Abstract
SWI/SNF is a multi-subunit chromatin remodeling complex that performs fundamental roles in gene regulation, cell lineage specification, and organismal development. Mutations that inactivate SWI/SNF subunits are found in nearly 20% of human cancers, which indicates that the proper functioning of this complex is necessary to prevent tumor formation in diverse tissues. Recent studies show that SWI/SNF-mutant cancers depend on residual SWI/SNF complexes for their aberrant growth, thus revealing synthetic lethal interactions that could be exploited for therapeutic purposes. Other studies show that certain acute leukemias and small cell lung cancers, which lack SWI/SNF mutations, can be vulnerable to inhibition of the SWI/SNF ATPase subunit BRG1, while several normal and malignant cell types lack this sensitivity. Here, we review the emerging evidence that implicates SWI/SNF as a tumor dependency and candidate drug target in human cancer.
Keywords: SWI/SNF, synthetic lethality, cancer therapy
The SWI/SNF chromatin remodeling complex
The task of deriving a large number of distinct gene expression programs from a single genome is accomplished in part through the regulation of chromatin structure. Hence, mechanisms to condense or loosen chromatin are an integral component of eukaryotic gene regulation. Prominent among such mechanisms are the activities of chromatin remodeling complexes, which use the energy derived from ATP hydrolysis to disrupt histone-DNA contacts, thereby controlling access of nuclear machinery to DNA [1]. Four classes of chromatin remodeling complexes have been defined (SWI/SNF, ISWI, CHD and INO80) that share a conserved ATPase domain but function in a largely non-redundant manner to influence discrete aspects of transcriptional regulation, DNA replication, and DNA repair in a chromatin environment [2, 3].
The multi-subunit SWI/SNF complex is one of the most thoroughly studied chromatin remodelers. Early studies in the budding yeast S. cerevisiae implicated the complex in a variety of transcriptional responses that are accompanied by changes in DNA accessibility at the associated promoter regions [4-7]. The requirement for SWI/SNF at these genes could be alleviated by reducing the expression of core histones, which first suggested that SWI/SNF acts to overcome the nucleosome barrier to allow transcription [7]. However, the influence of SWI/SNF activity on gene expression is highly contextual, in that it can repress some promoters while in other cases it promotes gene activation [8]. Biochemical purifications led to the description of yeast SWI/SNF as a 12 subunit complex that can disrupt histone-DNA contacts in an ATP-dependent manner on purified nucleosome templates, thereby allowing transcription factors to access their cognate DNA elements [9-11]. In vitro and in vivo, chromatin remodeling by yeast SWI/SNF can lead to a variety of different outcomes, including nucleosome sliding, nucleosome eviction, and selective removal of H2A/H2B dimers [12].
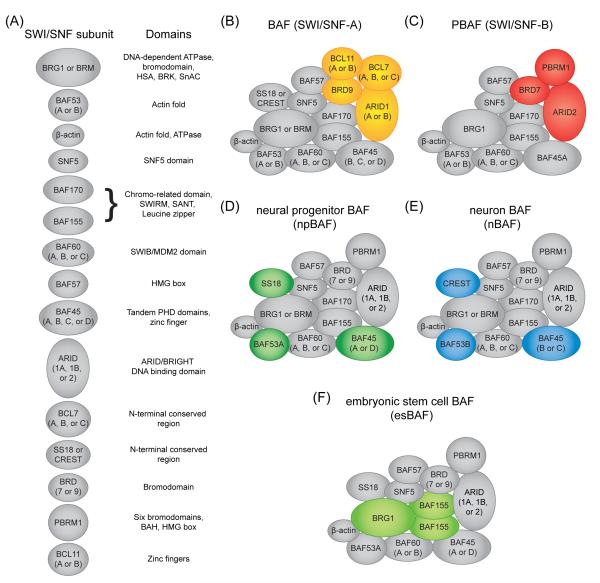
The mammalian SWI/SNF complex (also known as BAF) exhibits a similar nucleosome remodeling activity in vitro as its yeast counterpart [13-16]. This activity can be reconstituted with a set of four core subunits (BRG1/SMARCA4, SNF5/SMARCB1, BAF155/SMARCC1, and BAF170/SMARCC2), which have orthologs in the yeast complex [17]. However, mammalian SWI/SNF contains several subunits not found in the yeast counterpart, which can provide interaction surfaces for chromatin (e.g. acetyl-lysine recognition by bromodomains) or transcription factors and thus contribute to the genomic targeting of the complex (Figure 1A) [13, 18, 19]. A key attribute of mammalian SWI/SNF (hereafter referred to simply as SWI/SNF) is the heterogeneity of subunit configurations that can exist in different tissues and even in a single cell type (Figure 1B-F) [13, 18, 20]. Several individual SWI/SNF subunits are encoded by gene families, whose protein products are mutually exclusive in the complex [20]. Thus, only one paralog is incorporated in a given SWI/SNF assembly. Combinatorial association of SWI/SNF subunits could in principle give rise to hundreds of distinct complexes, although the exact number has yet to be determined (Figure 1) [20].
Figure 1.
Overview of mammalian SWI/SNF complexes. A) Subunits that comprise the mammalian SWI/SNF complex. Protein domains present in each subunit are listed on the right. Subunit nomenclature was chosen based on prevailing usage in the literature. A full list of protein/gene names of each subunit can be found in reference [3]. B-F) Examples of known SWI/SNF subunit configurations. B & C) BAF and PBAF represent two alternative subunit arrangements for SWI/SNF that can exist in the same cell type. D-F) Examples of cell type-specific SWI/SNF subunit configurations. The different coloring is used to highlight the most well established subunits that distinguish these different assemblies. The position of individual subunits within the diagram is not intended to imply direct interactions within the complex. Owing to space limitations we are unable to provide a full list of references that provide evidence for these different configurations.
Genetic evidence suggests that distinct subunit configurations of SWI/SNF are equipped to perform specialized functions. As an example, SWI/SNF contains one of two ATPase subunits, BRG1 or BRM/SMARCA2, which share 75% amino acid sequence identity [16]. While in certain cell types BRG1 and BRM can compensate for loss of the other subunit, in other contexts these two ATPases perform divergent functions [21-25]. In some cell types, BRG1 and BRM can even functionally oppose one another to regulate differentiation [26]. The functional specificity of BRG1 and BRM has been linked to sequence variations near their N-terminus, which have different interaction specificities for transcription factors [27]. Another example of paralogous subunits that form mutually exclusive SWI/SNF complexes are ARID1A/BAF250A, ARID1B/BAF250B, and ARID2/BAF200 [28-30]. ARID1A and ARID1B share 60% sequence identity, but yet can perform opposing functions in regulating the cell cycle, with MYC being an important downstream target of each paralog [31]. ARID2 has diverged considerably from ARID1A/ARID1B and exists in a unique SWI/SNF assembly known as PBAF (or SWI/SNF-B), which contains several unique subunits not found in ARID1A/B-containing complexes (Figure 1C) [13, 30]. The composition of SWI/SNF can also be dynamically reconfigured during cell fate transitions through cell type-specific expression patterns of certain subunits (Figure 1D-F). For example, BAF53A/ACTL6A is repressed and replaced by BAF53B/ACTL6B during neuronal differentiation, a switch that is essential for proper neuronal functions in vivo (Figure 1D-E) [32-35]. These studies stress that SWI/SNF in fact represents a collection of multi-subunit complexes whose integrated functions control diverse cellular processes.
Mutational inactivation of SWI/SNF subunits as a tumorigenic mechanism
Two recently published meta-analyses of cancer genome sequencing data estimate that nearly 20% of human cancers harbor mutations in one (or more) of the genes encoding SWI/SNF [36, 37]. Such mutations are generally loss-of-function, implicating SWI/SNF as a major tumor suppressor in diverse cancers. Specific SWI/SNF gene mutations are generally linked to a specific subset of cancer lineages: SNF5 is mutated in malignant rhabdoid tumors (MRT) [38, 39], PBRM1/BAF180 is frequently inactivated in renal carcinoma [40], and BRG1 is mutated in non-small cell lung cancer (NSCLC) and several other cancers [41-43]. The association of specific SWI/SNF subunit mutations with unique tumor spectra would imply that SWI/SNF performs multiple distinct tumor suppressor functions across these different malignancies, rather than a single common protective activity.
The role of SNF5 in the pathogenesis of MRT has been most extensively characterized to date, owing to its early discovery as a tumor suppressor in 1998 [38, 39]. Despite being a core subunit of SWI/SNF, loss of SNF5 does not disrupt the integrity of the complex, but instead leads to gene-specific alterations of transcription [44]. Loss of SNF5 has been shown to deregulate several oncogenic signaling pathways, including Hedgehog, WNT, and MYC [45-47]. In the absence of SNF5, SWI/SNF complexes are no longer fully capable of antagonizing the repressive function of EZH2, which is an enzymatic subunit of the histone H3K27 methyltransferase Polycomb complex, PRC2 [48]. Hence, MRTs lacking SNF5 exhibit robust PRC2-mediated repression, which modulates stem cell-associated gene expression programs to drive tumor growth [48].
The mechanisms underlying tumorigenesis provoked by mutations in SWI/SNF subunits other than SNF5 are less well understood, but are likely to differ between individual subunits. ARID1A and BRG1 have been implicated in preventing DNA entanglements during mitosis, hence their mutational inactivation could lead to genomic instability in addition to altered gene expression [49]. In several cellular contexts, loss of SWI/SNF function leads to impaired cell differentiation or even de-differentiation, which is a hallmark of many cancers [50, 51]. SWI/SNF mutations can be mutually exclusive with other tumor suppressor mutations (e.g. PTEN and p53) in certain tumor types, suggesting that SWI/SNF could perform tumor protective functions that overlap with known pathways [36, 37]. Several review articles provide a more comprehensive overview of the known tumors suppressor functions performed by various SWI/SNF subunits [52-54].
Synthetic lethal interactions involving SWI/SNF: vital roles for the residual complex
Targeting the aberrant molecular pathways of cancer cells is the central paradigm of modern cancer therapy. Restoring the lost functions of an inactivated tumor suppressor is, however, far more difficult than inhibiting the function of a hyperactive oncoprotein. An alternative to reviving inactivated tumor suppressors is to target dependencies created by their absence, thereby exploiting synthetic-lethal genetic interactions [55]. Synthetic lethality describes a scenario in which mutations in either of two (or more) individual genes are compatible with cell viability, while simultaneous mutation of both genes results in cell death. As an example, the antagonism between SNF5 and EZH2, described above, renders SNF5-mutant tumors dependent on EZH2 for disease maintenance in animal models [48]. Pharmacological inhibition of the methyltransferase activity of EZH2 selectively inhibits growth of MRT cell lines with SNF5 mutations but not those with wild type SNF5 [56]. These findings suggest a synthetic lethal interaction between SNF5 and EZH2 and consequently offer a promising therapeutic approach in this disease. While it remains to be investigated whether other SWI/SNF-mutant cancers will also be sensitive to EZH2 inhibition, this observation provides support for the concept that SWI/SNF mutations can create cancer-specific chromatin regulator dependencies.
When a mutated tumor suppressor is part of a multi-subunit protein complex, the question arises whether it is the lost function of the mutated subunit, the disassembly of the entire complex, or the deregulated activities of an aberrant residual complex that drive tumor growth. In the case of SNF5-mutant cancers it was found that the inactivation of this subunit is not equivalent to a complete loss of SWI/SNF function, as biochemical studies showed that SNF5 is dispensable for the integrity of SWI/SNF complexes and for specific cellular transcriptional functions of BRG1 [44]. One study investigated the functional significance of the residual SWI/SNF complexes that are present in SNF5-mutant MRT [22]. Using RNAi-based knockdown in human MRT cell lines, it was found that SNF5-deficient cells were dependent on BRG1 for their proliferation, whereas BRG1 was dispensable in various SNF5-proficient cancer lines [22]. This result was further verified in a genetically-engineered mouse model of SNF5-mutant lymphoma, which was also found to be dependent on BRG1 for disease progression in vivo [22]. This study suggested that loss of a single SWI/SNF subunit might drive tumorigenesis by unmasking an oncogenic function of residual BRG1-containing SWI/SNF complexes [22]. However, the observed dependence could also be explained by the residual SWI/SNF complex performing a lineage-specific function in allowing cell survival, as BRG1 is also essential for normal lymphoid development [57]. While elucidation of the underlying mechanism will require further investigation, these observations suggest that targeting residual SWI/SNF complexes could be a therapeutic strategy in SWI/SNF-mutant cancers. This possibility is particularly desirable as certain SWI/SNF subunits have domains with potential for targeting with small-molecule inhibitors (e.g. ATPase and bromdomains) (Figure 1A) [58, 59].
As SNF5 mutations are relatively rare outside of MRTs, a key question is whether other SWI/SNF-mutant tumors are likewise dependent on residual SWI/SNF activity. The SWI/SNF ATPase subunit BRG1 is mutationally inactivated or epigenetically silenced in diverse cancers, including NSCLC, medulloblastoma, and Burkitt’s lymphoma [43, 60, 61]. BRG1-deficient NSCLC generally lacks other targetable oncogenes (such as mutant EGFR), highlighting the urgent need for therapeutic targets in this form of cancer [62]. Prior studies suggested that BRG1 and its homolog BRM can compensate, at least in part, for one another’s essential functions, which raised the possibility that residual BRM-containing SWI/SNF complexes could be essential for the viability of BRG1-mutant cancers [24, 63]. A 2013 study investigated this question across a panel of human cancer cell lines and revealed that knockdown of BRM led to growth inhibition of all BRG1-deficient NSCLC lines, but not in wild type BRG1 proficient cancer or untransformed cells [62]. Targeting BRM led to suppression of colony formation in vitro and inhibition of tumor growth in vivo, associated with the induction of cellular senescence [62]. As a key control, the authors showed that reintroducing wild type BRG1 cDNA in BRG1-deficient cell lines could alleviate the BRM requirement, thus verifying the causal relationship between the genetic status of BRG1 and the level of addiction to BRM [62]. Importantly, this synthetic lethal interaction is likely to be relevant in only a subset of BRG1-mutant tumors, since BRG1-deficiency can often co-occur with epigenetic silencing of the BRM gene in primary NSCLC tumors [62, 64]. It remains an unanswered question whether residual SWI/SNF complexes are relevant in dual BRG1/BRM-deficient cancers. Nonetheless, the minimal phenotypic abnormalities of BRM-null mice highlight a remarkable selectivity of the BRM dependency for certain BRG1-mutant cancers [24].
The synthetic lethal interaction between BRG1 and BRM has been independently corroborated by two other studies that used unbiased negative-selection shRNA screens [25, 65]. Both screening strategies evaluated dependencies across a large panel of human cancer cell lines with known genetic backgrounds and revealed BRM as a top cancer dependency in cell lines with BRG1 mutations [25, 65]. These studies further showed that not only NSCLC, but also ovarian, liver, endometrial and skin cancer cell lines with complete loss of BRG1 were sensitive to BRM knockdown [25, 65].
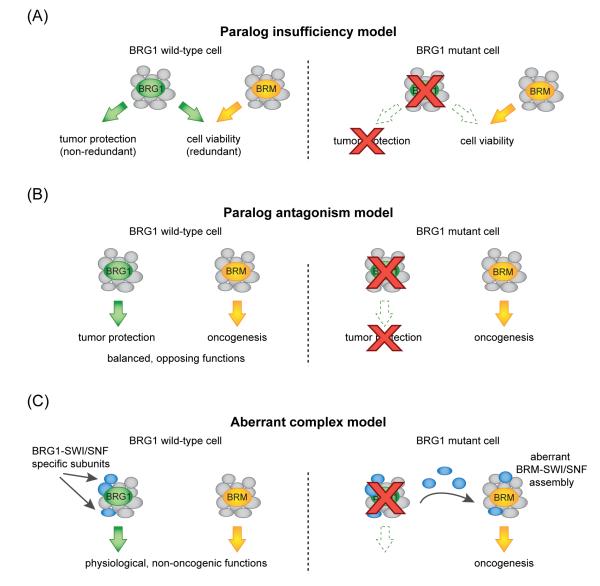
A key issue raised by these studies is how the residual BRM-SWI/SNF complex performs its vital function in BRG1-mutant cancers. In one scenario, BRG1 and BRM perform overlapping regulatory functions in the cell-of-origin that are partially redundant with one another (Figure 2A). Upon BRG1 inactivation, BRM would compensate and sustain a minimal degree of SWI/SNF functionality to support cellular viability, but would be unable to fulfill the tumor-protective BRG1 functions. Such a model has been termed ‘paralog insufficiency’ and would be consistent with known instances in which BRG1 and BRM partially compensate for loss of one another [21, 22, 24, 25]. A second mechanistic scenario would exist if BRG1 and BRM perform opposing functions in the cell-of-origin: cell proliferation being inhibited by BRG1 and facilitated by BRM (Figure 2B). Inactivation of BRG1 would disrupt the balanced state and lead to the unopposed oncogenic function of BRM-SWI/SNF complexes, thereby endowing cancer cells with tumorigenic capacities (Figure 2B). Such a model is supported by the prior observation that BRG1 and BRM can have opposing effects on differentiation of the osteoblast precursor cell line MC3T3-E1 [26]. A third hypothesis is that loss of BRG1 causes the release of some of its associated SWI/SNF subunits, which now become available to form aberrant SWI/SNF complexes with BRM with altered regulatory functions (Figure 2C). In support of this model, the PBAF assembly of SWI/SNF normally excludes BRM in favor of BRG1, thus calling into question the fate of PBAF-specific subunits (e.g. BRD7, PBRM1, and ARID2) upon BRG1 inactivation (Figure 1C) [23]. Identification of key downstream target genes of residual BRM-SWI/SNF complexes in NSCLC and comparing these with the known targets of BRG1 in this disease would be an important line of investigation to distinguish among these different models [66].
Figure 2.
Hypothetical models for the function of residual BRM-SWI/SNF complexes in BRG1-mutant cancers. A) Paralog insufficiency model. In the cancer cell-of-origin, BRG1 and BRM perform redundant functions in supporting cell viability while BRG1 performs a non-redundant tumor suppressor function. Loss of BRG1 would lead to tumorigenic effects while simultaneously rendering BRM the sole ATPase subunit responsible for supporting tumor cell viability. B) Paralog antagonism model. In the cancer cell-of-origin, BRG1 performs a specific function in tumor protection while BRM promotes oncogenesis, resulting in a balanced state of SWI/SNF functions. Loss of BRG1 would result in unopposed BRM-driven proliferation and tumorigenesis. C) Aberrant complex model. Loss of BRG1 would release specific subunits of its dedicated protein complex, which would form aberrant associations with BRM that deregulate cancer-relevant transcriptional programs.
A similar synthetic lethal interaction has also recently been uncovered between ARID1A and ARID1B [67]. ARID1A is one of the most commonly mutated subunits of SWI/SNF, which occurs in a broad spectrum of cancers [36]. ARID1B is also inactivated in certain cancers, albeit at a lower frequency [36]. Through large-scale negative selection shRNA screens, ARID1B was identified as the top differential dependency (among ~10,000 candidate genes) that was selectively required for growth of ARID1A-mutant as compared to wild type ARID1A lines [67]. Interestingly, the authors noted that ARID1B and ARID1A were often co-mutated in human cancer, but these tumors always retain one copy of a wild type ARID1B allele, which presumably is sufficient to preserve residual SWI/SNF complexes necessary for tumor viability [67]. This study raises the possibility that synthetic lethal interactions might exist more broadly between mutated SWI/SNF subunits and their wild type paralogs.
Another question yet to be fully answered is whether SWI/SNF-mutant cancers depend exclusively on the paralogs of the inactivated subunit or whether the entirety of the residual SWI/SNF complex represents a cancer-specific dependency. It was demonstrated that knockdown of SNF5 inhibited the proliferation of BRG1-deficient cancer cells, however the requirement for SNF5 in BRG1-proficient cell lines was not included for comparison in this analysis [65]. An important goal for future studies will be to determine whether targeting other residual SWI/SNF subunits in SWI/SNF-mutant cancers might block proliferation with equal efficiency.
A role for BRG1-SWI/SNF in acute leukemia maintenance
As SWI/SNF is an integral component of numerous transcriptional programs, cancers that are driven by aberrant transcriptional regulators could conceivably become reliant on SWI/SNF to sustain a transformed cellular state, even in the absence of genetic alterations in the complex. Acute myeloid leukemia (AML) is an example of a malignancy that is driven in large part by mutations in transcription factors, chromatin modifiers, and DNA methylation machinery [68]. However, SWI/SNF mutations are rarely found in this particular cancer, suggesting that the complex does not perform a significant tumor suppressor function in this malignancy [68].
Two recent studies have shown that AML mouse models and human cell lines are particularly dependent on BRG1 for disease progression [69, 70]. By means of an shRNA screen performed in cells derived from a mouse model of MLL-rearranged AML, BRG1 was identified as being among the top chromatin regulator dependencies in this cancer [71]. A ~4-fold reduction of BRG1 levels triggered leukemia cell apoptosis and terminal differentiation, while a similar degree of knockdown in non-hematopoietic cell lines (e.g. fibroblasts and various carcinomas) had no effect on cell proliferation or viability [69]. Using a conditional knockout allele, it was independently shown that BRG1 is essential for AML initiated by overexpression of the Hoxa9/Meis1 transcription factor oncoproteins, with BRG1-deficient leukemia cells undergoing cell cycle-arrest and apoptosis [70]. The authors further demonstrated that BRG1 deletion in normal bone marrow caused pronounced deficiencies in myeloid and lymphoid progenitors and mature white blood cells, however the hematopoietic stem cell compartment remained intact in BRG1-mutant animals [70]. Indeed, even dual inactivation of BRG1/BRM in adult mice was found to cause minimal effects on the abundance of hematopoietic stem cells and multipotent progenitors [63]. Collectively these observations suggest a therapeutic window for targeting BRG1 in leukemia, since hematopoietic stem cells and some progenitors would be expected to withstand BRG1 inactivation. Unlike BRM knockout mice, however, BRG1-deficiency leads to severe developmental abnormalities, hence the tolerability of BRG1 inhibition in a fully-developed animal remains an open question [72].
Mechanistically, it was found that BRG1 is critical for maintaining expression of specific genes within the transcriptional program induced by the MLL-AF9 oncoprotein, including Myc and Hoxa9 [69]. The role of BRG1 in maintaining MYC transcription appears to be unique to normal and malignant hematopoietic cells, as BRG1 knockdown in other cell lineages was found to have negligible effects on MYC expression [57, 69]. To account for this observation, it was shown that BRG1 occupies a cluster of 3′ enhancer elements at the MYC locus that are only activated in hematopoietic cells [69]. At these distal enhancers, BRG1 is critical to sustain occupancy of several hematopoietic transcription factors and for long-range enhancer-promoter looping interactions [69]. Furthermore, the ATPase activity of BRG1 is critical for its leukemia maintenance function, consistent with nucleosome remodeling of enhancer elements being essential for transcription factor occupancy [69]. These results suggest that leukemia cells promote MYC transcription through a unique enhancer-based mechanism, which is particularly sensitive to SWI/SNF perturbation. Other cell lineages also utilize SWI/SNF to repress or activate MYC expression, although these effects have generally been linked to promoter regulation [31, 50, 73].
In several non-leukemia cell line contexts highlighted above, suppression of BRG1 alone leads to minimal effects on cell proliferation, which has been attributed to compensation by BRM [22, 25]. It has been noted that SWI/SNF complexes isolated from leukemia cells predominantly contained BRG1 but only low amounts of BRM, suggesting an imbalance between these two ATPases in this lineage [70]. A deficiency in BRM-mediated compensation might explain, at least in part, the hypersensitivity of leukemia cells to BRG1 knockdown as compared to other cell types [69, 70]. Hence, a BRG1/BRM imbalance in leukemia would be analogous to the mutational mechanism that drives the imbalance between BRG1/BRM in solid tumors.
A tumor maintenance function for SWI/SNF in other cancer contexts
Synthetic lethal interactions involving SWI/SNF are not limited to the setting of mutations within the complex, but could involve other genetic drivers of human cancer. A recent study found that MAX, the MYC-associated factor X gene, is a tumor suppressor inactivated in a subset of small cell lung cancer (SCLC) [74]. Since prior studies indicated a link between MYC and BRG1 in lung cancer, the authors investigated the role of BRG1 in MAX-mutant SCLC [50, 74]. Remarkably, lung cancers harboring MAX mutations were selectively dependent on BRG1 for cell viability whereas growth of MAX wild type cancer lines was found to be unaffected by BRG1 depletion [74]. Although the precise mechanism underlying this observation remains to be determined, it may involve the regulation by BRG1 and MAX of a common set of transcriptional target genes linked to metabolism and differentiation [74]. This study suggests that other gene mutations found in cancer might also be associated with a reliance on SWI/SNF for tumor growth.
In the case of breast cancer, SWI/SNF has been linked to both tumor protection and tumor maintenance. SWI/SNF mutations occur at a significant frequency in breast cancer and BRG1 acts as a haploinsufficient mammary tumor suppressor in mice [36, 37, 75]. Conversely, high BRG1 expression in breast cancer biopsies has also been correlated with poor prognosis and knockdown of BRG1 in various breast cancer cell lines leads to diminished proliferation and invasion [76]. These observations raise the issue as to how SWI/SNF can perform these two opposing functions in one form of cancer. One study suggests that the oncogenic function SWI/SNF in breast cancer can be modulated through post-translational modification of a core subunit. BAF155 was found to be di-methylated at arginine 1064 by the CARM1 methyltransferase, an enzyme whose expression is often elevated in metastatic breast cancer [77]. Methylation of BAF155 was found to influence the genomic targeting of its associated SWI/SNF complex in breast cancer cells to drive an oncogenic transcriptional program that includes genes in the MYC pathway [77]. This study provides a unique mechanism by which global SWI/SNF functions could be dynamically regulated during the course of tumor evolution.
Concluding Remarks
A theme emerges from recent studies in which imbalances between alternative subunits within SWI/SNF can render cells more tumorigenic and simultaneously hypersensitive to targeting of the residual complex. This body of genetic evidence should, in principle, motivate efforts to pharmacologically validate SWI/SNF dependencies in appropriate pre-clinical cancer models to evaluate therapeutic efficacy and tolerability. Critical to such studies will be the availability of chemical probes that target the key functionalities of SWI/SNF, which are not widely available to the biomedical research community at present. Small-molecule ATPase inhibitors of BRG1/BRM would be essential to study the response of cancer cells to acute inactivation of the SWI/SNF remodeling function, an activity that has already been linked to cancer maintenance [69]. The prior identification of potent and selective inhibitors of the ATPase activity of Kinesin KIF11 would support the feasibility of such a drug discovery campaign [58]. Targeting the acetyl-lysine recognition function of SWI/SNF bromodomains (which are found in PBRM1, BRG1, BRM, BRD7, and BRD9 subunits) presents an additional possibility for more selective perturbations of the complex, although the significance of these domains to the cancer-relevant SWI/SNF functions is currently unknown. Crystal and/or NMR structures exist for all of the SWI/SNF bromodomains, which might aid efforts to target these domains pharmacologically [e.g. 78]. Genetic evidence would suggest that discrimination between paralogous subunits (e.g. BRG1 and BRM) would be an ideal property to minimize toxicity of a SWI/SNF-targeting small-molecule, particularly as carcinogenesis would be a potential on-target consequence of SWI/SNF perturbation. Furthermore, the developmental abnormalities associated with SWI/SNF-mutations in mice and in humans imply that highly selective modes of SWI/SNF perturbation would be most suitable for therapeutic intervention [72, 79]. In this regard, a comprehensive structure-function analysis that compares the tumor-protective, tumor-maintenance, and homeostatic SWI/SNF functions would be invaluable for defining the suitable routes for therapeutic targeting of this multi-functional chromatin remodeling complex.
Highlights.
Inactivating mutations of SWI/SNF are found in 20% of human cancers
SWI/SNF-mutant cancers rely on specific subunits of residual SWI/SNF complexes
The SWI/SNF subunit BRG1 is required for maintenance of acute myeloid leukemia
SWI/SNF is candidate drug target in human cancer
Acknowledgements
A.F.H. is supported by a Boehringer Ingelheim Fonds PhD Fellowship. C.R.V. is supported by NIH CA174793 and a Burroughs-Wellcome Fund Career Award for Medical Scientists.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annual review of biochemistry. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annual review of biochemistry. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 3.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell research. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern M, et al. Five SWI genes are required for expression of the HO gene in yeast. Journal of molecular biology. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 6.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn JN, et al. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes & development. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 8.Martens JA, Winston F. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes & development. 2002;16:2231–2236. doi: 10.1101/gad.1009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote J, et al. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 10.Cairns BR, et al. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson CL, et al. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narlikar GJ, et al. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. The EMBO journal. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 14.Imbalzano AN, et al. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 15.Kwon H, et al. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 16.Khavari PA, et al. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 17.Phelan ML, et al. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes & development. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 19.Nie Z, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Molecular and cellular biology. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu JI, et al. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strobeck MW, et al. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. The Journal of biological chemistry. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, et al. Oncogenesis caused by loss of the SNF5 tumor suppressor is dependent on activity of BRG1, the ATPase of the SWI/SNF chromatin remodeling complex. Cancer research. 2009;69:8094–8101. doi: 10.1158/0008-5472.CAN-09-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) The EMBO journal. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffman GR, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3128–3133. doi: 10.1073/pnas.1316793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flowers S, et al. Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. The Journal of biological chemistry. 2009;284:10067–10075. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Molecular cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, et al. Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. The Biochemical journal. 2004;383:319–325. doi: 10.1042/BJ20040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallas PB, et al. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Molecular and cellular biology. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes & development. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagl NG, Jr., et al. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. The EMBO journal. 2007;26:752–763. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo AS, et al. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JI, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 35.Olave I, et al. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes & development. 2002;16:2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PloS one. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 39.Biegel JA, et al. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer research. 1999;59:74–79. [PubMed] [Google Scholar]
- 40.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medina PP, et al. Frequent BRG1/SMARCA4-inactivating mutations in human lung cancer cell lines. Human mutation. 2008;29:617–622. doi: 10.1002/humu.20730. [DOI] [PubMed] [Google Scholar]
- 42.Wong AK, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer research. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 43.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doan DN, et al. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene. 2004;23:3462–3473. doi: 10.1038/sj.onc.1207472. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, et al. TCR-dependent transformation of mature memory phenotype T cells in mice. The Journal of clinical investigation. 2011;121:3834–3845. doi: 10.1172/JCI37210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mora-Blanco EL, et al. Activation of beta-catenin/TCF targets following loss of the tumor suppressor SNF5. Oncogene. 2014;33:933–938. doi: 10.1038/onc.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jagani Z, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nature medicine. 2010;16:1429–1433. doi: 10.1038/nm.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson BG, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dykhuizen EC, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIalpha. Nature. 2013;497:624–627. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero OA, et al. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO molecular medicine. 2012;4:603–616. doi: 10.1002/emmm.201200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eroglu E, et al. SWI/SNF Complex Prevents Lineage Reversion and Induces Temporal Patterning in Neural Stem Cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 52.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, et al. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer--mechanisms and potential therapeutic insights. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:21–27. doi: 10.1158/1078-0432.CCR-13-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer research. 2009;69:8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaelin WG., Jr. The concept of synthetic lethality in the context of anticancer therapy. Nature reviews. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 56.Knutson SK, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chi TH, et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 58.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 59.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love C, et al. The genetic landscape of mutations in Burkitt lymphoma. Nature genetics. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oike T, et al. A synthetic lethality-based strategy to treat cancers harboring a genetic deficiency in the chromatin remodeling factor BRG1. Cancer research. 2013;73:5508–5518. doi: 10.1158/0008-5472.CAN-12-4593. [DOI] [PubMed] [Google Scholar]
- 63.Willis MS, et al. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circulation research. 2012;111:e111–122. doi: 10.1161/CIRCRESAHA.112.265587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reisman DN, et al. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer research. 2003;63:560–566. [PubMed] [Google Scholar]
- 65.Wilson BG, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Molecular and cellular biology. 2014;34:1136–1144. doi: 10.1128/MCB.01372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medina PP, et al. Transcriptional targets of the chromatin-remodelling factor SMARCA4/BRG1 in lung cancer cells. Human molecular genetics. 2005;14:973–982. doi: 10.1093/hmg/ddi091. [DOI] [PubMed] [Google Scholar]
- 67.Helming KC, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nature medicine. 2014;20:251–254. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cancer Genome Atlas Research, N Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi J, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buscarlet M, et al. Essential role of BRG, the ATPase subunit of BAF chromatin remodeling complexes, in leukemia maintenance. Blood. 2014;123:1720–1728. doi: 10.1182/blood-2013-02-483495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Molecular cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 73.Nagl NG, Jr., et al. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer research. 2006;66:1289–1293. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 74.Romero OA, et al. MAX Inactivation in Small Cell Lung Cancer Disrupts MYC SWI/SNF Programs and Is Synthetic Lethal with BRG1. Cancer discovery. 2014;4:292–303. doi: 10.1158/2159-8290.CD-13-0799. [DOI] [PubMed] [Google Scholar]
- 75.Bultman SJ, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–468. doi: 10.1038/sj.onc.1210664. [DOI] [PubMed] [Google Scholar]
- 76.Bai J, et al. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PloS one. 2013;8:e59772. doi: 10.1371/journal.pone.0059772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang L, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsurusaki Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nature genetics. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]