Abstract
Objective
To assess the virological response, genotypic resistance profiles, and antiretroviral plasma concentrations in HIV-2 antiretroviral-treated (antiretroviral therapy, ART) patients in Côte d‘Ivoire.
Methods
A cross-sectional survey was conducted among HIV-2 patients receiving ART. Plasma HIV-2 viral load was performed using the Agence Nationale de Recherche sur le SIDA et les hépatites virales (ANRS) assay. Protease and reverse transcriptase sequencing was performed using in-house methods and antiretroviral plasma concentrations were assessed using ultra performance liquid chromatography combined with tandem mass spectrometry.
Results
One hundred and forty-five HIV-2-treated patients were enrolled with a median CD4+ cell count of 360 cells/µl (interquartile range, IQR = 215–528). Median duration of ART was 4 years (IQR = 2–7) and 74% of patients displayed viral load less than 50 copies/ml. Median plasma HIV-2 RNA among patients with viral load more than 50 copies/ml was 3016 copies/ml (IQR = 436–5156). Most patients (84%) received a lopinavir/ritonavir-based regimen. HIV-2 resistance mutations to nucleoside reverse transcriptase inhibitors and protease inhibitors were detected in 21 of 25 (84%) and 20 of 29 (69%) samples, respectively. The most prevalent nucleoside reverse transcriptase inhibitor resistance mutations were M184I/V (90%), Q151M (24%), and S215F/Y (24%). The most prevalent protease inhibitor resistance mutations were V47A (60%) and I54M (30%). Median CD4+ cell counts were 434 cells/µl (292–573) and 204 cells/µl (122–281) in patients with viral load less than 50 copies/ml and those exhibiting virological failure (P < 0.0001), respectively. The proportions of patients with adequate antiretroviral plasma concentrations were 81 and 93% in patients displaying virological failure and in those with viral load less than 50 copies/ml, respectively (P = 0.046), suggesting good treatment adherence.
Conclusion
We observed adequate drug plasma concentrations and virological suppression in a high proportion of HIV-2-infected patients. However, in cases of virological failure, the limited HIV-2 therapeutic arsenal and cross-resistance dramatically reduced treatment options.
Keywords: antiretroviral resistance, Côte d’Ivoire, HIV-2, plasma drug concentrations
Introduction
Between one and two million people are estimated to be infected with HIV-2, mainly in West Africa [1]. HIV-2 infection represents a unique model of attenuated retroviral infection, characterized by slow disease progression associated with prolonged maintenance of a normal CD4+ cell count [2]; low infectivity, with a high proportion of patients with a spontaneous undetectable plasma viral load in the absence of antiretroviral drugs [3]; and a low rate of transmission [4, 5].
HIV type 1 (HIV-1) and HIV-2 genomes differ by about 50–60% at the nucleotide level. These differences may be correlated with differential responses to some antiretrovirals, as observed with the natural resistance of HIV-2 to nonnucleoside reverse transcriptase inhibitors, the fusion inhibitor enfuvirtide, and the decreased susceptibility to some protease inhibitors (amprenavir, indinavir, nelfinavir, atazanavir, tipranavir) [6–9]. Owing to genetic diversity, HIV-2-specific assays are needed for virological monitoring: that is, viral load quantification assays and genotypic resistance tests. These processes complicate the biological follow-up of HIV-2-infected patients, particularly in West Africa, the endemic region of this virus. Data available in the literature among HIV-2 genotypic resistance profiles issued from patients in virological failure are mainly obtained from limited series [10–14], and plasma drug concentrations are usually not assessed.
The aim of this study was to assess the virological response, genotypic resistance profiles, and treatment adherence of HIV-2 antiretroviral-treated patients in Côte d‘Ivoire.
Patients and methods
From April 2012 to August 2012, a cross-sectional survey was conducted among HIV-2-infected patients receiving an antiretroviral-based treatment (antiretroviral therapy, ART) and followed up in six HIV clinics in Abidjan, Côte d’Ivoire, which were participating in the International epidemiologic Databases to Evaluate AIDS (IeDEA)West Africa collaboration [15, 16]. The HIV-2 West Africa cohort is a network of HIV-2 and dually HIV-1/HIV-2-seropositive patients of West Africa embedded in the IeDEA-West Africa collaboration with five countries. The IeDEA-West Africa HIV-2 cohort aims, first, to build and strengthen an operational and clinical research agenda to better describe HIV-2 case management and its short-term and then longer-term consequences in the West Africa region.
All HIV-2 patients registered in this database and on ART and who attended one of the participating clinics during the study period were proposed to participate in this survey. The National Bioethics Committee of the Ministry of Health of Côte d’Ivoire approved the study protocol.
National guidelines in Côte d’Ivoire are based on World Health Organization recommendations for HIV-2 treatment in resource-limited settings. These latter recommend in first-line regimen, the use of two nucleoside reverse transcriptase inhibitors (NRTIs) combined with boosted protease inhibitors (lopinavir, saquinavir, or indinavir: with lopinavir as the preferred option); or as an alternative first-line, for patients with a CD4+ cell count above 200 cells/µl, a triple-NRTI regimen [17].
Ultrasensitive plasma HIV-2 RNA quantification was performed using the new in-house real-time PCR assay, developed in the AC11 Quantification Group of the French National Agency for AIDS and Viral Hepatitis Research (Agence Nationale de Recherches sur le Sida et les hépatites virales, ANRS) and commercialized by Biocentric, using a quantification threshold of 10 copies/ml used in this study.
Genotypic resistance tests, that is, protease and reverse transcriptase sequencing, were performed in samples with a plasma HIV-2 viral load above 50 copies/ml using in-house methods. Briefly, a fragment of each region was amplified by reverse transcriptase-PCR according to the manufacturer’s instructions (Titan One Tube RT-PCR Kit; Roche, Mannheim, Germany) using the following primers: RTC/RT2 for reverse transcriptase [18], DP20 [19]/PRRT1 (5′ CYTGRGTYACCYTRTTTARYT) for protease, and INT1S/INT1AS for integrase [20]. Nested PCR reactions were performed with Taq DNA polymerase (Roche) following the manufacturer’s instructions and the primers: RT3/RT4 for reverse transcriptase [18], DP26/PRRT2 (5′ CTATYAGCATYCTCCATTTG) [19] for protease, and with two different pairs of primers for integrase region (INT1S/INT2SA and INT2RS/INT2R) [20]. Sequencing reactions were run on an ABI Prism BigDye Terminator kit using an automated sequencer (ABI Prism 3130 XL; Applied Biosystems, Foster City, California, USA). Sequences editing and alignment were performed using the SmartGene software.
In this study, HIV-2 resistance mutations were identified using the list generated by the ‘Collaborative HIV and Anti-HIV Drug Resistance Network’ [21], leading to the following mutations in reverse transcriptase – K65R, D67G/N, N69S/T, K70N/R, L74V, V111I, Y115F, M184I/V, Q151M, S215A/C/F/L/Y, K223R; and in protease – V47A, G48V, I50V, I54L/M, V62A, I82F, I84V, L90M, L99F. Among the protease inhibitor resistance mutations, the I50V, I54M, and I84V were associated with resistance to darunavir [21, 22]. HIV-2 subtype was evaluated by comparing the polymerase sequence to consensus sequences using the Los Alamos database (www.hiv.lanl.gov). Protease and reverse transcriptase sequences were submitted to GenBank with the following accession numbers: KJ131111-KJ131164 and KJ156622.
Estimation of adherence was based on information provided by the patients regarding the intake of medicine during three periods: the previous day; the previous 4 days; and the previous 30 days.
Antiretroviral plasma concentrations were measured in the same samples as those used for viral load assay, using ultra performance liquid chromatography combined with tandem mass spectrometry (UPLC-MS/MS; Waters Corporation Milford, Milford, Massachusetts, USA) [23]. Considering the lack of in-vivo data on HIV-2 threshold plasma concentrations and according to the respective daily dosage administered, plasma concentrations were considered adequate if lopinavir was at least 1000 ng/ml, indinavir at least 150 ng/ml, and saquinavir at least 100 ng/ml [24].
Qualitative variables were expressed as percentages. Quantitative variables were expressed as medians with their interquartile ranges (IQRs). Comparison of percentages was analyzed using the Pearson χ2 test, or Fisher’s exact test, and for comparisons of means, Student’s t-test was used. Comparisons of medians were made using the Mann–Whitney U-test.
Results
Patients’ characteristics
A total of 145 HIV-2-treated patients were enrolled: 54% men, median age of 45 years (IQR = 38–51; Table 1). The median time since initiation of the first ART was 4 years (IQR = 2–7), and the median pre-ART CD4+ cell count, available for 107 patients, was 171 cells/µl (IQR = 94–285). The regimens prescribed at initiation of ART were as follows: protease inhibitor-based regimens for 112 patients (77%), including 67 of lopinavir/ritonavir (60%), 37 (33%) of indinavir (including 17 boosted with ritonavir), and eight (7%) of nelfinavir; dual or triple NRTI regimens in 13 patients (9%); and NNRTI-based regimens, prescribed during several months before HIV type determination, in 20 patients (14%). These latter were all switched to a non-NNRTI-based regimen within a median delay of 9 months, including seven (35%) who were switched during the first month of ART. At the time of the study, 48 of the 145 patients (33%) were still receiving their first-line regimen, and 67% had switched to another ART.
Table 1.
Characteristics of 145 HIV-2-infected patients.
| Characteristic | Value |
|---|---|
| Men, n (%) | 79 (54) |
| Age, median years (IQR) | 45 (38–51) |
| Pre-ART CD4+ T-cell count, median cells/µl (IQR) | 171 (94–285) |
| Current CD4+ T-cell count, median cells/µl (IQR) | 360 (215–528) |
| Time since HIV-2 diagnosis, median years (IQR) | 5(2–7) |
| Time since first antiretroviral therapy, median years (IQR) | 4(2–7) |
| Patients in first-line regimen, n (%) | 48 (33) |
| First ART regimen, n (%) | |
| PI-based | 112 (77) |
| Lopinavir +/− ritonavir | 67 |
| Indinavir +/− ritonavir | 37 (17 with ritonavir) |
| Nelfinavir | 8 |
| Dual or triple NRTI | 13 (9) |
| NNRTI-based | 20 (14) |
| Patients still receiving their first-line ART regimen, n (%) | 48 (33) |
| Current ART regimen, n (%) | |
| PI-based | 137 (94) |
| Lopinavir/ritonavir | 131 |
| Darunavir/ritonavir | 2 |
| Saquinavir | 2 |
| Indinavir | 2 |
| Triple NRTI | 8 (6) |
| NNRTI-based | 0 (0) |
ART, antiretroviral therapy; IQR, interquartile range; NNRTI, nonnucleoside reverse transcriptase inhibitor; NRTI, nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
Immunovirological profiles
At the time of the study, patients were receiving the following ART: protease inhibitor-based regimen (n=137, 94%) including lopinavir/ritonavir (n=131), darunavir/ritonavir (n=2), saquinavir (n=2), and indinavir (n=2); and triple NRTI regimen (n=8, 6%).
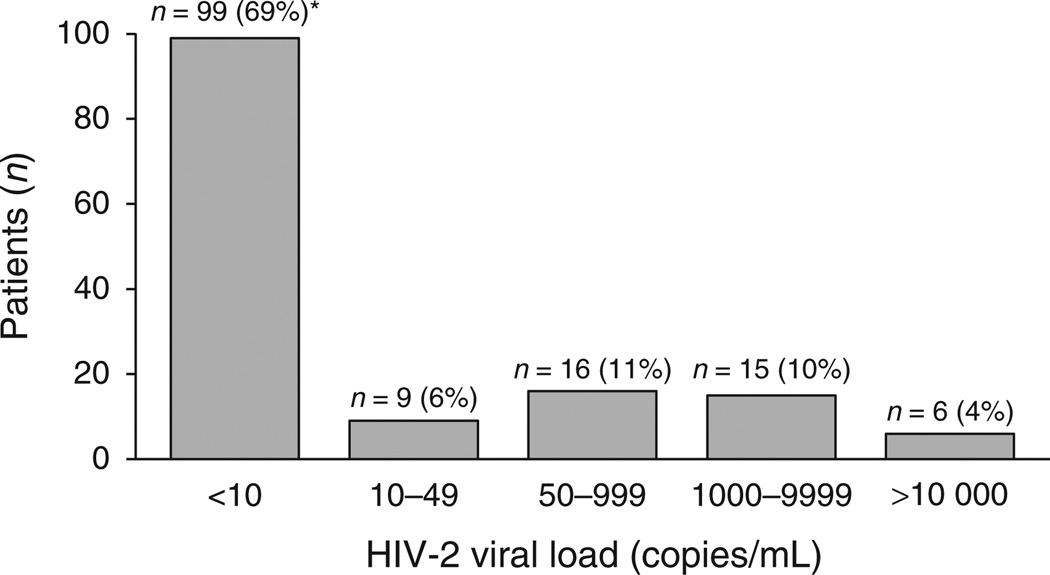
At the time of the study, median CD4+ cell count was 360 cells/µl (IQR = 215–528) and the distribution of HIV-2 plasma viral load was as follows: 99 patients had a viral load less than 10 copies/ml. Among these latter, 11 patients had detectable HIV-2 RNA (i.e. between 1 and 10 copies/ml), nine had a HIV-2 viral load between 10 and 49 copies/ml, 16 between 50 and 999 copies/ml, 15 between 1000 and 9999 copies/ml, and six had at least 10 000 copies/ml (Fig. 1). Thus, at a threshold value of 50 copies/ml, 108 patients (74%) displayed a plasma HIV-2 viral load less than 50 copies/ml, and 37 (26%) were in virological failure, that is, with a HIV-2 plasma viral load more than 50 copies/ml. Patients with viral load more than 50 copies/ml were treated for a longer time than those with viral load less than 50 copies/ml (76 vs. 42 months, P = 0.0001).
Fig. 1. Distribution of plasma HIV-2 viral load among 145 patients at the time of the study.
*11 with detectable HIV-2 RNA.
Among the latter, the median viral load was 3016 copies/ml (IQR = 454–5156). Most of these patients (n = 30, 81%) were receiving at least a second-line regimen. The ART received at the time of the study was as follows: protease inhibitor-based regimen (n = 31, 86%) with lopinavir/ritonavir in 29 cases, saquinavir/ritonavir in one case, and indinavir in one case; and triple NRTI regimen in four patients (11%). The two remaining patients received a dual protease inhibitor therapy (lopinavir/ritonavir+saquinavir) and a salvage therapy (tenofovir/emtricitabine+raltegravir+darunavir/ritonavir).
At the time of the study, median CD4+ cell count was 434 cells/µl (IQR = 292–573) in those patients with viral load less than 50 copies/ml, and 204 cells/µl (IQR = 122–281) in patients who had virological failure (P < 0.0001). Median change in CD4+ cell count between initiation of first ART and the time of the study was +172 cells/µl (IQR=+70 to +305) and +29 cells/µl (IQR = −65 to + 69) in patients with viral load less than 50 copies/ml and in those with virological failure, respectively (P < 0.0001).
Genotypic resistance tests
In the 37 patients with virological failure, protease and reverse transcriptase sequencing were successful in 29 (78%) and 25 (68%) of samples, respectively. Among the 31 samples with available sequences (protease or reverse transcriptase), 22 (71%) patients were infected with HIV-2 group B and nine (29%) with HIV-2 group A.
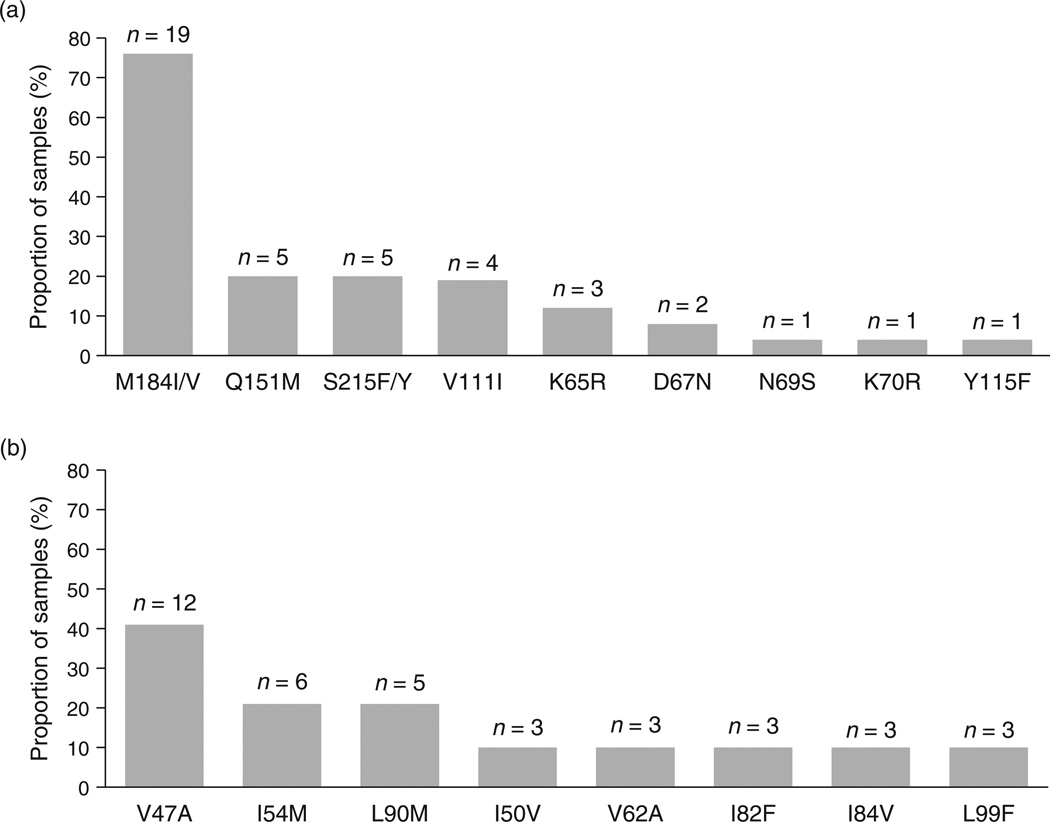
HIV-2 mutations associated with NRTI and protease inhibitors resistance were detected in 21 of 25 (84%) and 20 of 29 (69%) of samples, respectively. The most prevalent resistance mutations to NRTI were the following: M184V (n = 17, 81%), Q151M (n = 5, 24%), S215F/Y (n = 5, 24%), V111I (n = 4, 19%), K65R (n = 3, 14%), M184I (n = 2, 10%), and D67N (n = 2, 10%) (Fig. 2a). Each of the following mutations N69S, K70R, and Y115F were detected in one sample (5%). The most prevalent resistance mutations to protease inhibitors were as follows: V47A (n = 12, 60%), I54M (n = 6, 30%), and L90M (n = 5, 25%; Fig. 2b). Each of the following protease inhibitor resistance mutations I50V, I82F, I84V, V62A, and L99F were detected in three samples (15%). Among the protease inhibitor-resistant viruses, eight (40%) displayed at least one darunavir resistance-associated mutation [I54M(n = 5), I50V+I84V (n = 2), I50V+I54M (n = 1)]. Nine of the 12 patients harboring V47A-mutated viruses had previously received an indinavir-based regimen (boosted with ritonavir in five cases), and all but one were receiving lopinavir/ritonavir.
Fig. 2. Proportion of patients whose viruses showed resistance-associated mutations to nucleoside reverse transcriptase inhibitors (a) and to protease inhibitors (b).
Among patients exhibiting viruses with resistance mutations, 15 (65%) displayed resistance mutations to both NRTI and protease inhibitor drug classes.
No significant differences were observed between genotypic resistance profiles of group A and group B HIV-2-infected patients (data not shown).
Integrase region was sequenced in the patient in virological failure when receiving raltegravir-based regimen, showing the major resistance mutation N155H associated with the secondary mutations E92Q and T97A.
Plasma drug concentrations
Twenty out of the 145 patients (13.8%) self-reported missed doses of ART during the last day, and/or last 4 days, and/or last 30 days. Among these, there were 20 non-adherent patients, of whom three displayed drug concentrations below the limit of quantification (10 ng/ml).
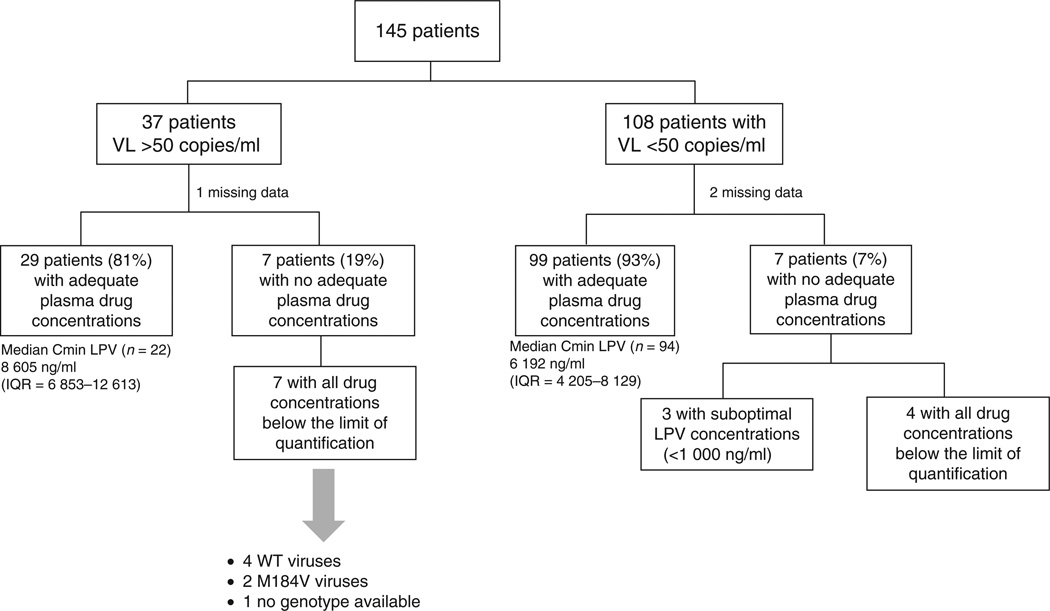
Overall, 128 of the 142 patients (90%) with available plasma drug concentrations showed adequate drugs concentrations, as previously defined in the Methods section. The proportions of patients with adequate plasma drug concentrations were 81% (29/36) in patients with virological failure and 93% (99/106) in those with viral load less than 50 copies/ml (P = 0.046; Fig. 3).
Fig. 3. Flow chart of patients with their plasma drug concentration results (n = 142).
LPV, lopinavir; VL, HIV-2 viral load; WT, wild-type.
All seven patients with virological failure and inadequate plasma drug concentrations displayed drug concentrations below the limit of quantification. Genotypic data were available for six of them, showing wild-type viruses in four cases and M184V-mutated viruses in two.
Among the seven patients with viral load less than 50 copies/ml and exhibiting inadequate plasma drug concentrations, three displayed suboptimal concentrations of lopinavir (<1000 ng/ml), and four had all drug concentrations below the limit of quantification. No significant differences were observed between patients with adequate and those with inadequate plasma drug concentrations regarding median CD4+ cell counts at the time of the study or the median gain in CD4+ cell counts since the initiation of first ART.
Discussion
In this study, based on 145 long-term HIV-2-treated patients from six HIV care centers in Abidjan, Côte d‘Ivoire, we have shown that 74% displayed a HIV-2 plasma viral load less than 50 copies/ml, the median CD4+ cell count was 360 cells/µl, and 90% had adequate plasma concentrations, suggesting adequate adherence to their ART regimen. In patients with virological failure, a high rate of NRTI and protease inhibitor drug resistance mutations were detected.
This cross-sectional observational study has some limitations. Pre-ART plasma HIV-2 viral load values were not available and participants had heterogeneous therapeutic histories in terms of drug exposure and duration of ART. However, to our knowledge, this is one of the largest HIV-2 studies (n = 145 patients) to assess the virological response and development of resistance in Western Africa.
In HIV-2 infection, an immunological response is a key criterion for assessing a response to ART as many patients display undetectable plasma HIV-2 RNA prior to initiation of ART [3]. Previous studies have reported a poorer immunological response to ART in HIV-2-infected patients when compared with HIV-1-infected patients [25, 26], particularly regarding triple-NRTI regimens and regimens that include first-generation protease inhibitors [25]. In our study, patients receiving a first-line ART containing lopinavir had a good immunological response, as previously reported [25, 26]. Moreover, we observed improved immune reconstitution in patients with viral load less than 50 copies/ml, with a significant gain in CD4+ cells since the initiation of ART compared with patients with plasma viral load more than 50 copies/ml (+ 172 vs. +29 cells/µl). Interestingly, median duration of ART in patients with viral load more than 50 copies/ml and in those with viral load less than 50 copies/ml is not comparable, showing a more advanced therapeutic history in patients with viral load more than 50 copies/ml.
In the 37 patients with HIV-2 RNA more than 50 copies/ml, a high prevalence of resistance was observed, with NRTI and protease inhibitor drug resistance mutations detected in 84 and 69% of samples, respectively. Regarding NRTI drug class, we found common profiles of genotypic resistance to HIV-2, with a high proportion of Q151M, K65R, and M184V mutations [27, 28]. Among the 21 patients exhibiting NRTI resistance-associated mutations, a decreased susceptibility to tenofovir and/or abacavir was predicted in 12 (57%) patients. In our study, the Q151M mutation was not associated with a specific dual or triple-NRTI regimen failure. Two of the four Q151M-mutated viruses also harbored the V111I mutation, and it has been previously shown that when Q151M was selected with the V111I mutation, a decrease in susceptibility to all NRTIs was observed [14, 29].
Regarding protease inhibitor drug class, the V47A resistance mutation was selected in one-third of virological failures that occurred in patients who received a first-line ART that contained lopinavir. These in-vivo findings confirm in-vitro phenotypic data, which show a significant increase in phenotypic resistance to lopinavir of the V47A site-directed mutant [22]. However, the V47A mutation does not decrease phenotypic susceptibility to darunavir [22]. The continuous viral replication under protease inhibitor drug pressure and protease inhibitor cross-resistance are both deleterious for subsequent therapeutic options. In our study, selection of lopinavir resistance may have impact, in some cases, on viral susceptibility to darunavir. Indeed, 40% of the patients harboring protease inhibitor-resistant viruses displayed at least one mutation associated with a decreased phenotypic susceptibility to darunavir [22]. We mainly observed the I54M darunavir resistance-associated mutation, which was detected in six cases, a single mutation that resulted in 6.2-fold increased resistance to darunavir [22]. In the other two cases, we observed a combination of I84V and L90M mutations, resulting in 3.3-fold increased resistance to darunavir [22]. Among the patients with a viral load more than 50 copies/ml, five (13%) exhibited dual class-resistant viruses with resistance to both tenofovir (and/or abacavir) and darunavir. This high rate of resistance prevalence observed in patients with virological failure, combined with the cross-resistance mechanism within protease inhibitor drug class, and also the reduced number of antiretroviral drugs active against HIV-2, is a critical issue.
With regard to the HIV-2 therapeutic arsenal, in cases of NRTI and protease inhibitor resistance, the only active drug class is integrase inhibitors [20, 30], and possibly the CCR5 inhibitor maraviroc [31]. However, the use of integrase inhibitors may be limited in pretreated patients who harbor NRTI-resistant viruses because of the low genetic barrier to resistance of this drug class, and who require a combination with fully active drugs.
Adequate drug concentrations were observed in a high proportion of patients, strongly suggesting overall good treatment adherence and this confirms the role of drug resistance-associated mutations in virological failure. Eleven patients in the study showed no antiretroviral drugs in their plasma, suggesting either nonadherence or intestinal malabsorption, or drug–drug interactions with a potent enzymatic inducer, such as rifampicin. These latter mechanisms might also explain that some of the patients had drug concentrations below the limit of quantification, despite a self-reported good adherence.
The findings of our study underline the need to try to define a sequencing of HIV-2 therapeutic strategies, especially in case of protease inhibitor-based first-line regimen. Lopinavir should be used as a first-line therapy to guarantee, in cases of a V47A mutation, the possibility of using a potent second-line therapy with darunavir or saquinavir associated with integrase inhibitors. Lopinavir cannot be recommended as a second-line therapy after failure of a first-generation protease inhibitor/ritonavir-based regimen (saquinavir or indinavir). Another clinical implication is the need to find an alternate boosted protease inhibitor-based regimen to initiate ART in HIV-2-infected patients. Randomized trials that evaluate triple-NRTI or integrase inhibitor-based regimen, compared to protease inhibitor/ritonavir-based regimen, are future options to consider [32].
Among 145 HIV-2-infected patients on ART in our study, we observed a high proportion of patients with viral load less than 50 copies/ml and adequate drug plasma concentrations. However, in cases of virological failure, the limited HIV-2 therapeutic arsenal and cross-resistance dramatically reduced treatment options.
Acknowledgements
The study is funded by The National Cancer Institute (NCI), the Eunice Kennedy Shriver National Institute of Child Health & Human Development (NICHD), and the National Institute of Allergy and Infectious Diseases (NIAID) of the US National Institutes of Health (NIH), as part of the International Epidemiologic Databases to Evaluate AIDS (IeDEA) under Award Number U01AI069919. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Appendix
The IeDEA West African Collaboration Study Group (as of June, 2013): Executive Committee: François Dabis (Principal Investigator, Bordeaux, France), Emmanuel Bissagnene (Co-Principal Investigator, Abidjan, Côte d’Ivoire), Elise Arrivé (Bordeaux, France), Patrick Coffie (Abidjan, Côte d’Ivoire), Didier Ekouevi (Abidjan, Côte d’Ivoire), Antoine Jaquet (Bordeaux, France), Valériane Leroy (Bordeaux, France), Charlotte Lewden (Bordeaux, France), Annie J Sasco (Bordeaux, France).
Participating sites (*members of the Steering Committee, §members of the Executive Committee):
Benin, Cotonou:
Adults: Djimon Marcel Zannou*, Carin Ahouada, Jocelyn Akakpo, Christelle Ahomadegbé, Jules Bashi, Alice Gougounon-Houéto, Angèle Azon-Kouanou, Fabien Houngbé, Jean Sehonou (CNHU Hubert Maga).
Pediatrics: Sikiratou Koumakpaï*§, Florence Alihonou, Marcelline d’Almeida, Irvine Hodonou, Ghislaine Hounhoui, Gracien Sagbo, Leïla Tossa-Bagnan, Herman Adjide (CNHU Hubert Maga).
Burkina Faso:
Adults: Joseph Drabo*, René Bognounou, Arnaud Dienderé, Eliezer Traore, Lassane Zoungrana, Béatrice Zerbo (CHU Yalgado, Ouagadougou), Adrien Bruno Sawadogo*§, Jacques Zoungrana, Arsène Héma, Ibrahim Soré, Guillaume Bado, Achille Tapsoba (CHU Souro Sanou, Bobo Dioulasso).
Pediatrics: Diarra Yé*, Fla Kouéta, Sylvie Ouedraogo, Rasmata Ouédraogo, William Hiembo, Mady Gansonré (CH Charles de Gaulle, Ouagadougou).
Côte d’Ivoire, Abidjan:
Adults: Eugène Messou*, Joachim Charles Gnokoro, Mamadou Koné, Guillaume Martial Kouakou, (ACONDA-CePReF); Clarisse Amani Bosse*, Kouakou Brou, Achi Isidore Assi (ACONDA-MTCT-Plus); Henri Chenal*, Denise Hawerlander, Franck Soppi (CIRBA); Albert Minga*, Yao Abo, Jean-Michel Yoboue (CMSDS/CNTS); Serge Paul Eholié*§, Mensah Deborah Noelly Amego, Viviane Andavi, Zelica Diallo, Frédéric Ello, Aristophane Koffi Tanon (SMIT, CHU de Treichville), Serge Olivier Koule*, Koffi Charles Anzan, Calixte Guehi (USAC, CHU de Treichville).
Pediatrics: Edmond Addi Aka*, Koffi Ladji Issouf, Jean-Claude Kouakou, Marie-Sylvie N’Gbeche, (ACONDA-CePReF); Touré Pety*, Divine Avit-Edi (ACONDA-MTCT-Plus); Kouadio Kouakou*, Magloire Moh, Valérie Andoblé Yao (CIRBA); Madeleine Amorissani Folquet*, Marie-Evelyne Dainguy, Cyrille Kouakou, Véronique Tanoh Méa-Assande, Gladys Oka-Berete, Nathalie Zobo, Patrick Acquah, Marie-Berthe Kokora (CHU Cocody); Tanoh François Eboua*, Marguerite Timité-Konan, Lucrèce Diecket Ahoussou, Julie Kebé Assouan, Mabéa Flora Sami, Clémence Kouadio (CHU Yopougon).
Ghana, Accra:
Pediatrics: Lorna Renner*§, Bamenla Goka, Jennifer Welbeck, Adziri Sackey, Seth Ntiri Owiafe (Korle Bu TH).
Guinea-Bissau:
Adults: Christian Wejse*§, Zacarias José Da Silva*, Joao Paulo (Bandim Health Project), The Bissau HIV cohort study group: Amabelia Rodrigues (Bandim Health Project), David da Silva (National HIV program Bissau), Candida Medina (Hospital National Simao Mendes, Bissau), Ines Oliviera-Souto (Bandim Health Project), Lars Østergaard (Dept of Infectious Diseases, Aarhus University Hospital), Alex Laursen (Dept of Infectious Diseases, Aarhus University Hospital), Morten Sodemann (Dept of Infectious Diseases, Odense University Hospital), Peter Aaby (Bandim Health Project), Anders Fomsgaard (Dept. of Virology, Statens Serum Institut, Copenhagen), Christian Erikstrup (Dept. of Clinical Immunology), Jesper Eugen-Olsen (Dept. of Infectious Diseases, Hvidovre Hospital, Copenhagen).
Mali, Bamako:
Adults: Moussa Y Maïga*§, Fatoumata Fofana Diakité, Abdoulaye Kalle, Drissa Katile (CH Gabriel Toure), Hamar Alassane Traore*, Daouda Minta*, Tidiani Cissé, Mamadou Dembelé, Mohammed Doumbia, Mahamadou Fomba, Assétou Soukho Kaya, Abdoulaye M Traoré, Hamady Traoré, Amadou Abathina Toure (CH Point G).
Pediatrics: Fatoumata Dicko*, Mariam Sylla, Alima Berthé, Hadizatou Coulibaly Traoré, Anta Koïta, Niaboula Koné, Clémentine N’Diaye, Safiatou Touré Coulibaly, Mamadou Traoré, Naïchata Traoré (CH Gabriel Toure).
Nigeria:
Adults: Man Charurat* (UMB/IHV), Samuel Ajayi*, Georgina Alim, Stephen Dapiap, Otu (UATH, Abuja), Festus Igbinoba (National Hospital Abuja), Okwara Benson*, Clément Adebamowo*, Jesse James, Obaseki, Philip Osakede (UBTH, Benin City), John Olasode (OATH, Ile-Ife).
Senegal, Dakar:
Adults: Moussa Seydi*, Papa Salif Sow, Bernard Diop, Noël Magloire Manga, Judicael Malick Tine§, Coumba Cissé Bassabi (SMIT, CHU Fann).
Pediatrics: Haby Signate Sy*, Abou Ba, Aida Diagne, Hélène Dior, Malick Faye, Ramatoulaye Diagne Gueye, Aminata Diack Mbaye (CH Albert Royer).
Togo, Lomé:
Adults: Akessiwe Patassi*, Awèrou Kotosso, Benjamin Goilibe Kariyare, Gafarou Gbadamassi, Agbo Komi, Kankoé Edem Mensah-Zukong, Pinuwe Pakpame (CHU Tokoin/Sylvanus Olympio).
Pediatrics: Koko Lawson-Evi*§, Yawo Atakouma, Elom Takassi, Améyo Djeha, Ayoko Ephoévi-gah, Sherifa El-Hadj Djibril (CHU Tokoin/Sylvanus Olympio).
Operational and Statistical Team: Dieudonné Amani (Abidjan, Côte d’Ivoire), Jean-Claude Azani (Abidjan, Côte d’Ivoire), Eric Balestre (Bordeaux, France), Serge Bessekon (Abidjan, Côte d’Ivoire), Franck Bohossou (Abidjan, Côte d’Ivoire), Camille Gilbert (Bordeaux, France), Sophie Karcher (Bordeaux, France), Jules Mahan Gonsan (Abidjan, Côte d’Ivoire), Jérôme Le Carrou (Bordeaux, France), Séverin Lenaud (Abidjan, Côte d’Ivoire), Célestin Nchot (Abidjan, Côte d’Ivoire), Karen Malateste (Bordeaux, France), Amon Roseamonde Yao (Abidjan, Côte d’Ivoire), Bertine Siloué (Abidjan, Côte d’Ivoire).
Administrative Team: Gwenaelle Clouet (Bordeaux, France), Madikona Dosso (Abidjan, Côte d’Ivoire), Alexandra Doring§ (Bordeaux, France), Adrienne Kouakou (Abidjan, Côte d’Ivoire), Elodie Rabourdin (Bordeaux, France), Jean Rivenc (Pessac, France).
Consultants/Working Groups: Xavier Anglaret (Bordeaux, France), Boubacar Ba (Bamako, Mali), Renaud Becquet (Bordeaux, France), Jean Bosco Essanin (Abidjan), Andrea Ciaranello (Boston, USA), Sébastien Datté (Abidjan, Côte d’Ivoire), Sophie Desmonde (Bordeaux, France), Jean-Serge Elvis Diby (Abidjan, Côte d’Ivoire), Geoffrey S.Gottlieb* (Seattle, USA), Apollinaire Gninlgninrin Horo (Abidjan, Côte d’Ivoire), Serge N’zoré Kangah (Abidjan, Côte d’Ivoire), Denis Malvy (Bordeaux, France), David Meless (Abidjan, Côte d’Ivoire), Aida Mounkaila-Harouna (Bordeaux, France), Camille Ndondoki (Bordeaux, France), Caroline Shiboski (San Francisco USA), Boris Tchounga (Abidjan, Côte d’Ivoire), Rodolphe Thiébaut (Bordeaux, France), Gilles Wandeler (Dakar, Senegal).
Coordinating Centre: ISPED, Univ Bordeaux Segalen, Bordeaux, France
Regional Office: PAC-CI, Abidjan, Côte d’Ivoire
Methodologic Support: MEREVA, Bordeaux, France
Website: http://www.mereva.net/iedea
Footnotes
S.E., D.K.E., X.A., F.rD., and F.B.V. contributed to this study’s concept. C.C. and M.B. performed the virological analyses. J.C.P., F.l.D., V.A.F., and C.R. designed and performed the viral load assays. G.P. performed the drug plasma concentrations. E.M., A.M., and S.E. followed up patients enrolled in the study. C.C., D.K.E., S.E., X.A., G.P., F.r.D., and F.B.V. contributed to the analysis and interpretation of the data. C.C., D.K.E., S.E., X.A., G.P., F.r.D., and F.B.V. contributed to writing the article. All authors contributed to the critical review of the article.
Conflicts of interest
There are no conflicts of interest.
This study is presented in part at the International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies, Toronto, Canada, June 2013 [abstract 58].
References
- 1.UNAIDS AIDS epidemic update 2006. Geneva, Switzerland: Joint United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO); 2004; 2006. pp. 1–94. [Google Scholar]
- 2.Marlink R, Kanki P, Thior I, Travers K, Eisen G, Siby T, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–1590. doi: 10.1126/science.7915856. [DOI] [PubMed] [Google Scholar]
- 3.Popper SJ, Sarr AD, Travers KU, Guèye-Ndiaye A, Mboup S, Essex ME, et al. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J Infect Dis. 1999;180 doi: 10.1086/315010. 1116-01121. [DOI] [PubMed] [Google Scholar]
- 4.Kanki PJ, Travers KU, MBoup S, Hsieh CC, Marlink RG, Gueye-NDiaye A, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–946. doi: 10.1016/s0140-6736(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 5.Burgard M, Jasseron C, Matheron S, Damond F, Hamrene K, Blanche S, et al. Mother-to-child transmission of HIV-2 infection from 1986 to 2007 in the ANRS French Perinatal Cohort EPF-CO1. Clin Infect Dis. 2010;51:833–843. doi: 10.1086/656284. [DOI] [PubMed] [Google Scholar]
- 6.Ren J, Bird LE, Chamberlain PP, Stewart-Jones GB, Stuart DI, Stammers DK. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to nonnucleoside inhibitors. Proc Natl Acad Sci U S A. 2002;99:14410–14415. doi: 10.1073/pnas.222366699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. Susceptibility of HIV-2, SIV and SHIV to various anti-HIV-1 compounds: implications for treatment and postexposure prophylaxis. Antivir Ther. 2004;9:57–65. [PubMed] [Google Scholar]
- 8.Desbois D, Roquebert B, Peytavin G, Damond F, Collin G, Bénard A, et al. In vitro phenotypic susceptibility of human immunodeficiency virus type 2 clinical isolates to protease inhibitors. Antimicrob Agents Chemother. 2008;52:1545–1548. doi: 10.1128/AAC.01284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poveda E, Briz V, Soriano V. Enfuvirtide, the first fusion inhibitor to treat HIV infection. AIDS Rev. 2005;7:139–147. [PubMed] [Google Scholar]
- 10.Ntemgwa ML, d’Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob Agents Chemother. 2009;53:3611–3619. doi: 10.1128/AAC.00154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho RJ. Special aspects of the treatment of HIV-2-infected patients. Intervirology. 2012;55:179–183. doi: 10.1159/000332025. [DOI] [PubMed] [Google Scholar]
- 12.Adjé-Touré CA, Cheingsong R, García-Lerma JG, Eholié S, Borget MY, Bouchez JM, et al. Antiretroviral therapy in HIV-2-infected patients: changes in plasma viral load, CD4+ cell counts, and drug resistance profiles of patients treated in Abidjan, Côte d’Ivoire. AIDS. 2003;17:S49–S54. [PubMed] [Google Scholar]
- 13.Jallow S, Alabi A, Sarge-Njie R, Peterson K, Whittle H, Corrah T, et al. Virological response to highly active antiretroviral therapy in patients infected with human immunodeficiency virus type 2 (HIV-2) and in patients dually infected with HIV-1 and HIV-2 in the Gambia and emergence of drug-resistant variants. J Clin Microbiol. 2009;47:2200–2208. doi: 10.1128/JCM.01654-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottlieb GS, Badiane NM, Hawes SE, Fortes L, Toure M, Ndour CT, et al. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resource-limited West Africa. Clin Infect Dis. 2009;48:476–483. doi: 10.1086/596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekouevi DK, Balestre E, Coffie PA, Minta D, Messou E, Sawadogo A, et al. Characteristics of HIV-2 and HIV-1/HIV-2 dually seropositive adults in West Africa presenting for care and antiretroviral therapy: the IeDEA-West Africa HIV-2 cohort study. PLoS One. 2013;8:e66135. doi: 10.1371/journal.pone.0066135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drylewicz J, Eholie S, Maiga M, Zannou DM, Sow PS, Ekouevi DK, et al. First-year lymphocyte T CD4+ response to antiretroviral therapy according to the HIV type in the IeDEA West Africa collaboration. AIDS. 2010;24:1043–1050. doi: 10.1097/qad.0b013e3283377a06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. [Accessed 24 February 2014];World Health Organization, antiretroviral therapy for HIV infections in adults and adolescents, Recommendations. 2010 http://www.who.int. [PubMed]
- 18.Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, et al. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–7447. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pieniazek D, Ellenberger D, Janini LM, Ramos AC, Nkengasong J, Sassan-Morokro M, et al. Predominance of human immunodeficiency virus type 2 subtype B in Abidjan, Ivory Coast. AIDS Res Hum Retroviruses. 1999;15:603–608. doi: 10.1089/088922299311132. [DOI] [PubMed] [Google Scholar]
- 20.Roquebert B, Damond F, Collin G, Matheron S, Peytavin G, Bénard A, et al. HIV-2 integrase gene polymorphism and phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitors raltegravir and elvitegravir in vitro. J Antimicrob Chemother. 2008;62:914–920. doi: 10.1093/jac/dkn335. [DOI] [PubMed] [Google Scholar]
- 21.Charpentier C, Camacho R, Ruelle J, Kaiser R, Eberle J, Gürtler L, et al. HIV-2EU: supporting standardized HIV-2 drug resistance interpretation in Europe. Clin Infect Dis. 2013;56:1654–1658. doi: 10.1093/cid/cit104. [DOI] [PubMed] [Google Scholar]
- 22.Raugi DN, Smith RA, Ba S, Toure M, Traore F, Sall F, et al. Complex patterns of protease inhibitor resistance among antiretroviral treatment-experienced HIV-2 patients from Senegal: implications for second-line therapy. Antimicrob Agents Chemother. 2013;57:2751–2760. doi: 10.1128/AAC.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung BH, Rezk NL, Bridges AS, Corbett AH, Kashuba AD. Simultaneous determination of 17 antiretroviral drugs in human plasma for quantitative analysis with liquid chromatography-tandem mass spectrometry. Biomed Chromatogr. 2007;21:1095–1104. doi: 10.1002/bmc.865. [DOI] [PubMed] [Google Scholar]
- 24.Prise en charge médicale des personnes vivant avec le VIH. Recommandations du groupe d’experts. [Accessed 24 February 2014];Rapport 2013. Sous la direction du Pr P. Morlat. http://www.sante.gouv.fr/IMG/pdf/Rapport_Morlat_2013_Mise_en_ligne.pdf.
- 25.Matheron S, Damond F, Benard A, Taieb A, Campa P, Peytavin G, et al. CD4 cell recovery in treated HIV-2-infected adults is lower than expected: results from the French ANRS CO5 HIV-2 cohort. AIDS. 2006;20:459–462. doi: 10.1097/01.aids.0000199829.57112.2f. [DOI] [PubMed] [Google Scholar]
- 26.Benard A, van Sighem A, Taieb A, Valadas E, Ruelle J, Soriano V, et al. Immunovirological response to triple nucleotide reverse-transcriptase inhibitors and ritonavir-boosted protease inhibitors in treatment-naive HIV-2-infected patients: the ACHIEV2E Collaboration Study Group. Clin Infect Dis. 2011;52:1257–1266. doi: 10.1093/cid/cir123. [DOI] [PubMed] [Google Scholar]
- 27.Smith RA, Anderson DJ, Pyrak CL, Preston BD, Gottlieb GS. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J Infect Dis. 2009;199:1323–1326. doi: 10.1086/597802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Descamps D, Damond F, Matheron S, Collin G, Campa P, Delzarue S, et al. High frequency of selection of K65R and Q151M mutations in HIV-2 infected patients receiving nucleoside reverse transcriptase inhibitors containing regimen. J Med Virol. 2004;74:197–201. doi: 10.1002/jmv.20174. [DOI] [PubMed] [Google Scholar]
- 29.Damond F, Collin G, Matheron S, Peytavin G, Campa P, Delarue S, et al. In vitro phenotypic susceptibility to nucleoside reverse transcriptase inhibitors of HIV-2 isolates with the Q151M mutation in the reverse transcriptase gene. Antivir Ther. 2005;10:861–865. [PubMed] [Google Scholar]
- 30.Charpentier C, Larrouy L, Collin G, Damond F, Matheron S, Chêne G, et al. In vitro phenotypic susceptibility of HIV-2 clinical isolates to the integrase inhibitor S/GSK1349572. AIDS. 2010;24:2753–2755. doi: 10.1097/QAD.0b013e32833f9e36. [DOI] [PubMed] [Google Scholar]
- 31.Visseaux B, Charpentier C, Hurtado-Nedelec M, Storto A, Antoine R, Peytavin G, et al. In vitro phenotypic susceptibility of HIV-2 clinical isolates to CCR5 inhibitors. Antimicrob Agents Chemother. 2012;56:137–139. doi: 10.1128/AAC.05313-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottlieb GS, Eholié SP, Nkengasong JN, Jallow S, Rowland-Jones S, Whittle HC, et al. A call for randomized controlled trials of antiretroviral therapy for HIV-2 infection in West Africa. AIDS. 2008;22:2069–2072. doi: 10.1097/QAD.0b013e32830edd44. [DOI] [PMC free article] [PubMed] [Google Scholar]