (HNPCC) (Kolodner et al., 1994). Acquired defects in MMR are seen in 15% to 25% of sporadic cancers of the colon and other sites. In most cases, the MMR defects result from silencing of MLH1, a central factor in the MMR pathway (Esteller et al., 1998; Herman et al., 1998).
Hypoxia is a key microenvironmental stress in solid tumors that is associated with poor prognosis (Jubb et al., 2010). Hypoxia also induces genetic instability in the form of elevated point mutations (Reynolds et al., 1996), gene amplification (Young et al., 1988), and fragile-site induction (Coquelle et al., 1998). We have shown that BRCA1 and RAD51, in the homology dependent repair (HDR) pathway, and MLH1 and MSH2, components of MMR, are transiently down-regulated at the transcriptional level in response to hypoxia via the action of specific transcription factors, including p130/E2F4 and Myc/Max/Mnt/Mad, respectively (Bindra and Glazer, 2006; Bindra and Glazer, 2007; Bindra et al., 2004).
Epigenetic gene regulation, defined as heritable changes in gene activity that are not caused by changes in DNA sequence, has emerged as a major driver of the cancer phenotype. Epigenetic regulation can be mediated by both DNA methylation and histone modifications (Chi et al., 2010; Elsässer et al., 2011). Recently, we found that hypoxia induces epigenetic modification and silencing of the BRCA1 promoter (Lu et al., 2011). This raised the possibility that, more broadly, hypoxia may play a key role in the aberrant silencing of other tumor suppressor genes. To test this, we have focused on MLH1, because, like BRCA1, it is down-regulated at the transcriptional level in response hypoxia (Mihaylova et al., 2003) and is silenced in sporadic tumors.
Although we had previously found that hypoxia induces transient repression of MLH1 via a shift in promoter occupancy from activating c-Myc/Max to repressive Mad1/Max and Mnt/Max complexes (Bindra and Glazer, 2007), this represents a short-term, reversible effect of hypoxia. We sought to test for a role of hypoxia with respect to durable, long-term silencing of MLH1 that would persist even when the hypoxic stimulus was no longer present.
Here, we report that hypoxic stress induces MLH1 durable promoter silencing in a pathway that is dependent on the histone demethylase, LSD1. We find that LSD1, plus its co-repressor, CoREST, is necessary for MLH1 silencing. The results indicate that hypoxia is major driver of epigenetic silencing of MLH1 gene and suggest a novel mechanism by which hypoxia promotes a mutator phenotype in cancer. The results also suggest that hypoxia may be a key factor in the silencing of other tumor suppressor genes in human malignancies.
RESULTS
Hypoxia induces repressive histone modifications at the MLH1 promoter
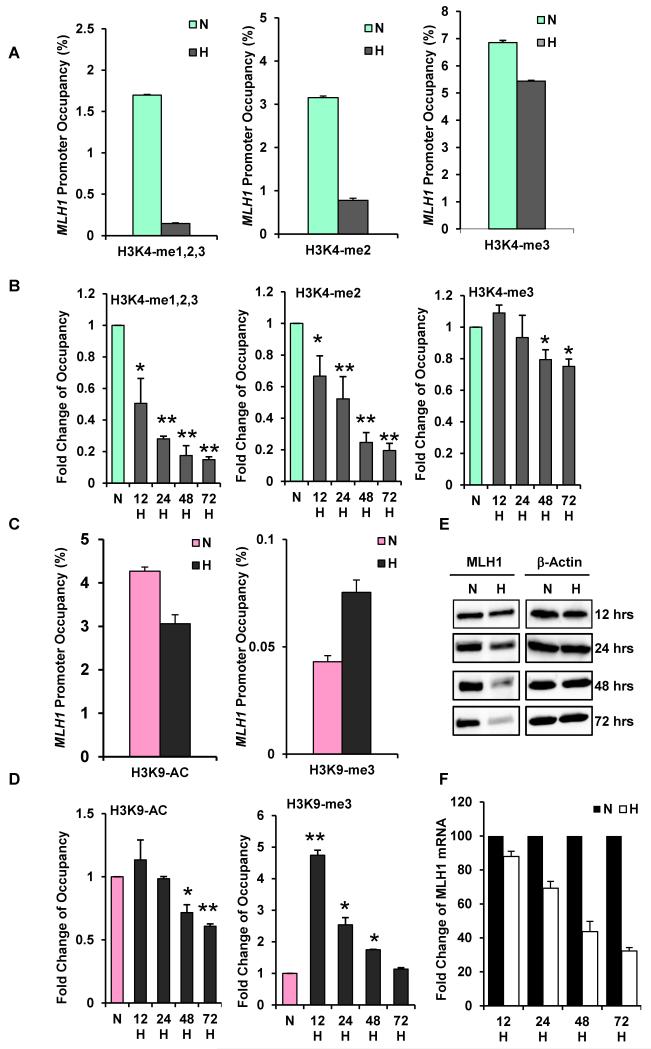
As one measure of epigenetic regulation of MLH1, we probed for hypoxia-induced histone modifications at the MLH1 promoter. Because MLH1 is silenced in sporadic breast as well as colon cancers (Herman et al., 1998; Naqvi et al., 2008), we examined both a breast cancer line (MCF-7) and a colon cancer line (SW480) to examine histone changes at the MLH1 promoter in response to hypoxia as measured by quantitative chromatin immunoprecipitation (qChIP). In MCF-7, hypoxia caused a 90% decrease in the levels of H3K4 me1,2,3 (the combined mono-, di-, and tri- methylated forms of H3K4) at the MLH1 promoter after 48 h (Fig. 1A). Levels of H3K4me2 and H3K4me3 were decreased 75% and 20%, respectively (Fig. 1A). Agarose gel images corresponding to Fig. 1A are shown in Fig. S1A. 1. A time-course study revealed that H3K4 demethylation at the MLH1 promoter is evident by 12 h and persists through 72 h (Fig. 1B).
Fig. 1. Hypoxia-induced histone modifications at the MLH1 promoter, accompanied by down-regulated MLH1 expression.
MCF-7 cells were exposed to normoxia (N) or to hypoxia at 0.01% O2 (H) for various times. Cells were collected for quantitative ChIP analyses using specific antibodies to determine H3K4 methylation levels and H3K9 acetylation and methylation levels at the MLH1 promoters. (A) Decreased H3K4 methylation levels at the MLH1 promoter in response to 48 h of hypoxia. Specific antibodies were used to determine specific H3K4 methylation forms. Relative promoter occupancies (% input) are shown with error bars based on standard errors (SEs) calculated from at least three replicates. The input signal is set as 100% (not depicted in graphs) for each assay. (B) Time-course assay of H3K4 methylation changes at the MLH1 promoter. MCF-7 cells placed under hypoxia were collected at the indicated times for qChIP analysis. Promoter occupancy levels are expressed as the fold change relative to normoxia, based on three independent ChIP assays, with error bars based on SEs. Significant differences were identified as p<0.05 (as indicated by *) or p<0.01 (as indicated by **) compared to normoxic levels. (C) Hypoxia increases H3K9 methylation and decreases H3K9 acetylation at the MLH1 promoter. qChIP analysis of H3K9 acetylation (left) and methylation (right) levels at the MLH1 promoter after 48 h normoxia or hypoxia exposure. Relative promoter occupancies (% input) are shown with error bars calculated as above. (D) Time course of hypoxia-induced H3K9 acetylation (left) and methylation (right) at the MLH1 promoter by qChIP analysis. MCF-7 cells were exposed to normoxia or hypoxia for the indicated times, and H3K9 acetylation and methylation levels at the MLH1 promoter were analyzed as above. (E) Time-course of MLH1 expression at the protein level determined by western blot analysis in MCF-7 cells. Cells were exposed to normoxia (N) or hypoxia (H) for the indicated times, and were collected for western blot analysis. (F) Time-course of MLH1 mRNA expression by quantitative real-time PCR analysis (qRT-PCR). mRNA levels are expressed as the fold change relative to those of the corresponding normoxic control cells at each time point.
Histone modification at H3K9 has dual effects on gene transcription: H3K9 acetylation is a marker of activation, while H3K9 methylation is repressive, and it is known that hypoxia alters H3K9 modification at various gene promoters (Chen et al., 2006; Johnson et al., 2008). We detected a 30% decrease in H3K9 acetylation and 70% increase in H3K9 me3 levels at the MLH1 promoter in response to 48 h hypoxic exposure (Fig.1C). Over time, we found decreased H3K9 acetylation beginning at 48 h; however, the hypoxia-induced increase in H3K9 methylation peaked by 12 h, then gradually returned back to the normoxic level by 72 h (Fig. 1D), suggesting that increased H3K9 methylation is an early modification at the MLH1 promoter that may be upstream of H3K9 deacetylation and H3K4 demethylation under hypoxic stress.
In SW480 cells, we observed a 90% decrease in H3K4 me1,2,3 levels and an 80% decrease in H3K4 me2 levels at the MLH1 promoter in response to hypoxia (Fig. S1B & C), a pattern similar to that in MCF-7 cells.
For comparison, we examined global H3K4 methylation levels by western blot of total chromatin in both MCF-7 and SW480 cells in normoxia versus hypoxia, and we found that global H3K4 methylation levels are not decreased (Fig. S2A). Hence the decreased methylation of H3K4 seen at the MLH1 promoter does not simply reflect global changes in H3K4 methylation (since overall levels of H3K4 methylation do not go down). Rather, it likely reflects promoter-specific effects. However, this does not mean that the effect is unique to the MLH1 promoter, as many other sites may be targeted for H3K4 demethylation in hypoxia. In fact, we previously observed hypoxia-induced H3K4 demethylation at the BRCA1 promoter (Lu et al., 2011). In that same work, we also found increased H3K4 methylation at the VEGF promoter, showing that hypoxia-mediated H3K4 methylation changes can vary from gene to gene reflecting specific differences in regulation.
Next, we evaluated MLH1 protein and mRNA levels by western blot and quantitative real-time PCR (qRT-PCR) in the MCF-7 and SW480 cell lines. We found that MLH1 protein levels and mRNA levels are both reduced in conjunction with the changes in chromatin marks at the promoter in response to hypoxia in MCF-7 cells (Figs. 1 E and F) and in SW480 cells (Fig. S1D and E).
To provide an in vivo correlation with the reduced MLH1 expression in hypoxic cells, we examined SW480 xenograft tumors in nude mice. By immune fluorescence on tumor sections, we found that MLH1 expression is inversely correlated with expression of carbonic anhydrase IX (CAIX), a marker of hypoxia (Fig. S3A). Although this analysis reflects just one snapshot in time within the tumor and therefore cannot demonstrate long-term silencing, it does provide a useful in vivo data point corroborating the influence of hypoxia on MLH1 expression.
We also tested for the presence of cytosine methylation in SW480 cells after short-term hypoxia as in Fig. 1. In prior work, we had found no cytosine methylation at the MLH1 promoter after just 48 h of hypoxia (Mihaylova et al., 2003), and we had the same results again using two different methods (Herman et al., 1998; Xiong and Laird, 1997) (data not shown). Given the very short-term nature of this exposure, the lack of detectable DNA methylation is not surprising.
Because MLH1 plays a major role in DNA mismatch repair, a decrease in MLH1 levels would be predicted to yield an increase in genetic instability. We had previously shown that decreased MLH1 expression in hypoxia confers a mutator phenotype, characterized by reporter transgene mutagenesis (Mihaylova et al., 2003). To further confirm this, we assayed for the impact of hypoxia on the stability of a (CA)29 dinucleotide insert within the β-galactosidase gene in an episomal vector. Frameshift mutations can put β-galactosidase back into frame, as measured by β-galactosidase activity in cell lysates. We found that hypoxia causes an increase in β-galactosidase activity and that this increase can be suppressed by forced expression of MLH1 via a heterologous promoter (Fig. S3B). In addition, TSA, a histone deacetylase inhibitor that was previously found to prevent MLH1 mRNA down-regulation in short-term hypoxia (Mihaylova et al., 2003), was also able to attenuate the increased mutator phenotype (Fig. S3B). These data confirm prior findings that hypoxia induces mutagenesis and provide a further link to altered MLH1 levels.
The lysine-specific demethylases, LSD1 and PLU-1, mediate hypoxia-induced H3K4 demethylation at the MLH1 promoter
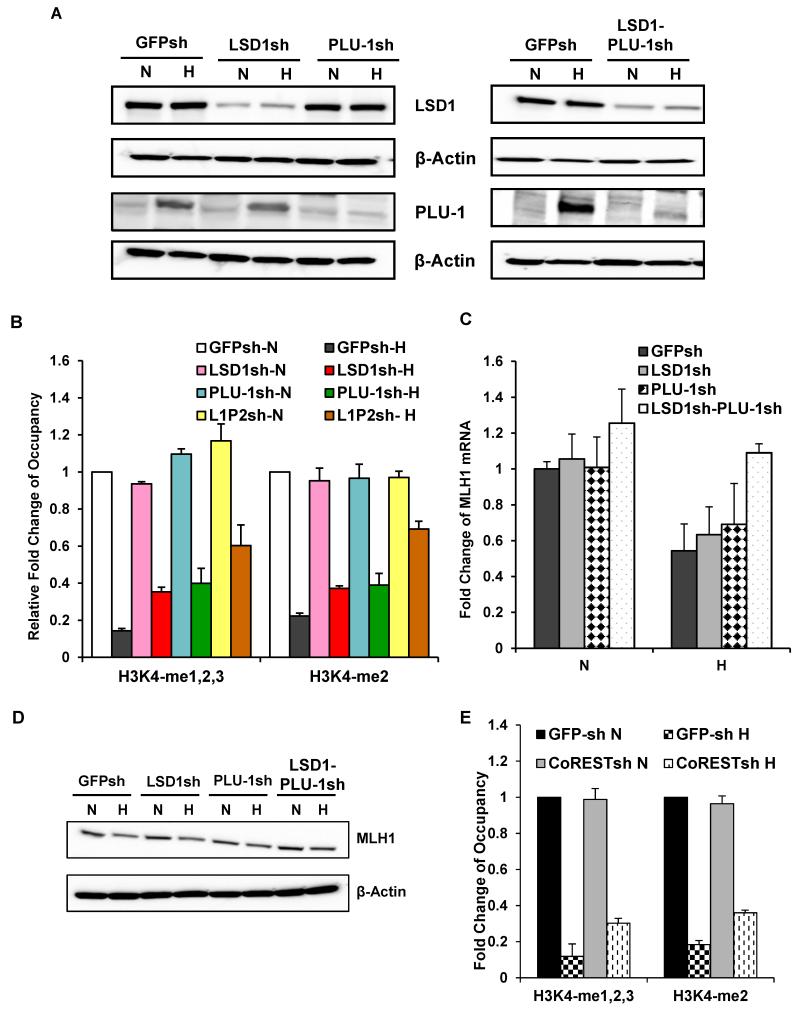
In prior work, we demonstrated that LSD1 mediates repression of BRCA1 and RAD51 in response to hypoxia (Lu et al., 2011). Therefore, we tested whether LSD1 is also required for H3K4 demethylation at the MLH1 promoter in response to hypoxia. We established SW480 sub-clones with stable shRNA knockdown of LSD1, SW480LSD1sh (Fig. 2A) along with a control line with shRNA targeting of GFP (Fig. 2A). The cells were exposed or not to hypoxia for 48 h and analyzed for H3K4 methylation status. We found that knockdown of LSD1 partially attenuated hypoxia-induced decreases in H3K4 methylation levels at the MLH1 promoter (Fig. 2B). However, these effects were less than we expected based on prior work with BRCA1 and RAD51, and so we went on to analyze the impact of other histone demethylases. Because it is induced by hypoxia, we focused on the possible role of PLU-1 (Lu et al., 2011; Xia et al., 2009). As above, we established stable shRNA knockdown of PLU-1 in an SW480 sub-clone (Fig. 2A). Although PLU-1 is normally expressed at low baseline levels in SW480 cells, we confirmed that it is inducible in response to hypoxia and that this induction is blocked in the PLU-1 shRNA-expressing line (Fig. 2A). Using these cells, we found that knockdown of PLU-1 also partially attenuates hypoxia-induced H3K4 demethylation at the MLH1 promoter (Fig. 2B), with a level of attenuation similar to that in SW480 LSD1sh cells.
Fig. 2. The histone demethylases, LSD1 and PLU-1, and the LSD1 partner CoREST, together mediate hypoxia-induced H3K4 demethylation at the MLH1 promoter.
SW480 cells with LSD1 knockdown, with PLU-1 knockdown, or with double knockdown of both LSD1 and PLU-1 were established using lentiviral shRNAs constructs targeting LSD1 or PLU-1. Control cells were transduced with a lentiviral expression construct for a GFP shRNA. (A) Western blot analyses to determine LSD1 and PLU-1 expression levels in the SW480-GFPsh, SW480-LSD1sh, SW480-PLU-1sh and double knockdown SW480-LSD1-PLU-1sh cells under normoxic and hypoxic conditions. (B) qChIP analyses of H3K4 methylation levels at the MLH1 promoter following 48 h exposure to normoxia or hypoxia in SW480-GFPsh cells; SW480-LSD1sh cells; SW480-PLU-1sh cells; and double knockdown SW480-LSD1-PLU-1sh cells (indicated as SW-L1P2). Promoter occupancy levels are expressed as the fold change relative to the normoxic control SW480 GFPsh cells. Standard errors are indicated. (C). Quantitative real-time PCR analysis of MLH1 mRNA levels in SW480 GFPsh, SW480 LSD1sh, SW480 PLU-1sh and SW480 LSD1-PLU-1sh cells after 48 h of normoxic or hypoxic exposure. mRNA levels are expressed as the fold change relative to normoxic control SW480 GFPsh cells. (D). Western blot analysis of MLH1 protein levels in the same cell lines as in (C), above, after 48 h of normoxic or hypoxic exposure (E). CoREST also plays a role in hypoxia-induced H3K4 demethylation at the MLH1 promoter in SW480 cells. Quantification of ChIP analyses of H3K4 methylation levels at the MLH1 promoter following 48 h exposure to normoxia or hypoxia in the SW480 GFPsh cells compared to the SW480 CoRESTsh cells. Promoter occupancy levels are expressed as the fold change relative to the normoxic SW480 GFPsh cells, based on three independent ChIP assays with error bars based on SEs.
Since knockdown of LSD1 and PLU-1 individually showed partial reduction of hypoxia-induced H3K4 demethylation, we tested simultaneous knockdown of both (Fig.2A). We found that the dual knockdown in SW480 cells yielded greater inhibition of hypoxia-induced H3K4 demethylation than in the single knockdown lines (Fig. 2B; in the comparison of cells with double knockdown of LSD1 and PLU-1 to GFPsh control cells in hypoxia, the p-values are p= 0.011 for H3K4-me1,2,3 levels and p=0.015 for H3K4-me2 levels). Similar results were seen in MCF7 cells (data not shown). These results indicate that LSD1 and PLU-1 are non-redundant and that both are involved in hypoxia-induced H3K4 demethylation at the MLH1 promoter. This result differs from our previous work implicating LSD1, by itself, in hypoxia-induced H3K4 demethylation at the BRCA1 and RAD51 promoters (Lu et al., 2011); PLU-1 was not seen to play any role in that prior work.
We next examined the changes in MLH1 expression at both the protein and mRNA levels in response to hypoxia in control SW480 GFPsh cell line compared to LSD1 knockdown cells (SW480 LSD1sh), PLU-1 knockdown cells (SW480 PLU-1sh), or double knockdown cells (SW480 LSD1sh-PLU-1sh). We found that knockdown of either LSD1 or PLU-1 alone did not have statistically significant effects on the hypoxia-induced reduction in MLH1 mRNA (Fig. 2C; p = 0.12 in comparison between LSD1 knockdown to GFPsh control, and p = 0.09 in comparison of PLU-1 knockdown to GFPsh control) or protein levels (Fig. 2D). However, double knockdown of LSD1 and PLU-1 did substantially prevent the down-regulation of MLH1 by hypoxia at both the mRNA (Fig. 2C; p=0.0053) and protein levels (Fig. 2D), in keeping with the ChIP data (Fig. 2B).
To further probe the mechanism of the MLH1 down-regulation, we tested whether hypoxia induces LSD1 occupancy at MLH1 promoter. ChIP assays were performed on chromatin from SW480 cells following exposure to normoxia or hypoxia. We found almost no LSD1 binding to the MLH1 promoter in normoxic cells; however, in hypoxic cells, we observed increased LSD1 binding to the MLH1 promoter (Fig. S4). The ACTB-2 promoter was used as a positive control for LSD1 binding (Fig. S4).
The demethylase activity of LSD1 requires its heterodimer partner, CoREST (Lee et al., 2005; Shi et al., 2005), and so we examined the role of CoREST in the hypoxia induced histone modifications at the MLH1 promoter. In SW480 cells with shRNA-mediated knockdown of CoREST (SW480 CoRESTsh), ChIP assays revealed that CoREST knockdown, like LSD1 knockdown, attenuated hypoxia-induced H3K4 demethylation at the MLH1 promoter in SW480 cells (Fig. 2E).
Hypoxia induces MLH1 promoter silencing in a pathway dependent on LSD1
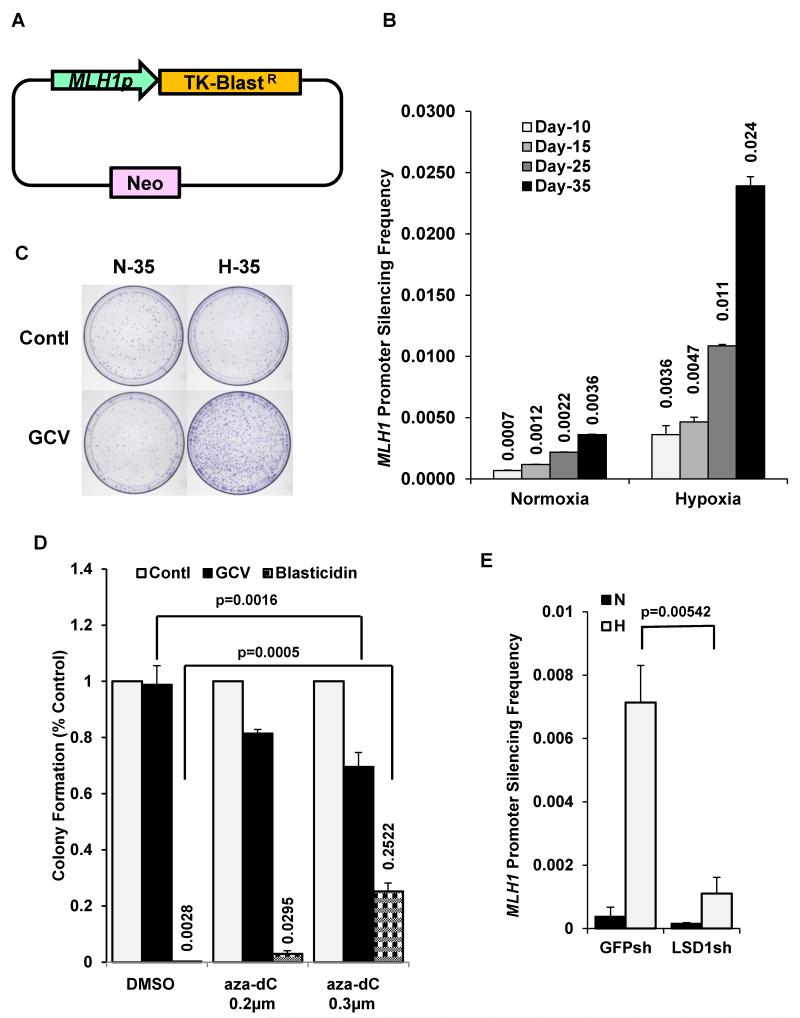
Next, we sought to test whether hypoxia can induce the durable, long-term silencing of the MLH1 promoter, an endpoint distinct from short-term, reversible repression. We engineered an assay system to select for cells in which the MLH1 promoter had undergone silencing. We used a construct containing the 1.7 kb MLH1 promoter driving the expression of the thymidine kinase (TK) gene fused to blasticidin resistance (BlastR) gene (Fig. 3A). This construct was transfected into RKO cells and the resultant stable cell line, designated RKO MLH1p-TK-BlastR, and was resistant to blasticidin but sensitive to ganciclovir (GCV), reflecting expression of both selectable markers in the expression cassette. To test whether hypoxia could silence the MLH1 promoter in this construct, the RKO MLH1p-TK-BlastR cells were exposed to hypoxia for 10, 15, 25, or 35 days at a moderate hypoxia level of 1% O2. The hypoxia-exposed cell populations at each time point (along with normoxic control cells grown in parallel) were subjected to selection (under normoxic conditions) for GCV resistance (and therefore lack of TK expression) by incubation for 10 days in medium containing GCV, which is toxic to cells with functional TK. GCV-resistant clones represent cells in which the MLH1 promoter has been silenced. We found that as the duration of hypoxic exposure increased, so did the frequency of GCV-resistant colonies (Fig. 3B). After 35 days of exposure to 1% O2, the cells gave rise to GCV-resistant clones at a frequency of about 0.024%, 8-fold more than the background frequency in cells grown in normoxic conditions (Figs. 3B and 3C). Consistent with promoter silencing in the expression cassette, the clones were blasticidin sensitive and lacked transcription from the cassette, but still retained the construct (data not shown). Beyond the 10 day selection period, we confirmed that the silencing was durable, as randomly selected clones were grown in normoxic conditions for over 4 months and still showed silencing of the MLH1 promoter (data not shown). These results show that hypoxia can enforce MLH1 promoter silencing and that the silencing persists even after the cells are no longer in hypoxic conditions.
Fig. 3. Hypoxia induces silencing of the MLH1 promoter in a pathway dependent on LSD1.
(A) Schematic of the MLH1-TK-BlastR dNheI pDisplay construct used to select for clones undergoing MLH1 promoter silencing. (B) Frequency of ganciclovir (GCV)-resistant clones (indicative of silencing of MLH1-TK-BlastR expression) following exposure of RKO cells to normoxia or hypoxia (1% O2) for the indicated number of days. Selection in the presence of GCV was performed under normoxic conditions for 10 additional days. Error bars represent SEs from three replicates. (C) Image of representative cell culture wells showing differential GCV-resistant colony formation following growth in normoxia or hypoxia and subsequent GCV selection. (D) Treatment of MLH1-silenced clones with the DNA methylation inhibitor, 5-azadC, reactivates the silenced MLH1 promoters to yield GCV sensitivity (due to reactivated TK expression) and blasticidin resistance (due to BlastR expression). GCV-resistant clones induced by hypoxia were pooled and treated with DMSO, 0.2 μm 5-aza-dC, or 0.3 μm 5-aza-dC for 48 h. Colony formation in the presence of GCV (blue bar) or blasticidin (red bar) was quantified relative to the DMSO control as shown. (E) Knockdown of LSD1 inhibits the hypoxia-induced silencing of the MLH1 promoter construct. RKO cells containing the MLH1- TKR-BlastR construct and expressing shRNA to either GFP or LSD1 were exposed to normoxia or 1% O2 for 35 days and then selected in GCV for 10 additional days under normoxic conditions. The frequency of the resulting GCV resistant clones indicative of MLH1 promoter silencing is shown.
Because RKO cells are a colon cancer cell line in which the endogenous MLH1 gene is silenced, we considered the possibility that they might have a special susceptibility to silencing of the MLH1 promoter in the MLH1p-TK-BlastR reporter construct. Therefore, we also tested for hypoxia-induced silencing of the MLH1p-TK-BlastR construct in SW480 cells, since they are a colon cancer cell line without endogenous MLH1 silencing. The stable transfectant cell line, designated SW480 MLH1p-TK-BlastR, was exposed to normoxia or hypoxia (0.5% O2) for 5 weeks. We then selected for GCV-resistant cells as a measure of MLH1 promoter silencing, as above for RKO cells. We observed hypoxia-induced GCV-resistant SW480 cells (Fig. S5), indicating that hypoxia-mediated silencing of the MLH1p-TK-BlastR construct had occurred in the SW480 cells and showing that this effect is generalizable to other cell lines besides RKO. Although the frequency of silenced clones in SW480 was lower than in RKO, it was still substantially more than the background in the normoxic cells, and this difference was statistically significant (p=0.019).
Because gene silencing is frequently associated with DNA hypermethylation at promoter CpG sites (including in the MLH1 promoter in sporadic colon cancers), we probed the role of DNA methylation at cytosines in the observed hypoxia-induced MLH1 silencing. We asked whether treatment with the DNA methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-aza-dC), could reactivate the silenced MLH1 promoter in the hypoxia-induced GCV resistant cells. Reactivation of the silenced MLH1 promoter would be expected to yield TK and BlastR gene expression so that cells would become sensitive to GCV and resistant to blasticidin. We pooled the GCV-resistant RKO MLH1p-TK-BlastR clones and exposed the pooled cells to 5-aza-dC for 48 h. Following a dose of 0.3 μM 5-aza-dC, we observed that approximately 30% of the cells had become sensitive to GCV again (Fig. 3D), and a corresponding number (approximately 25%) had simultaneously become resistant to blasticidin (Fig. 3D). In contrast, in the DMSO treated control group, essentially all of the cells remained resistant to GCV and sensitive to blasticidin (Fig. 3D). These results demonstrate that hypoxia-induced MLH1 promoter silencing is partially reversible with 5-aza-dC treatment, and thus was associated with DNA methylation. However, in the absence of 5-aza-dC treatment, the silencing is otherwise stable because, as noted above, it persists long-term after the cells are returned to normoxic conditions.
To test the putative role of LSD1 in the hypoxia-induced MLH1 silencing, we established a sub-line of the RKO MLH1p-TK-BlastR cells with stable knockdown of LSD1 and exposed the cells to hypoxia (1% O2) for 35 days. We found that LSD1 knockdown substantially inhibited the hypoxia-induced MLH1 silencing, reducing the frequency to less than one fifth of that in the control RKO MLH1p-TK-BlastR cells containing a GFPsh vector (Fig. 3E). (There was a small reduction in silenced clones in normoxia by LSD1 knockdown compared to GFP knockdown, but this was not statistically significant; p=0.367). The substantial reduction in silencing in hypoxia by LSD1 knockdown was not simply due to LSD1 knockdown affecting plating efficiency. LSD1 knockdown, by itself, had no effect on colony formation by cells under normoxic conditions, and only a minimal effect under hypoxic conditions, with less than a 10% reduction (data not shown). Importantly, this small effect on colony formation was taken into account in analyzing the results, as the data presented in Fig. 3E are normalized to the plating efficiency control. It should be mentioned that there was a 20% reduction in the rate of cell proliferation in the LSD1 knockdown cell line, compared to the GFPsh control cell line (as judged by serial cell counts; data not shown), consistent with reports showing that LSD1 inhibition can impact tumor cell growth (Ding et al., 2013; Huang et al., 2009). But this did not translate into an effect on clonogenicity as measured by the frequency of ultimate colony formation from a specific number of seeded cells, which was minimally impacted. Overall, these results indicate that LSD1 is necessary for MLH1 silencing in response to hypoxia.
LSD1 and its co-repressor, Co-REST, mediate MLH1 re-silencing following reactivation by 5-aza-dC treatment in RKO cells
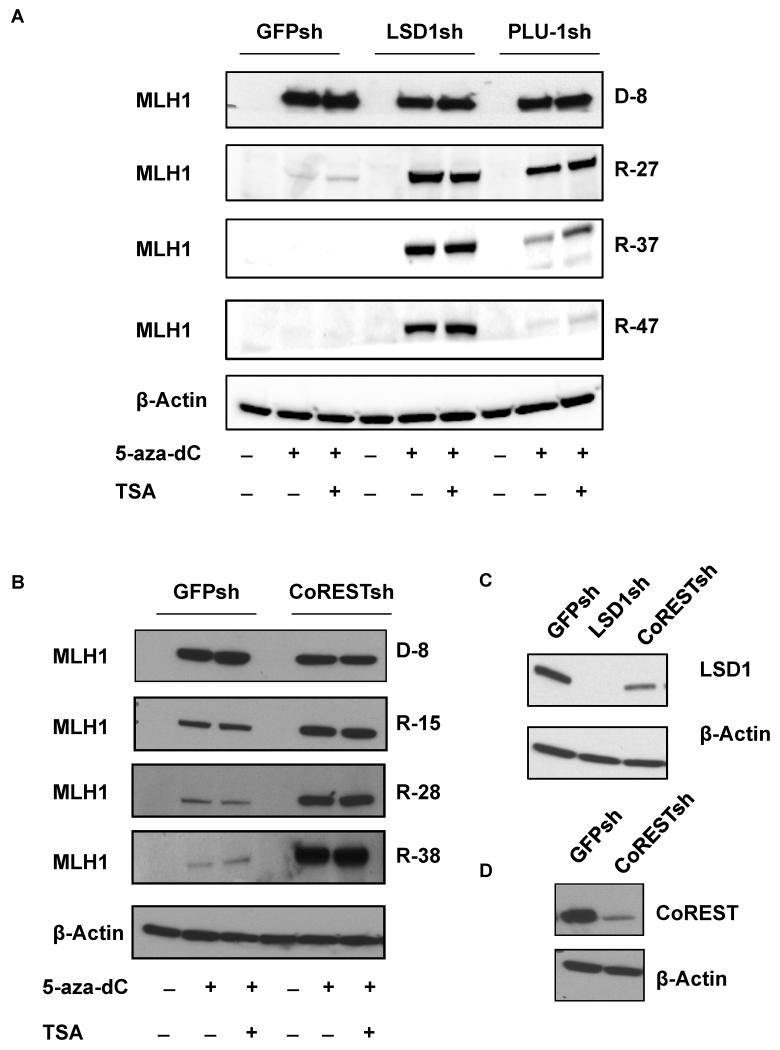
Inhibition of LSD1 by small molecules or knockdown by RNA interference can reactivate certain aberrantly silenced genes in cancer cell lines, in particular secreted frizzled-related proteins (SFRPs) (Huang et al., 2007; Wang et al., 2011). Consequently, we asked whether knockdown of LSD1 (or other lysine demethylases) might similarly reactivate silenced MLH1 in RKO cells. We established shRNA expressing cell lines by transducing lentiviral expression constructs for shRNAs targeting LSD1, RBP2, or PLU-1 in RKO cells, designated as RKO LSD1sh cells, RKO RBP2sh cells, and RKO PLU-1sh cells, respectively. In contrast to the published results for the SFRP genes, MLH1 could not be reactivated with LSD1 knockdown (Fig. S6; compare lanes without 5-aza-dC or TSA treatment). There was also no MLH1 reactivation seen with knockdown of PLU-1 or RBP2 (Fig. S6). However, treatment of the RKO cells with 5-aza-dC at 5 μm for 72h was able to reactivate MLH1 expression (Fig. S6), consistent with other published work (Fahrner et al., 2002). Interestingly, there was no additional increase in MLH1 expression when 5-aza-dC treatment was combined with knockdown of any of the histone demethylases or treatment with trichostatin A (TSA), a histone deacetylase inhibitor.
Although 5-aza-dC treatment of cancer cells in culture can reactivate DNA-hypermethylated genes, gene silencing is typically reestablished once cells are released from 5-aza-dC inhibition (McGarvey et al., 2006). Specifically, in RKO cells, MLH1 can be reactivated by 5-aza-dC treatment, but it becomes silenced again once the agent is removed (McGarvey et al., 2006). We sought to determine what roles, if any, LSD1 or PLU-1 might play in the MLH1 re-silencing after removal of 5-aza-dC treatment in RKO cells. Again, stable H3K4 demethylase knockdown cell lines, RKO LSD1sh and RKO PLU-1sh, were tested in comparison to RKO GFPsh as a control (Fig. 4C). Cells were treated with 5-aza-dC with or without TSA for 8 days, and then placed in regular growth conditions for up to 47 days. Immediately after the 8-day exposure to 5-aza-dC, all cell lines showed re-expression of MLH1 at similar protein levels (Fig. 4A). The addition of TSA did not produce any additional increased expression. In keeping with prior reports, after the cells were removed from 5-aza-dC and grown in standard conditions for 27 days, MLH1 became undetectable at the protein level in the control cell line, RKO GFPsh cells, consistent with re-silencing of the gene (Fig. 4A). However, MLH1 expression was maintained in RKO LSD1sh cells. In the RKO PLU-1sh cells, re-silencing of MLH1 did eventually occur by day 47, but this process was delayed and attenuated at earlier time points compared to controls (Fig. 4A). The results indicate that knockdown of LSD1 can inhibit MLH1 re-silencing in RKO cells and that knockdown of PLU-1 can partially disrupt and slow MLH1 re-silencing.
Fig. 4. LSD1 and CoREST are required for MLH1 re-silencing in RKO cells following reactivation by transient 5-aza-dC exposure.
RKO GFPsh cells, RKO LSD1sh cells, RKO PLU-1sh cells and RKO CoRESTsh cells were treated with 5-aza-dC at 5 μm for 8 days. The cells were then placed in standard conditions for 42 additional days. MLH1 expression was analyzed at the indicated times. (A) Western blot analyses to determine MLH1 expression levels in RKO GFPsh cells, RKO LSD1sh cells, and RKO PLU-1sh immediately after 8 days of 5-azadC treatment (indicated as D-8) or after replacement in standard medium without of 5-aza-dC for 27 days (indicated as R-27), or 33 days (indicated as R-33) or 42 days (indicated as R-42). (B) Western blot analyses to determine MLH1 expression levels in RKO GFPsh cells, RKO CoRESTsh cells immediately after 5-aza-dC treatment for 8 days (indicated as D-8) or after replacement in standard medium for 15 days (indicated as R-15), 28 days (indicated as R-28) or 38 days (indicated as R-38). (C) Western blot analyses to determine LSD1 expression levels in RKO GFPsh, RKO LSD1sh, and RKO CoRESTsh cells. (D) Western blot analyses to determine CoREST expression levels in RKO GFPsh and RKO CoRESTsh cells.
We next examined if the re-silencing of the MLH1 promoter in the RKO cells would be influenced by hypoxia. RKO GFPsh, RKO LSD1sh and RKO PLU-1sh cells were treated with 5-aza-dC for 8 days in normoxia as same as above (Fig. S7A). After release from 5-aza-dC, the cells were maintained either in normoxic or hypoxic conditions (0.5% O2) for the indicated times (Fig. S 7B). Hypoxia exposure to the RKO cells with reactivated MLH1 promoters did not increase the rate at which re-silencing occurred. As above, MLH1 re-silencing was blocked by LSD1 knockdown under both normoxic and hypoxic condition (Fig. S7B), and, again, PLU-1 knockdown delayed the re-silencing under both conditions (Fig. S7B).
The demethylase activity of LSD1 requires the CoREST protein (Lee et al., 2005; Shi et al., 2005), and so we examined whether CoREST participates with LSD1 in mediating MLH1 silencing. We established stable CoREST knockdown in RKO cells (RKO CoRESTsh cells; Fig.4D.) and tested the impact of CoREST on MLH1 silencing (Fig. 4B). Similar to what we observed in the LSD1sh cell line (Fig. 4A), we found that knockdown of CoREST, by itself, did not reactivate the silenced MLH1 (see lanes without 5-aza-dC treatment), However, knockdown of CoREST did block MLH1 re-silencing in RKO cells after 5-aza-dC removal (Fig. 4B), similar to the effect of LSD1 knockdown (Fig. 4A). Interestingly, we observed that knockdown of CoREST resulted in partial knockdown of LSD1 (Fig. 4C), consistent with a prior report showing that CoREST protects LSD1 from degradation (Shi et al., 2005). Hence, some of the effects of CoREST knockdown may also reflect the consequent decrease in LSD1 levels. Nonetheless, these results indicate that the LSD1/CoREST complex is critical for establishment of MLH1 silencing but that knockdown of either of the components of this complex is not sufficient, by itself, to reactivate the gene once it has been silenced. The additional factors required for stable MLH1 reactivation remain to be determined.
H3K4 demethylation occurs at the MLH1 promoter during re-silencing in RKO cells and is mediated by LSD1
DNA methyltransferase inhibitors not only directly inhibit DNA methylation but also can influence histone modifications (Fahrner et al., 2002; McGarvey et al., 2006). We conducted qChIP analyses to evaluate the dynamics of histone marks upon 5-aza-dC-mediated activation of MLH1 and then during re-silencing following cessation of 5-aza-dC treatment, as well as to probe the role of LSD1 in this process. Consistent with previous work (Fahrner et al., 2002), we found increased H3K9 acetylation and increased H3K4 methylation at MLH1 immediately after 5-aza-dC treatment in both the RKO GFPsh and RKO LSD1sh cells (Figs. 5A and B). Following release from 5-aza-dC and growth in standard conditions for 28 days, qChIP analyses revealed that, in the control RKO GFPsh cells, H3K9 acetylation and H3K4 methylation returned back to the levels seen in cells that were not treated with 5-aza-dC (Figs. 5C and D); however, in the RKO LSD1sh cells, the levels of H3K4 methylation were still elevated at the MLH1 promoter (Figs. 5C and D; p = 0.008 for H3K4-me1,2,3 levels and p = 0.001 for H3K4-me2 levels). The H3K9 acetylation levels also appeared to remain elevated (Figs. 5C and D), but the differences in H3K9 acetylation levels did not reach statistical significance (p = 0.07)]. Nonetheless, the H3K4 modification patterns are consistent with the MLH1 protein expression data in Fig. 4A and show that LSD1 serves as a key regulatory factor to enforce repressive histone marks during MLH1 re-silencing.
Fig. 5. The histone demethylase, LSD1, mediates H3K4 demethylation at the MLH1 promoter during MLH1 silencing in RKO cells.
RKO GFPsh and RKO LSD1sh cells were treated with 5-aza-dC at 5 μm for 8 days. The cells were then replaced in normal culture medium for 28 additional days. ChIP analyses were performed to determine H3K4 methylation and H3K9 acetylation levels at the MLH1 promoter at the indicated times. (A) Agarose gel image of ChIP analyses of H3K4 methylation and H3K9 acetylation levels at the MLH1 promoter following 8 days (D-8) of 5-aza-dC treatment in RKO GFPsh cells and RKO LSD1sh cells. PCR amplification products corresponding to the MLH1 promoter region are shown. (B) Quantification of H3K4 methylation and H3K9 acetylation levels by real-time PCR at the MLH1 promoter under the same conditions as in (A). Relative promoter occupancies (% input) are shown with error bars based on standard errors (SEs) calculated from at least three replicates. (C) Agarose gel image of ChIP analyses of H3K4 methylation and H3K9 acetylation levels at the MLH1 following 28 days recovery (R-28) after 5-aza-dC treatment in RKO GFPsh cell line versus RKO LSD1sh cell line. (D) Quantification of H3K4 methylation and H3K9 acetylation levels by real-time PCR at the MLH1 promoter in the same condition as (C).
LSD1 and CoREST are required for MLH1 promoter DNA methylation after release from 5-aza-dC treatment
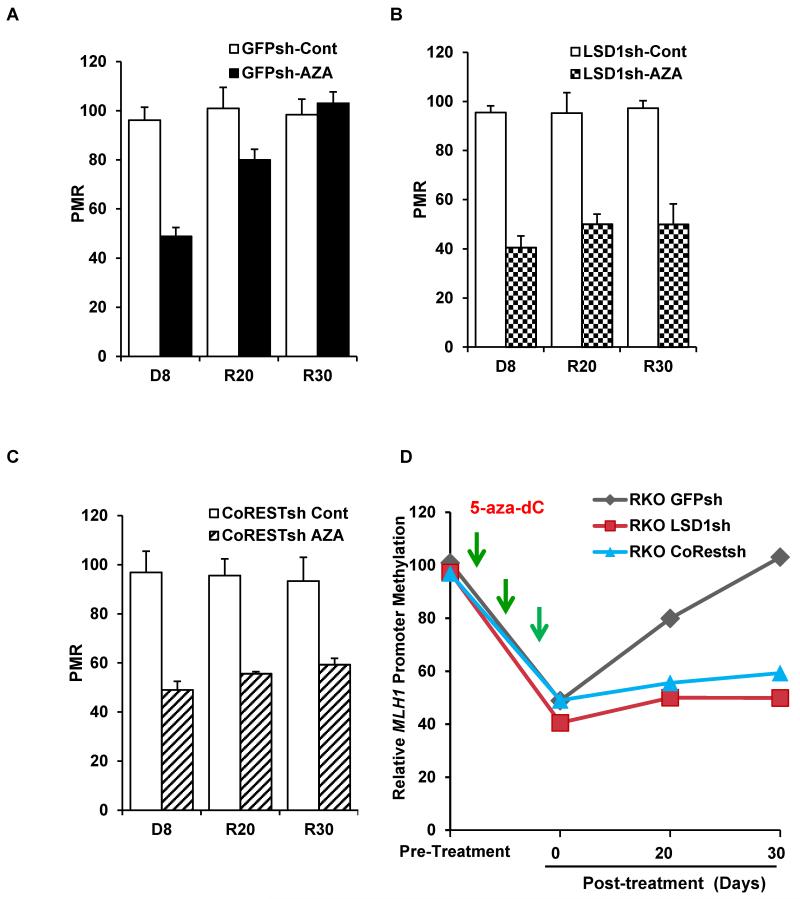
To test whether the LSD1/CoREST complex impacts MLH1 promoter DNA re-methylation after release of RKO cells from 5-aza-dC, we used a real-time PCR-based assay to quantify MLH1 promoter DNA methylation levels following bisulfite-induced conversion of unmethylated C into U, leaving methylated C intact. We treated RKO GFPsh cells, RKO LSDsh cells, and RKO CoRESTsh cells with 5-aza-dC for 8 days, followed by release into standard growth medium. Immediately after the 8-day treatment with 5-aza-dC, MLH1 promoter DNA methylation levels in all three cell lines (RKO GFPsh, RKO LSD1sh, and RKO CoRESTsh) were decreased to about 50% of untreated controls, as expected. In the RKO GFPsh cells, the MLH1 promoter DNA methylation levels gradually returned back to pre-treatment levels by 30 days after release (Fig. 6A and D). In contrast, in both the LSD1 and CoREST knockdown cell lines, MLH1 promoter DNA methylation levels remained low even 30 days after release from 5-aza-dC (Fig. 6B, C, D; p=0.011, and p=0.017 for LSD1 knockdown and CoREST knockdown RKO cells, respectively, in comparison to control GFPsh cells at 30 days post treatment). These results strongly suggest that histone modification patterns mediated by LSD1 and CoREST are needed for remethylation of the MLH1 promoter during the re-silencing process.
Fig. 6. LSD1 and CoREST prevent MLH1 promoter re-methylation after cessation of 5-aza-dC treatment in RKO cells.
RKO GFPsh, RKO LSD1sh, and RKO CoRESTsh cells were treated with 5-aza-dC at 5 μm for 8 days. Then cells were released from 5-aza-dC by placement in standard culture medium for 20 or 30 additional days. Real-time PCR to quantify DNA methylation levels at MLH1 promoter was performed using the Methylight assay. PMR (Percentage of Methylation Reference) value was used to quantify the methylation levels at MLH1 promoter. (A) Methylight analysis of DNA methylation levels in the MLH1 promoter in RKO-GFPsh cells immediately after 5-aza-dC treatment (indicated as D-8), 20 days post treatment (indicated as R20), or 30 days post treatment (indicated as R30). (B) Methylight analysis of DNA methylation levels at the MLH1 promoter in RKO-LSD1sh cells as above. (C) Methylight analysis of DNA methylation levels at the MLH1 promoter in RKO-CoRESTsh cells. (D) Graphical summary of the data in (A), (B), and (C).
The repressive complexes Max/Mad1 and Max/Mnt pathway are not required for hypoxia-induced histone modifications at the MLH1 promoter
Our previous work demonstrated that hypoxia induces short-term, reversible down-regulation of MLH1 via increased binding of repressive Max/Mad1 and Max/Mnt complexes at the proximal promoter of the MLH1 gene. To test if these repressive complexes are also required for hypoxia-induced epigenetic modifications at the MLH1 promoter, we established an SW480-derived cell line with stable shRNA-mediated knockdown of Max, SW480Maxsh-551 (Fig. S8A). ChIP assays were performed following exposure of SW480 Maxsh and the control cell line, SW480GFPsh, to normoxia or hypoxia. We found that knockdown of Max had no effect on hypoxia-induced H3K4 demethylation, compared to the control cell line (Fig. S8B). Hence, while Max/Mad1 and Max/Mnt complexes may mediate short-term, transient repression of MLH1, they are not needed to mediate the histone modifications that mark the locus for long-term silencing, a process that instead depends of LSD1/CoREST. Hence, in the case of hypoxia and regulation of MLH1, short-term, reversible repression and long-term, durable silencing depend on separate pathways.
DISCUSSION
In this work, we show that hypoxia induces MLH1 silencing through an orchestrated pattern of epigenetic modulation. Using a reporter construct with the MLH1 promoter driving selectable TK and BlastR genes, we established that hypoxia leads to silencing of the MLH1 promoter and determined that the H3K4 demethylase, LSD1, is required for this process. The silencing was seen to persist even after the cells were no longer in hypoxic conditions, demonstrating durable, long-term epigenetic change produced by exposure to hypoxic stress.
Mechanistically, we further show that the hypoxia causes H3K4 demethylation at the MLH1 promoter via the H3K4 demethylases, LSD1 and PLU1. We also show that LSD1 and CoREST are required for the re-silencing of the endogenous MLH1 promoter in RKO cells that occurs following transient reactivation by 5-aza-dC treatment. These results point to the LSD1/CoREST complex as a possible therapeutic target to inhibit the silencing and/or re-silencing of MLH1 and possibly other tumor suppressor genes.
The finding that hypoxia promotes silencing of the MLH1 promoter extends our previous work showing that hypoxia drives epigenetic silencing of BRCA1 (Lu et al., 2011). In the BRCA1 work, we showed that silencing of the promoter could be induced by hypoxia and that this could be prevented by treatment of the cells with the HDAC inhibitor, trichostatin A (Lu et al., 2011). However, we had not determined the histone modifying factors that were necessary to bring about the silencing. Here, we have identified LSD1 and CoREST as the key factors required for MLH1 silencing.
In earlier work, we also identified the hypoxic tumor microenvironment as a driver of genetic instability (Bindra and Glazer, 2005; Reynolds et al., 1996; Yuan and Glazer, 1998). As one mechanism for this effect, we determined that hypoxia causes transient transcriptional down-regulation of the MMR pathway by provoking a shift in MLH1 promoter occupancy from activating c-Myc/Max to repressive Mad1/Max and Mnt/Max complexes (Bindra and Glazer, 2007). We now provide another mechanism for hypoxia-induced genetic instability by directly linking hypoxia with epigenetic regulation and durable long-term silencing of MLH1.
More broadly, the results also suggest the possibility that hypoxia may play a central role in epigenetic silencing not only of MLH1 but also of other important tumor suppressor genes that contribute to human cancers, such as p16, VHL, or IL-2Rγ (Baylin and Ohm, 2006; Jones and Baylin, 2007). Our results may therefore provide new mechanistic insights to suggest yet another role for hypoxia in cancer progression: the induction of durable epigenetic change causing gene silencing and consequent inactivation of critical tumor suppressor pathways.
Since tumor hypoxia is a dynamic process, with fluctuating regions of acute and chronic hypoxia reflecting a range of vascular abnormalities and perfusion defects, the impact of hypoxia on both the genome and epigenome of malignant cells can be profound in its scope. This highlights hypoxia as a major factor in generating tumor heterogeneity, a phenomenon that threatens to confound efforts at personalized medicine and the development of targeted therapies.
LSD1 was one of the first histone demethylases identified (Shi et al., 2004) and has been implicated in many cellular processes (Forneris et al., 2008; Wang et al., 2007). Here, we have identified a critical role for LSD1 in hypoxia-induced MLH1 silencing and in re-silencing following transient reactivation (by 5-aza-dC treatment) in RKO cells. However, we found that knockdown of LSD1, by itself, is not sufficient to reactivate the silenced MLH1 allele in RKO cells, most likely because DNA methylation persists and retains repressive complexes at the promoter.
Nonetheless, our work does provide a link between the LSD1/CoREST and DNA methylation. Our results show that knockdown of LSD1 or CoREST not only blocks H3K4 demethylation at the MLH1 promoter, but also blocks promoter DNA methylation after cessation of 5-aza-dC exposure. These results are consistent with previous studies showing that H3K4 methylation status plays an important role in preventing the establishment of DNA methylation (Ooi et al., 2007). This is also in keeping with the observations that LSD1 and the related demethylase, LSD2, are important in maintaining global DNA methylation (Wang et al., 2009) and in establishing maternal DNA genomic imprints (Ciccone et al., 2009), respectively.
As knowledge of the cancer epigenome accumulates, there is increasing interest in the development of epigenetic therapy for cancer treatment. The DNA methyltransferase inhibitors, azacitidine and decitabine, and the HDAC inhibitor, vorinostat, are already in clinical use. It has been suggested that the success of such agents may lie with their ability to reactivate silenced genes over long periods of time; however, after demethylation and activation by 5-aza-dC treatment, many genes eventually return to a silenced state once the agent is removed (McGarvey et al., 2006). One approach to prevent this is the use of a combination of DNMT and HDAC inhibitors (Gore et al., 2006; Mossman and Scott, 2011). Our work suggests that targeting LSD1 may represent another promising approach that could be used alone or in combination with other such agents.
Several studies have reported overexpression of LSD1 in a number of human cancers (Hayami et al., 2011; Kahl et al., 2006), and LSD1 inhibitors have shown anticancer activity in preclinical studies (Schenk et al., 2012; Willmann et al., 2012). Our work demonstrating the role of LSD1/CoREST in MLH1 silencing provides a further rationale to support the use of LSD1 inhibitors in cancer therapy and suggests that cancers with over-expression of LSD1 may be particularly prone to tumor suppressor gene silencing.
In the particular case of MLH1, prevention of silencing or reactivation of a silenced allele would not only serve to suppress genetic instability but also to restore the pro-apoptotic role of MLH1 in the DNA damage response, as cells deficient in MLH1 show a damage tolerance phenotype (Buermeyer et al., 1999; Meyers et al., 2001) and are resistant to cisplatin and temozolomide, among others (Aebi et al., 1996; Drummond et al., 1996; Francia et al., 2005). Hence, a pharmacologic strategy to inhibit or reverse MLH1 silencing would be a valuable tool to render cancer cells more sensitive to conventional chemotherapy.
Materials and Methods
Cells
HeLa, MCF-7, RKO, and SW480 cells were obtained from the ATCC (Manassas, VA) and grown according to supplier’s instructions.
Constructs
Lentivirus vectors for shRNAs against KDM5A RBP2-sh-1 and RBP2-sh-3 and control vector LLP were obtained from Dr. Marie Classon (Massachusetts General Hospital). Lentivirus shRNA vectors for LSD1 knock down were obtained from Sigma-Aldrich (LSD1-1: TRCN0000046068). The lentivirus shRNA vector for PLU-1 was obtained from Dr. Qin Yan (Yale University). Lentivirus shRNA vectors for CoREST knockdown were obtained from Sigma-Aldrich (CoREST: TRCN0000128260 ). The MLH1-TK-BlastR construct was produced by cloning the human 1.7 Kb MLH1 promoter into NheI pDisplay TK- BlastR vector (Palakurthy et al., 2009) .
Hypoxia
For severe hypoxia, cells were maintained in culture under a continuous flow of a humidified mixture of 95%N2 and 5%CO2 gas certified to <10 ppm, O2 for 48 h at 37°C as previously described (Reynolds et al., 1996). For moderate hypoxia (1% O2), an incubator was equipped with a PRO-OX 710 sensor (Biospherix, Redfield, NY) to regulate the flow of 100% N2 at low pressure (<25 lb/in2) in order to achieve a constant O2 concentration within the entire incubator for the indicated times. The CO2 level was maintained at 5% using an internal CO2-regulation system.
Chromatin immunoprecipitation assays
ChIP assays were performed as described (Bindra and Glazer, 2007). The primer sequences for the MLH1 promoter have been reported (Bindra and Glazer, 2007). Antibodies used for ChIP assays are listed in Supplemental Experimental Procedures.
Assays for MLH1 promoter silencing
RKO cells were transfected by lipofectin® 2000 with 12μg of MLH1-TK-BlastR plasmid, and a stable clones expressing thymidine kinase (TK) and blasticidin resistant (BlastR) genes (designated RKO MLH1-TK-BlastR ) were established by selection first with medium containing 1.5 μg/ml puromycin and one week later with medium containing 10 μg/ml blasticidin. The RKO MLH1-TK-BlastR cells were then tested for sensitivity to ganciclovir (GCV) to confirm the functional expression of the full MLH1-TK-BlastR cassette.
To test the impact of hypoxia on silencing of the MLH1 promoter in the RKO MLH1-TKBlastR cells, cells were plated under 1% O2 (or normoxic conditions) for 15, 25, or 35 days with passage once or twice per week. Cells at 100,000 cells per 100-mm dish were then subject to selection in presence of 10 μg/ml GCV. Colonies formed were counted for each condition tested and normalized to plating efficiencies.
To measure reactivation of the MLH1 promoter in the silenced MLH1-TK-BlastR cassette by 5-aza-dC treatment, selected clones containing silenced MLH1 promoters (GCV resistant subclones of the RKO MLH1-TK-BlastR cells) were incubated in medium containing either DMSO, 0.2 μM 5-aza-dC or 0.3μM 5-aza-dC for 48 h to inhibit DNA methylation. The cells were then plated in 100-mm dishes and the next day were exposed to medium containing either 10 μg/ml GCV or 10 μg/ml blasticidin to quantify cells that had regained TK or BlastR expression. Silencing or reactivation frequencies were calculated by dividing the number of clones growing under selection by the effective number of cells plated (as determined by cloning efficiency).
Western Blots
Cells were lysed in RIPA buffer (25 mM Tris·HCl PH 7.6, 150 mM NaCl, 1% Igepal CA-630, 1% sodium deoxycholate, 0.1% SDS) with protease inhibitor cocktail (Clontech). The primary antibodies used for Western blotting are listed in Supplemental Experimental Procedures.
Analysis of DNA methylation
Real-time PCR (MethyLight) was used for quantitative DNA methylation analysis (Eads et al., 2000). It is based on bisulfite-induced conversion of unmethylated C into U, leaving methylated C intact. Bisulfite conversion was performed using the EpiTect Bisufite Kit (Qiagen). Two sets of primers and probes, designed specifically to assay bisulfite-converted DNA, were used: a methylated set for MLH1 gene and a reference set, COL2A1 (the collagen 2A1 gene), to normalize the amount of input bisulfite DNA (Pérez-Carbonell et al., 2010). After bisulfite conversion, genomic DNA was amplified by fluorescence-based, real-time quantitative PCR. PMR (Percentage of fully methylated reference) value was used to calculate the amount of methylated DNA at the MLH1 promoter. The PMR calculation method is presented in in Supplemental Experimental Procedures.
Supplementary Material
Hypoxia induces long-term MLH1 silencing through specific histone modifications.
LSD1 is required for hypoxia-induced MLH1 silencing.
The LSD1/CoREST complex is required for MLH1 re-silencing after 5-aza-dC treatment.
LSD1 may be an attractive target for cancer therapy.
SIGNIFICANCE.
Epigenetic silencing of tumor suppressor MLH1 is common in sporadic cancers of the colon and other sites. However, the mechanism of MLH1 silencing remains elusive. We demonstrate that hypoxia induces epigenetic silencing of MLH1 and that the LSD1/CoREST complex is necessary for this process. The results also reveal a novel mechanism by which hypoxia causes a mutator phenotype during tumor progression. The findings suggest that LSD1/CoREST acts as an oncogene by epigenetically silencing MLH1 and identify the LSD1/CoREST complex as a potential target for epigenetic-based therapy.
Acknowledgements
We thank Dr. Zhong Yun for use of equipment and Denise Hegan for assistance. This work was supported by a grant from the National Institutes of Health (R01ES005775) to P.M.G.
Footnotes
Author contributions. Y.L. designed some and performed all the experiments. N.W. and M.S.T. contributed to the experimental design and provided reagents. P.M.G. designed most of the experiments. Y.L. and P.M.G. wrote and edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen RD, Boland CR, Koi M, Fishel R, Howell SB. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Genetic instability and the tumor microenvironment: towards the concept of microenvironment-induced mutagenesis. Mutat Res. 2005;569:75–85. doi: 10.1016/j.mrfmmm.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4//p130 complexes in hypoxia. Oncogene. 2006;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: Role of the Myc/Max network. Cancer Letters. 2007;252:93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annual review of genetics. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- Chen H, Yan Y, Davidson TL, Shinkai Y, Costa M. Hypoxic Stress Induces Dimethylated Histone H3 Lysine 9 through Histone Methyltransferase G9a in Mammalian Cells. Cancer Research. 2006;66:9009–9016. doi: 10.1158/0008-5472.CAN-06-0101. [DOI] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications — miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- Coquelle A, Toledo F, Stern S, Bieth A, Debatisse M. A New Role for Hypoxia in Tumor Progression: Induction of Fragile Site Triggering Genomic Rearrangements and Formation of Complex DMs and HSRs. Molecular Cell. 1998;2:259–265. doi: 10.1016/s1097-2765(00)80137-9. [DOI] [PubMed] [Google Scholar]
- Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS. LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer. 2013;109:994–1003. doi: 10.1038/bjc.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JT, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLalpha and mismatch repair deficiency in an ovarian tumor cell line. The Journal of biological chemistry. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Research. 2000;28:e32–00. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Allis CD, Lewis PW. New Epigenetic Drivers of Cancers. Science. 2011;331:1145–1146. doi: 10.1126/science.1203280. [DOI] [PubMed] [Google Scholar]
- Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of Histone Modifications and Gene Expression on DNA Hypermethylation in Cancer. Cancer Research. 2002;62:7213–7218. [PubMed] [Google Scholar]
- Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends in Biochemical Sciences. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Francia G, Green SK, Bocci G, Man S, Emmenegger U, Ebos JML, Weinerman A, Shaked Y, Kerbel RS. Down-regulation of DNA mismatch repair proteins in human and murine tumor spheroids: implications for multicellular resistance to alkylating agents. Molecular Cancer Therapeutics. 2005;4:1484–1494. doi: 10.1158/1535-7163.MCT-04-0214. [DOI] [PubMed] [Google Scholar]
- Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Combined DNA Methyltransferase and Histone Deacetylase Inhibition in the Treatment of Myeloid Neoplasms. Cancer Research. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- Hayami S, Kelly JD, Cho H-S, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BAJ, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. International Journal of Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa J-PJ, Markowitz S, Willson JKV, Hamilton SR, Kinzler KW, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proceedings of the National Academy of Sciences. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Greene E, Murray Stewart T, Goodwin AC, Baylin SB, Woster PM, Casero RA. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proceedings of the National Academy of Sciences. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Stewart TM, Wu Y, Baylin SB, Marton LJ, Perkins B, Jones RJ, Woster PM, Casero RA. Novel Oligoamine Analogues Inhibit Lysine-Specific Demethylase 1 and Induce Reexpression of Epigenetically Silenced Genes. Clinical Cancer Research. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AB, Denko N, Barton MC. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2008;640:174–179. doi: 10.1016/j.mrfmmm.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb AM, Buffa FM, Harris AL. Assessment of tumour hypoxia for prediction of response to therapy and cancer prognosis. Journal of Cellular and Molecular Medicine. 2010;14:18–29. doi: 10.1111/j.1582-4934.2009.00944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl P, Gullotti L, Heukamp LC, Wolf S, Friedrichs N, Vorreuther R, Solleder G, Bastian PJ, Ellinger J, Metzger E, et al. Androgen Receptor Coactivators Lysine-Specific Histone Demethylase 1 and Four and a Half LIM Domain Protein 2 Predict Risk of Prostate Cancer Recurrence. Cancer Research. 2006;66:11341–11347. doi: 10.1158/0008-5472.CAN-06-1570. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Hall NR, Lipford J, Kane MF, Rao MR, Morrison P, Wirth L, Finan PJ, Burn J, Chapman P, et al. Human mismatch repair genes and their association with hereditary non-polyposis colon cancer. Cold Spring Harbor symposia on quantitative biology. 1994;59:331–338. doi: 10.1101/sqb.1994.059.01.037. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Mol Cell Biol. 2011;31:3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced Tumor Suppressor Genes Reactivated by DNA Demethylation Do Not Return to a Fully Euchromatic Chromatin State. Cancer Research. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Meyers M, Wagner MW, Hwang H-S, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA Mismatch Repair Protein in Fluoropyrimidine-mediated Cell Death and Cell Cycle Responses. Cancer Research. 2001;61:5193–5201. [PubMed] [Google Scholar]
- Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman D, Scott RJ. Long Term Transcriptional Reactivation of Epigenetically Silenced Genes in Colorectal Cancer Cells Requires DNA Hypomethylation and Histone Acetylation. PLoS ONE. 2011;6:e23127. doi: 10.1371/journal.pone.0023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi RA, Hussain A, Deo SSV, Kukreti H, Chauhan M, Sarin R, Saxena A, Asim M, Shukla NK, Husain SA, et al. Hypermethylation analysis of mismatch repair genes (hmlh1 and hmsh2) in locally advanced breast cancers in Indian women. Human Pathology. 2008;39:672–680. doi: 10.1016/j.humpath.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palakurthy RK, Wajapeyee N, Santra MK, Gazin C, Lin L, Gobeil S, Green MR. Epigenetic Silencing of the RASSF1A Tumor Suppressor Gene through HOXB3-Mediated Induction of DNMT3B Expression. Molecular Cell. 2009;36:219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Carbonell L, Alenda C, Payá A, Castillejo A, Barberá VM, Guillén C, Rojas E, Acame N, Gutiérrez-Aviñó FJ, Castells A, et al. Methylation Analysis of MLH1 Improves the Selection of Patients for Genetic Testing in Lynch Syndrome. The Journal of Molecular Diagnostics. 2010;12:498–504. doi: 10.2353/jmoldx.2010.090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TY, Rockwell S, Glazer PM. Genetic Instability Induced by the Tumor Microenvironment. Cancer Research. 1996;56:5754–5757. [PubMed] [Google Scholar]
- Schenk T, Chen WC, Gollner S, Howell L, Jin L, Hebestreit K, Klein H-U, Popescu AC, Burnett A, Mills K, et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nature medicine. 2012;18:605–611. doi: 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y-J, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 Histone Demethylase Activity by Its Associated Factors. Molecular Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, Zhang H. Novel Histone Demethylase LSD1 Inhibitors Selectively Target Cancer Cells with Pluripotent Stem Cell Properties. Cancer Research. 2011;71:7238–7249. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, et al. Impairment of prostate cancer cell growth by a selective and reversible lysine-specific demethylase 1 inhibitor. International Journal of Cancer. 2012;131:2704–2709. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proceedings of the National Academy of Sciences. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Research. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SD, Marshall RS, Hill RP. Hypoxia induces DNA overreplication and enhances metastatic potential of murine tumor cells. Proceedings of the National Academy of Sciences. 1988;85:9533–9537. doi: 10.1073/pnas.85.24.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Glazer PM. Mutagenesis induced by the tumor microenvironment. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1998;400:439–446. doi: 10.1016/s0027-5107(98)00042-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.