Abstract
Liquid chromatography/mass spectrometry-based untargeted metabolomics is now an established experimental approach that is being broadly applied by many laboratories worldwide. Interpreting untargeted metabolomic data, however, remains a challenge and limits the translation of results into biologically relevant conclusions. Here we review emerging technologies that can be applied after untargeted profiling to extend biological interpretation of metabolomic data. These technologies include advances in bioinformatic software that enable identification of isotopes and adducts, comprehensive pathway mapping, deconvolution of MS2 data, and tracking of isotopically labeled compounds. There are also opportunities to gain additional biological insight by complementing the metabolomic analysis of homogenized samples with recently developed technologies for metabolite imaging of intact tissues. To maximize the value of these emerging technologies, a unified workflow is discussed that builds on the traditional untargeted metabolomic pipeline. Particularly when integrated together, the combination of the advances highlighted in this review helps transform lists of masses and fold changes characteristic of untargeted profiling results into structures, absolute concentrations, pathway fluxes, and localization patterns that are typically needed to understand biology.
Introduction
Liquid chromatography/mass spectrometry (LC/MS) provides a robust analytical platform to assay a physiochemically diverse group of small molecules and is therefore widely used to study the metabolome.[1] By using reversed-phase and hydrophilic interaction liquid chromatography together with quadropole time-of-flight or Orbitrap mass spectrometers, thousands of peaks are detected in the metabolic extract of biological samples.[2] Each of these peaks, often referred to as a “feature”, has a unique pair of retention-time and mass-to-charge ratios. Although experimental strategies to optimize metabolome coverage are still being developed, the process of measuring thousands of metabolite features in a biological specimen is now routine and has been discussed in detail.[3] In contrast, the interpretation of untargeted metabolomic data remains a challenge for many laboratories. This review focuses on emerging technologies that can be applied downstream of untargeted metabolite profiling to drive biological discovery.
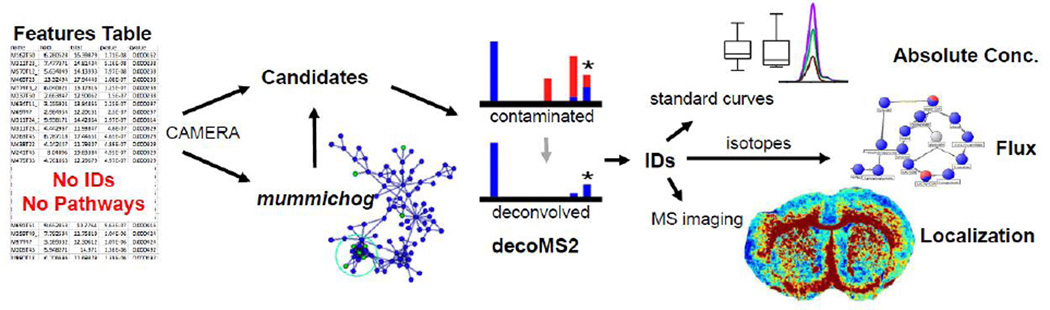
Traditionally, untargeted metabolomics is performed by analyzing metabolic extracts derived from two or more sample groups in MS1 mode. These raw data are then processed with bioinformatic software and a “features table” containing all detected compounds is produced. The most popular software for processing untargeted metabolomic data is the freely accessible and platform-independent XCMS, but other programs are also available.[4–6] A features table includes mass-to-charge ratios, retention time, statistical comparisons, and relative peak intensities.[7] Current software, however, does not provide metabolite identifications. Therefore, while the features table can be used to identify potential biomarkers or to broadly compare the similarity of samples, the value of the features table is relatively limited.[8,9] The question that inevitably arises is what the next step is after this initial processing of untargeted metabolomic data. Most investigators perform targeted MS2 analysis on peaks of interest with the objective of making structural identifications.[10] Given the time required for metabolite characterization and quantitation by LC/MS, generally only a small number of features are pursued. When comparing samples in which there are many metabolic differences, choosing the most relevant peaks to target for identification is a challenge. Moreover, even once structures are determined, biological interpretation is complicated because untargeted metabolomics only provides a relative comparison of metabolite levels. Additionally, untargeted metabolomics does not provide insight into pathway dynamics or spatial information with respect to tissue, cell type, or organelles. Here we review technologies that can be readily integrated with the untargeted metabolomic workflow to address these issues and facilitate data interpretation (Figure 1).
Figure 1.
Schematic showing the possible integration of emerging, mass spectrometry-based technologies into the untargeted metabolomic pipeline. The workflow starts with a features table that is output from LC/MS-based untargeted metabolomics. Features likely representing naturally occurring isotopes, adducts, and fragments of a single compound can be grouped by using the CAMERA software. After data reduction, mass-to-charge ratios can be searched in databases and interesting candidates chosen as targets for structural identification. Alternatively, the data can be analyzed by mummichog to find candidates that map onto related pathways. After acquiring MS2 data for features of interest, the spectra can be deconvolved by decoMS2 and matched against the MS2 of authentic standards. These pathway enrichments and identifications can be used to guide further experimentation that involve construction of standard curves to determine absolute concentrations, stable-isotope analysis to calculate metabolic flux, and mass spectrometry-based imaging to localize metabolites within biological tissues.
Post-Processing of Untargeted Metabolomic Data
A common strategy applied when prioritizing features to target for structural characterization is data filtering. Often, for example, features that are not changing within defined statistical thresholds are discarded. Additionally, features that have too weak of a signal intensity to obtain high-quality MS2 data or features that are not of biological origin can be removed.[11,12] In experiments where the sample groups being compared differ greatly, there may still be thousands of features that meet the specified criteria. Here, recently introduced software such as CAMERA and mummichog may facilitate further feature refinement.[7,13] CAMERA is a Bioconductor package that is designed to accept untargeted profiling data initially processed by XCMS. From the list of features detected by XCMS and the raw LC/MS data, CAMERA identifies features that likely correspond to the same metabolite. Given the tendency of metabolites to be detected as multiple features due to naturally occurring isotopes, in-source fragmentation, and adduct formation, CAMERA enables significant data reduction. In some cases, CAMERA reduces the number of features by ~50%.[14] It is important to emphasize that removal of these features from the data improves the likelihood of structurally characterizing a molecular ion with a fragmentation pattern that is in a metabolomic MS2 database. Although here we have described the filtering of a features table by manual inspection of statistical values and signal intensity followed by CAMERA processing, a freely available software package called MetShot was introduced last year to automate these steps.[15]
Even after this filtering, the remaining list of features is often too large for targeted MS2 analysis. The next post-processing step is to search each feature’s mass-to-charge ratio in metabolite databases. Databases with the largest number of mass-to-charge ratios for metabolites include METLIN, the Human Metabolome Database, LIPID MAPS, and the Madison Metabolomics Consortium Database.[16–20] These repositories can be manually searched on an individual basis or searched simultaneously by using a recently developed resource called MetaboSearch.[21] Database hits provide only putative feature assignments that must be validated by subsequent MS2 analysis, but these candidate matches can be assessed based on user interest and biochemical relevance. One strategy to assess features based on biochemical relevance is to prioritize features that have candidates belonging to the same metabolic pathway, a process that can be performed computationally by a program called mummichog. In brief, the input of mummichog is the mass-to-charge ratios of features determined to be unaltered between sample groups as well as those found to be statistically different. The mummichog software then determines possible candidate matches, maps all candidate matches onto a metabolic network (derived from KEGG, Recon1, and Edinburgh human metabolic network), and searches unique pathways for enrichment. Features within enriched pathways of interest may then be selectively targeted for MS2 analysis.
Taken together, the combination of post-processing technologies described above offers a powerful approach to refine the features table that is output from LC/MS-based untargeted metabolomics. The disadvantage of such a strategy is that it involves multiple software packages that require familiarity with programming languages such as R and Python. As an alternative, a web-based version of XCMS has been established called XCMS Online that allows users to freely upload and process raw untargeted metabolomic data.[22] XCMS Online generates a features table that is directly linked to the METLIN metabolite database. Then putative candidates can be viewed by clicking on features of interest. Data analyzed by XCMS Online is also processed by CAMERA and the features table is annotated accordingly. As another bioinformatic solution with post-processing functionality, Agilent Technologies has developed software called Mass Profiler Professional that can be used to process untargeted metabolomic data. Mass Profiler Professional incorporates algorithms similar to both CAMERA and mummichog, with pathway-enrichment analysis enhanced by the ability of the software to map both transcriptomic and proteomic data onto the same network.
Processing of MS2 data
After features are selected by using the post-processing described above, targeted MS2 experiments are typically performed on a single sample in which the peak of interest is above an acceptable intensity threshold. The threshold of intensity that provides high-quality MS2 data may vary and is determined by instrument sensitivity and settings. Ideally, MS2 spectra are acquired by using as narrow an ion isolation window as possible to reduce the possibility of detecting fragments from multiple precursors simultaneously. In practice, however, isolation windows greater than 1 Da are sometimes needed to analyze low-concentration compounds and can result in “mixed” MS2 spectra that contain fragments from multiple co-eluting precursors.[23] In some cases where compound separation is limited, even 1 Da isolation windows can lead to mixed MS2 spectra. Given that reference MS2 data are unavailable for an overwhelming number of naturally occurring metabolites and that fragmentation patterns are generally difficult to predict for small molecules, mixed MS2 spectra in metabolomics are challenging to interpret intuitively. Currently, metabolite identifications are validated by manually matching the MS2 data from a biological sample to reference MS2 data from a purified model compound in databases such as METLIN, MassBank, and mzCloud.[16,17,24,25] The presence of fragments from multiple precursors can therefore prevent metabolite identification. A computational strategy for deconvolving mixed MS2 spectra was recently developed called decoMS2.[23] While decoMS2 uses chromatographic deconvolution approaches that have been successfully applied in GC/MS-based metabolomics, it also deconvolves based on correlations between fragment and precursor intensities as the mass-to-charge ratio targeted for MS2 analysis is shifted.[26]
Once targeted MS2 data are acquired and deconvolved, they can be searched against reference MS2 databases. Although metabolomic MS2 databases have fragmentation spectra for more than 11,000 standard metabolites and continue to grow rapidly, their coverage is still incomplete.[17] Consequently, many queried MS2 spectra will return no database matches. In an effort to increase the number of reference MS2 spectra, in silico fragmentation approaches have been applied by software such as Mass Frontier, MetFrag, and LipidBlast to generate hypothetical fragmentation patterns that can be used for metabolite identification.[27–29] While these in silico approaches are less robust than matching to standard experimental data, they greatly expand the number of metabolites that can be searched with MS2 data and provide valuable leads that can be subsequently confirmed with authentic standard compounds.
Targeted Quantitation and Pathway Dynamics with Stable Isotopes
Untargeted metabolomics only reports relative quantitation in the form of a fold change, namely the average change in peak intensity between sample groups. Fold-change values reported in a features table are not equivalent to changes in concentration. The ionization efficiency of most metabolites is non-linear and matrix dependent, meaning that changes in signal intensity do not necessarily correspond to equal changes in concentration. A large fold change may represent only a small change in concentration or vice versa. Yet, the biological effects of metabolites are concentration dependent. If glucose concentrations are outside of a normal range in the blood, for example, the physiological consequences can be severe and lead to the dysfunction of multiple organs. Therefore, in a biological context, it is important to translate changes in signal intensity as reported in the untargeted metabolomic features table to changes in concentration. The most reliable approach to determine absolute concentration is to spike samples with a range of concentrations of a stable-isotope standard and then construct a standard curve.[30] Historically, this method for absolute quantitation has been performed by using triple quadrupole mass spectrometry. By the time that post-processing of the features table is completed and MS2 matching performed, typically less than 100 metabolites are structurally identified. From this list, researchers can perform a targeted analysis to determine the absolute concentration of selected metabolites key to pathway regulation or fundamentally related to the biological hypotheses being investigated.
Interpreting untargeted metabolomic data in a biological context is also complicated by the absence of information about pathway flux. As an example, consider a metabolite that is identified to be strongly up-regulated in a group of samples representing a unique phenotype. From this result, it is not possible to determine if the metabolite’s elevation reflects increased utilization or decreased production/excretion.[31] Additionally, because metabolites are consumed/produced by many reactions and many of those reactions have substrates that may not be measured by untargeted metabolomics, identifying dysregulated pathways can be challenging. To assess the flux of potentially dysregulated pathways as implicated by untargeted metabolomics, a follow-up experiment involving stable isotopes may be performed in which biological systems are provided with an isotopically enriched nutrient and the distribution of the isotope into downstream products is subsequently measured.[32,33] Several variations of flux analysis are commonly applied that involve examining positional labeling patterns in isotopomers and/or the distribution of label in isotopologues with time.[34–37] The latter approach is readily accomplished by the same types of LC/MS instrumentation that is used for untargeted metabolomics and platform-independent, freely available software called MAVEN has recently been introduced to facilitate the associated data analysis.[38] Although MAVEN does not calculate metabolic flux directly, flux values can be derived for targeted pathways by using the MAVEN output as recently described.[39] It is now also possible to process stable-isotope LC/MS data in an untargeted context by using the X13CMS software.[40] X13CMS does not report flux, but it may be used to monitor how the distribution of isotope changes among all detected compounds (including potential unknowns) after metabolic perturbation.
In Situ Imaging of Metabolites
When analyzing biological tissues by LC/MS-based metabolomics, the first step in the workflow is metabolite extraction by sample homogenization.[41] This procedure mixes the metabolome of all cells within the sample and therefore leads to an averaging effect that prevents cell-specific quantitation.[42] To demonstrate the challenges of interpreting LC/MS-based metabolomic data from intact tissue we refer to a recently published study in which Caenorhabditis elegans worms with extended lifespan were compared to wildtype controls.[43] Although the levels of multiple metabolites were determined to be altered in long-lived worms, the cell types or tissues in the worm contributing to this metabolic dysregulation could not be determined by LC/MS-based metabolomics. Here, the application of technological advances in mass spectrometry-based imaging have great potential for determining metabolite localization patterns.[44]
Mass spectrometry-based imaging is often accomplished by using matrix-assisted laser desorption ionization (MALDI). Tissues are sectioned, transferred to a MALDI plate, a matrix is applied, and mass spectra are acquired after systematic laser rastering. The signal intensity of analytes of interest can then be plotted spatially to construct a two-dimensional image. This approach has been successfully applied to determine the distribution pattern of neuropeptides within nervous tissue, to discover that phosphatidylcholine species increase in the brain following ischemic insult, and to determine that triacylglycerol biosynthesis varies across the tissues of a cotton embryo.[45–47] While MALDI has been effective for imaging peptides and lipids, matrix interference in the low-mass region has complicated its application to the measurement of some metabolites.[48] To image metabolites in the low-mass region a technology has emerged known as nanostructure-imaging mass spectrometry (NIMS, also referred to as nanostructure-initiator mass spectrometry).[49] NIMS is compatible with MALDI instrumentation, but is a matrix-free approach and therefore enables imaging of small molecules without background interference. NIMS has been applied to study sugars, glycolytic intermediates, nucleotides, amino acids, and xenobiotics as well as lipids.[50–52] In the example referenced above, the application of NIMS suggested that metabolites altered in long-lived worms are localized to muscle tissue.[43] While this level of anatomical localization provided by metabolomic imaging provides important insight, future technological developments facilitating single-cell imaging within a tissue or sub-cellular localization of metabolites will be of immense biological utility.
Conclusions
Currently, a major challenge in the field of metabolomics is interpreting untargeted profiling results in the context of a biological problem. LC/MS-based untargeted metabolomics is now an established technology that is routinely used to study biological systems. The output of these experiments is a mass-to-charge ratios and relative intensity for each of the thousands of peaks typically detected. One approach to data analysis is to consider the metabolomic output as providing potentially interesting biological leads that can be pursued with additional targeted experiments. In this review, we have described emerging mass spectrometry-based technologies that facilitate the identification and prioritization of leads as well as characterization of their structure, concentration, flux, and anatomical localization. The integration of these technologies into the untargeted profiling pipeline downstream of the conventional LC/MS steps will provide a much richer metabolic picture to drive biological discovery. Ultimately, however, biological validation of metabolomic data involves altering the phenotype of cell culture, animal models, or patients by pathway manipulation. While in some instances investigators may be able to move to pathway manipulation directly from untargeted metabolomic results, the technologies highlighted here are likely to refine the biological hypotheses being tested. In summary, untargeted metabolomics assays a broad portion of the metabolome for relative changes across biological systems and therefore has a unique capacity to generate unanticipated leads and biological questions. A particularly exciting application of this unbiased approach is the ability to generate biological questions related to unknown metabolites, namely compounds whose structures and pathways have not yet been discovered.[53] As with any scientific question, however, additional experiments are necessary for hypothesis testing and development. Of course, the specific experiments needed will be highly dependent upon the system studied and the questions being investigated. Yet, the innovative advances highlighted in this review provide an excellent framework of mass spectrometry-based technologies that are available downstream to leverage untargeted profiling results and drive biological discovery.
Metabolomic data are information rich, but challenging to interpret biologically.
New software can guide hypothesis generation and direct further experiments.
Technologies for isotope analysis & metabolite imaging drive biological discovery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of Special/Outstanding Interest
- 1.Baker M. Metabolomics: from small molecules to big ideas. Nat Meth. 2011;8:117–121. [Google Scholar]
- 2.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Expanding coverage of the metabolome for global metabolite profiling. Analytical chemistry. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milne SB, Mathews TP, Myers DS, Ivanova PT, Brown HA. Sum of the parts: mass spectrometry-based metabolomics. Biochemistry. 2013;52:3829–3840. doi: 10.1021/bi400060e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 5.Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- 6.Katajamaa M, Miettinen J, Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- 7. Kuhl C, Tautenhahn R, Bottcher C, Larson TR, Neumann S. CAMERA: An Integrated Strategy for Compound Spectra Extraction and Annotation of Liquid Chromatography/Mass Spectrometry Data Sets. Anal Chem. 2012;84:283–289. doi: 10.1021/ac202450g. Introduction of CAMERA software to identify features likely produced by the same metabolite.
- 8.Beltran A, Suarez M, Rodriguez MA, Vinaixa M, Samino S, Arola L, Correig X, Yanes O. Assessment of compatibility between extraction methods for NMR- and LC/MS-based metabolomics. Anal Chem. 2012;84:5838–5844. doi: 10.1021/ac3005567. [DOI] [PubMed] [Google Scholar]
- 9.Weisenberg SA, Butterfield TR, Fischer SM, Rhee KY. Suitability of silica hydride stationary phase, aqueous normal phase chromatography for untargeted metabolomic profiling of Enterococcus faecium and Staphylococcus aureus. J Sep Sci. 2009;32:2262–2265. doi: 10.1002/jssc.200900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stupp GS, Clendinen CS, Ajredini R, Szewc MA, Garrett T, Menger RF, Yost RA, Beecher C, Edison AS. Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans. Anal Chem. 2013;85:11858–11865. doi: 10.1021/ac4025413. Innovative use of stable isotopes to remove LC/MS signals that are not of biological origin.
- 12. Bueschl C, Kluger B, Berthiller F, Lirk G, Winkler S, Krska R, Schuhmacher R. MetExtract: a new software tool for the automated comprehensive extraction of metabolite-derived LC/MS signals in metabolomics research. Bioinformatics. 2012;28:736–738. doi: 10.1093/bioinformatics/bts012. Innovative use of stable isotopes to remove LC/MS signals that are not of biological origin.
- 13. Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9:e1003123. doi: 10.1371/journal.pcbi.1003123. Introduction of mummichog software to map candidate metabolite assignments onto metabolic networks.
- 14.Bottcher C, von Roepenack-Lahaye E, Schmidt J, Schmotz C, Neumann S, Scheel D, Clemens S. Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 2008;147:2107–2120. doi: 10.1104/pp.108.117754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neumann S, Thum A, Böttcher C. Nearline acquisition and processing of liquid chromatography-tandem mass spectrometry data. Metabolomics. 2013;9:84–91. Description of an approach and software to automate post-processing of untargeted metabolomic data.
- 16.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 17.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sud M, Fahy E, Cotter D, Brown A, Dennis EA, Glass CK, Merrill AH, Jr, Murphy RC, Raetz CR, Russell DW, et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 21. Zhou B, Wang J, Ressom HW. MetaboSearch: Tool for Mass-Based Metabolite Identification Using Multiple Databases. PLoS One. 2012;7:e40096. doi: 10.1371/journal.pone.0040096. Tool that enables searching of mass-to-charge ratios from multiple metabolite databases simultaneously.
- 22. Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. Introduction of a web-based version of XCMS that enables users to upload, process, and visualize data through an intuitive graphical user interface.
- 23. Nikolskiy I, Mahieu NG, Chen YJ, Tautenhahn R, Patti GJ. An untargeted metabolomic workflow to improve structural characterization of metabolites. Anal Chem. 2013;85:7713–7719. doi: 10.1021/ac400751j. Deconvolution of metabolomic MS2 data based on retention time and shifting mass-to-charge ratios isolated for fragmentation. Deconvolved MS2 spectra facilitate metabolite identifications.
- 24.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Weber RJ, Allwood JW, Mistrik R, Zhu Z, Ji Z, Chen S, Dunn WB, He S, Viant MR. HAMMER: automated operation of mass frontier to construct in silico mass spectral fragmentation libraries. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiller K, Hangebrauk J, Jager C, Spura J, Schreiber K, Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81:3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 27.Wolf S, Schmidt S, Muller-Hannemann M, Neumann S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinformatics. 2010;11:148. doi: 10.1186/1471-2105-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kind T, Liu KH, Lee do Y, DeFelice B, Meissen JK, Fiehn O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. Release of computer-generated MS2 library containing 212,516 spectra covering 119,200 compounds from 26 lipid compound classes.
- 29.Kasper PT, Rojas-Cherto M, Mistrik R, Reijmers T, Hankemeier T, Vreeken RJ. Fragmentation trees for the structural characterisation of metabolites. Rapid Commun Mass Spectrom. 2012;26:2275–2286. doi: 10.1002/rcm.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeBerardinis Ralph J, Thompson Craig B. Cellular Metabolism and Disease: What Do Metabolic Outliers Teach Us? Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woolston BM, Edgar S, Stephanopoulos G. Metabolic engineering: past and future. Annu Rev Chem Biomol Eng. 2013;4:259–288. doi: 10.1146/annurev-chembioeng-061312-103312. [DOI] [PubMed] [Google Scholar]
- 33.Metallo CM, Vander Heiden MG. Understanding metabolic regulation and its influence on cell physiology. Mol Cell. 2013;49:388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamboni N. 13C metabolic flux analysis in complex systems. Curr Opin Biotechnol. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat Protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weitzel M, Noh K, Dalman T, Niedenfuhr S, Stute B, Wiechert W. 13CFLUX2--high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics. 2013;29:143–145. doi: 10.1093/bioinformatics/bts646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics. 2012;Chapter 14(Unit14):11. doi: 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Chen YJ, Cho K, Nikolskiy I, Crawford PA, Patti GJ. XCMS: Global Tracking of Isotopic Labels in Untargeted Metabolomics. Anal Chem. 2014 doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheikh KD, Khanna S, Byers SW, Fornace A, Jr, Cheema AK. Small molecule metabolite extraction strategy for improving LC/MS detection of cancer cell metabolome. J Biomol Tech. 2011;22:1–4. [PMC free article] [PubMed] [Google Scholar]
- 42.Yanes O. Metabolomics: Playing pinata with single cells. Nat Chem Biol. 2013;9:471–473. doi: 10.1038/nchembio.1297. [DOI] [PubMed] [Google Scholar]
- 43. Patti GJ, Tautenhahn R, Johannsen D, Kalisiak E, Ravussin E, Bruning JC, Dillin A, Siuzdak G. Meta-Analysis of Global Metabolomic Data Identifies Metabolites Associated with Life-Span Extension. Metabolomics. 2013 doi: 10.1007/s11306-013-0608-8. in press. Data reduction is performed at the feature level by identifying shared alterations in multiple long-lived mutants. In situ metabolite imaging of these dysregulated compounds suggests that alterations are localized to worm muscle.
- 44.Lanni EJ, Rubakhin SS, Sweedler JV. Mass spectrometry imaging and profiling of single cells. J Proteomics. 2012;75:5036–5051. doi: 10.1016/j.jprot.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmerman TA, Monroe EB, Tucker KR, Rubakhin SS, Sweedler JV. Chapter 13: Imaging of cells and tissues with mass spectrometry: adding chemical information to imaging. Methods Cell Biol. 2008;89:361–390. doi: 10.1016/S0091-679X(08)00613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koizumi S, Yamamoto S, Hayasaka T, Konishi Y, Yamaguchi-Okada M, Goto-Inoue N, Sugiura Y, Setou M, Namba H. Imaging mass spectrometry revealed the production of lyso-phosphatidylcholine in the injured ischemic rat brain. Neuroscience. 2010;168:219–225. doi: 10.1016/j.neuroscience.2010.03.056. [DOI] [PubMed] [Google Scholar]
- 47.Horn PJ, Korte AR, Neogi PB, Love E, Fuchs J, Strupat K, Borisjuk L, Shulaev V, Lee YJ, Chapman KD. Spatial mapping of lipids at cellular resolution in embryos of cotton. Plant Cell. 2012;24:622–636. doi: 10.1105/tpc.111.094581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forsythe JG, Broussard JA, Lawrie JL, Kliman M, Jiao Y, Weiss SM, Webb DJ, McLean JA. Semitransparent nanostructured films for imaging mass spectrometry and optical microscopy. Anal Chem. 2012;84:10665–10670. doi: 10.1021/ac3022967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 50.Patti GJ, Woo HK, Yanes O, Shriver L, Thomas D, Uritboonthai W, Apon JV, Steenwyk R, Manchester M, Siuzdak G. Detection of carbohydrates and steroids by cation-enhanced nanostructure-initiator mass spectrometry (NIMS) for biofluid analysis and tissue imaging. Anal Chem. 2010;82:121–128. doi: 10.1021/ac9014353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amantonico A, Flamigni L, Glaus R, Zenobi R. Negative mode nanostructure-initiator mass spectrometry for detection of phosphorylated metabolites. Metabolomics. 2009;5:346–353. [Google Scholar]
- 52.Yanes O, Woo HK, Northen TR, Oppenheimer SR, Shriver L, Apon J, Estrada MN, Potchoiba MJ, Steenwyk R, Manchester M, et al. Nanostructure initiator mass spectrometry: tissue imaging and direct biofluid analysis. Anal Chem. 2009;81:2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patti GJ, Yanes O, Shriver LP, Courade JP, Tautenhahn R, Manchester M, Siuzdak G. Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin. Nat Chem Biol. 2012;8:232–234. doi: 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]